Reference Gene Selection for qPCR Analysis in Schima superba under Abiotic Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Reference Gene Selection and Primer Design
2.3. qPCR Analysis
2.4. Statistical Data Analysis
3. Results
3.1. Candidate Reference Genes and PCR Amplification
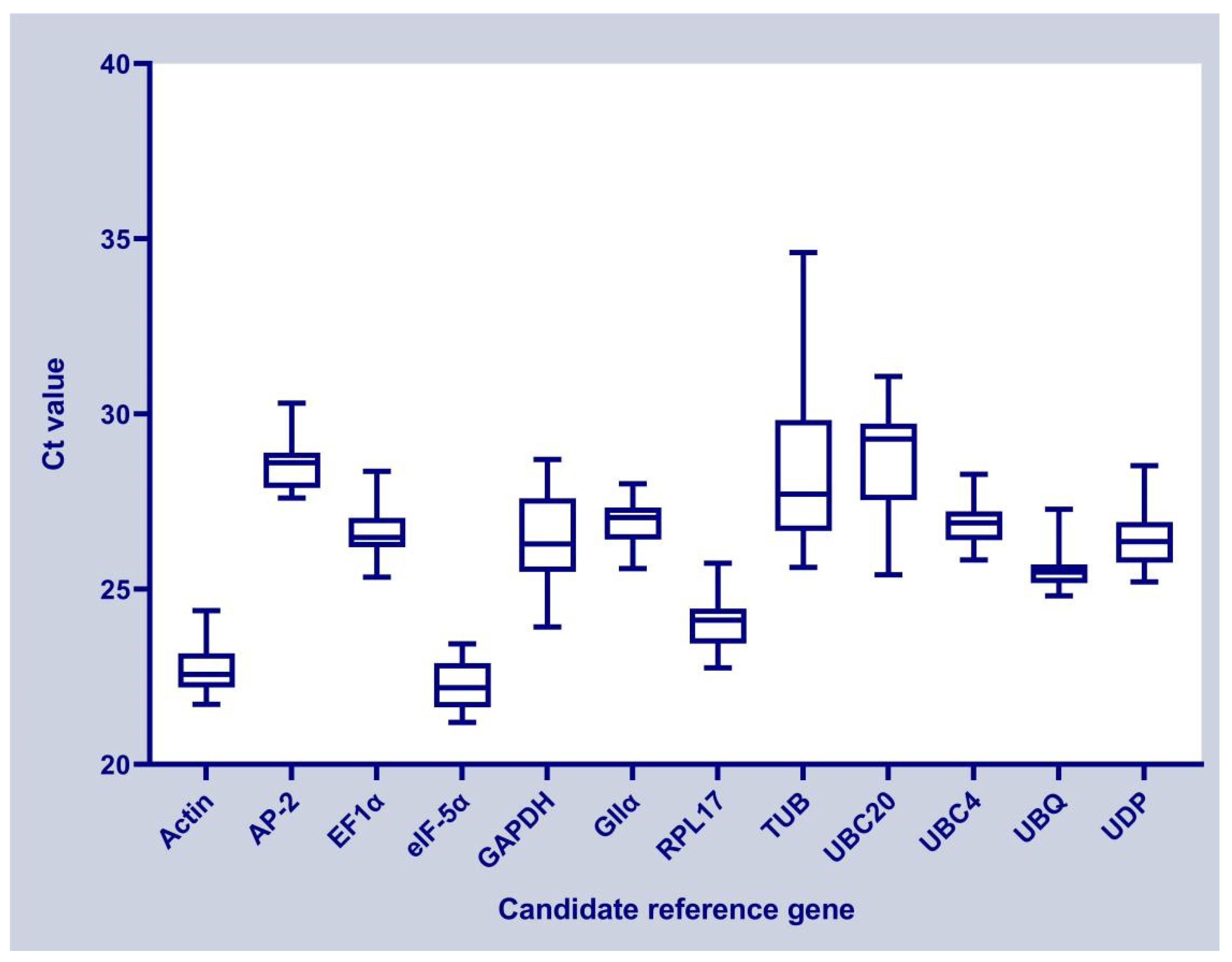
3.2. Ct Values of Candidate Reference Genes
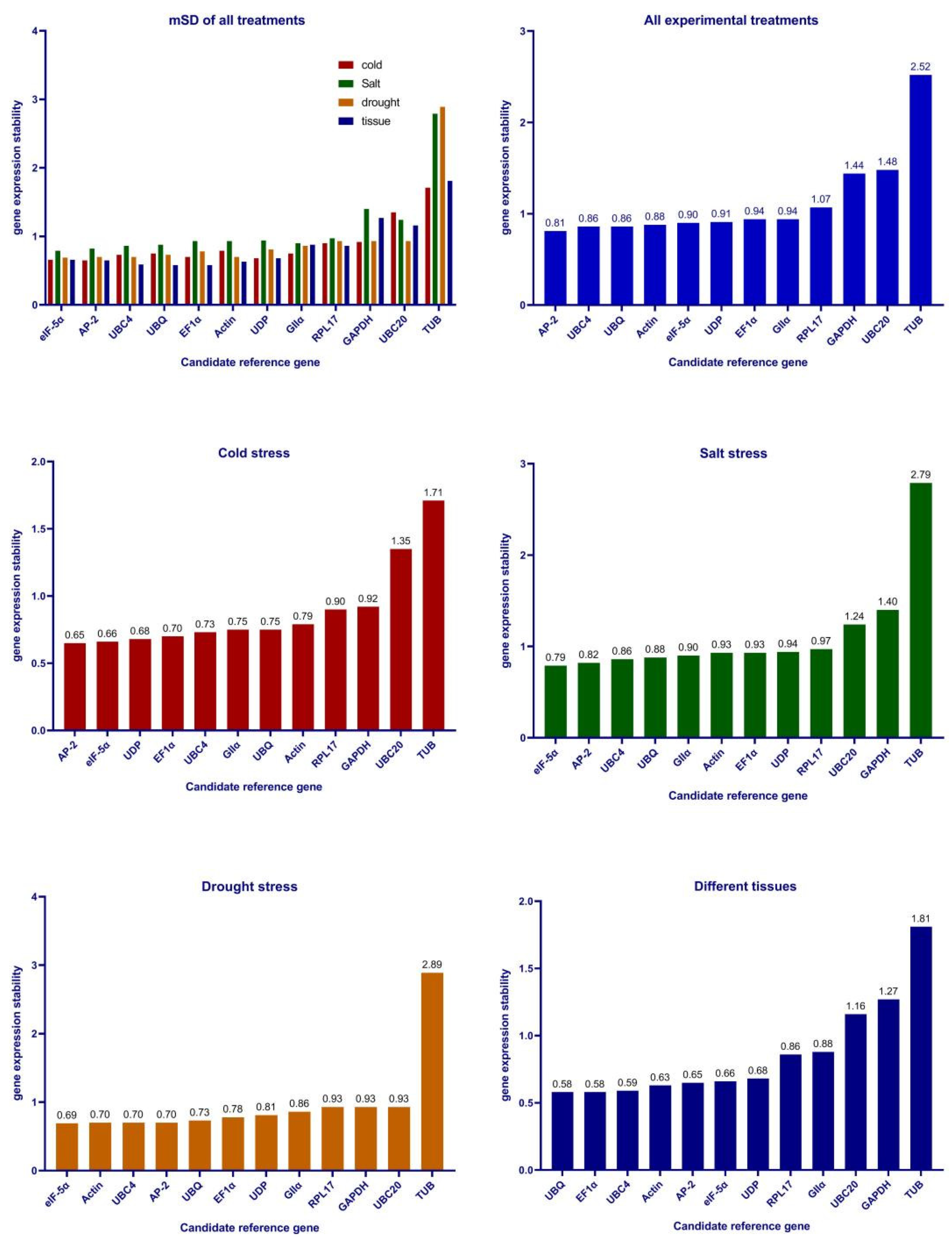
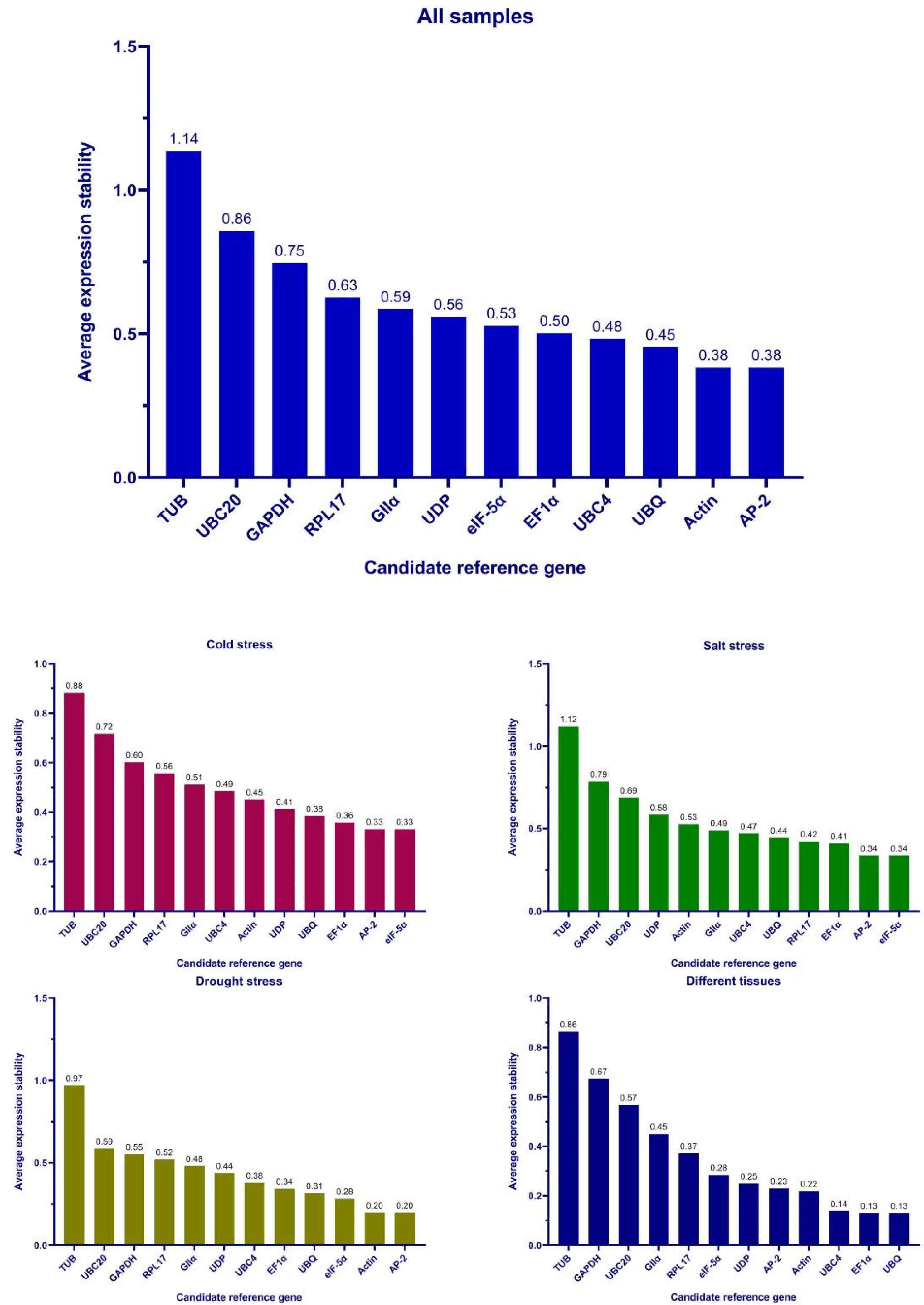
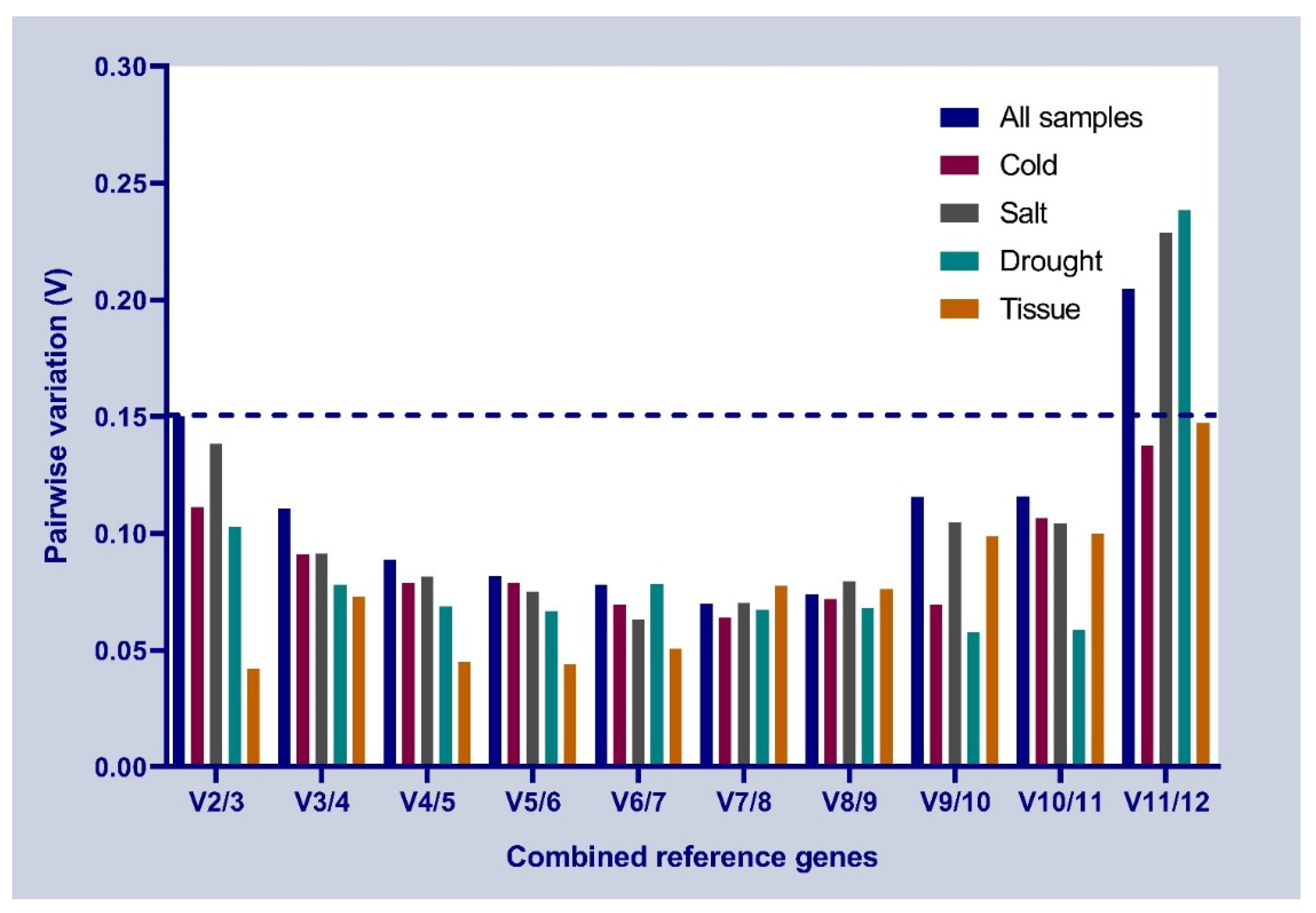
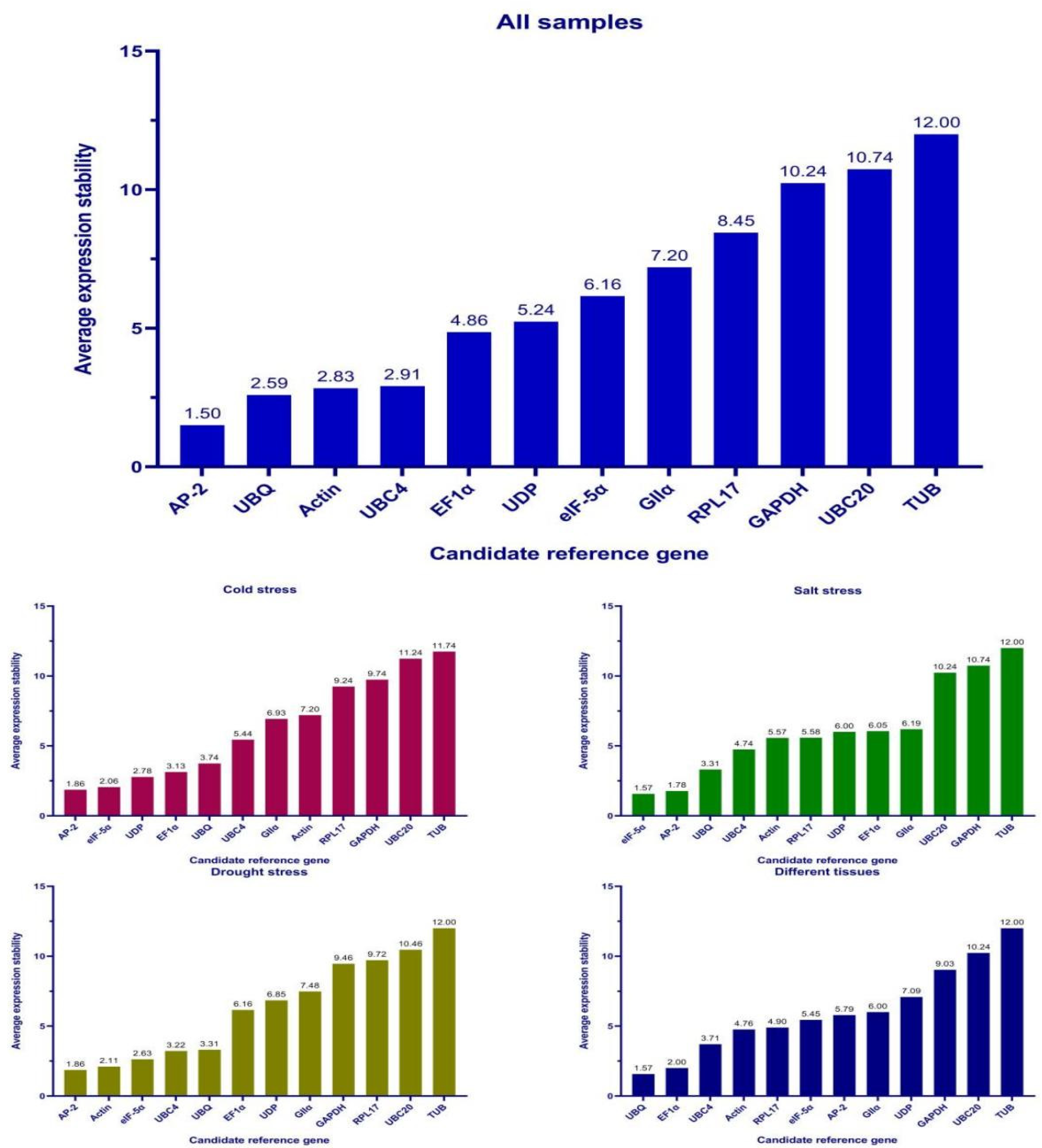
3.3. Stability of Candidate Reference Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kou, X.; Zhang, L.; Yang, S.; Li, G.; Ye, J. Selection and validation of reference genes for quantitative RT-PCR analysis in peach fruit under different experimental conditions. Sci. Hortic. 2017, 225, 195–203. [Google Scholar] [CrossRef]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Xu, S.; Zhao, Y.; Xia, B.; Wang, R. Selection and validation of appropriate reference genes for quantitative real-time PCR analysis of gene expression in Lycoris aurea. Front. Plant Sci. 2016, 7, 536. [Google Scholar] [CrossRef]
- Xiao, Z.; Sun, X.; Liu, X.; Li, C.; He, L.; Chen, S.; Su, J. Selection of reliable reference genes for gene expression studies on Rhododendron molle G. Don. Front. Plant Sci. 2016, 7, 1547. [Google Scholar] [CrossRef]
- Wei, C.; Qin, W.; Zhang, D.; Yan, L.; Wang, F. The Growth Rhythm of Schima Superba Plantation in Nanning, Guangxi. For. Environ. Sci. 2020, 36, 48–54. [Google Scholar]
- Zhang, P.; Zhou, Z.; Jin, G.; Wu, Y.; Fan, H. Provenance Differences for Fire-assistant and Fire-resistant Chemical Components in Fresh Leaf of Schima superba. For. Res. 2005, 18, 80. [Google Scholar]
- Li, Z.; Li, B.; Qi, C.; Yu, X.; Wu, Y. Studies on important of valuable wood species and its development strategy. Zhongnan Linye Keji Daxue Xuebao 2013, 32, 1–8. [Google Scholar]
- Huang, Y.; Zhuang, X. South China Native Tree Nursery Technology; China Forestry Publishing House: Beijing, China, 2007; pp. 341–342. [Google Scholar]
- Yang, Z.; Zhang, R.; Zhou, Z. Identification and validation of reference genes for gene expression analysis in Schima superba. Genes 2021, 12, 732. [Google Scholar] [CrossRef]
- Pabuayon, I.M.; Yamamoto, N.; Trinidad, J.L.; Longkumer, T.; Raorane, M.L.; Kohli, A. Reference genes for accurate gene expression analyses across different tissues, developmental stages and genotypes in rice for drought tolerance. Rice 2016, 9, 1–8. [Google Scholar] [CrossRef]
- Chen, M.D.; Wang, B.; Li, Y.P.; Zeng, M.J.; Liu, J.T.; Ye, X.R.; Zhu, H.S.; Wen, Q.F. Reference gene selection for qRT-PCR analyses of luffa (Luffa cylindrica) plants under abiotic stress conditions. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Sudhakar Reddy, P.; Srinivas Reddy, D.; Sivasakthi, K.; Bhatnagar-Mathur, P.; Vadez, V.; Sharma, K.K. Evaluation of sorghum [Sorghum bicolor (L.)] reference genes in various tissues and under abiotic stress conditions for quantitative real-time PCR data normalization. Front. Plant Sci. 2016, 7, 529. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 1–9. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Li, Q.F.; Sun, S.S.; Yuan, D.Y.; Yu, H.X.; Gu, M.H.; Liu, Q.Q. Validation of candidate reference genes for the accurate normalization of real-time quantitative RT-PCR data in rice during seed development. Plant Mol. Biol. Rep. 2010, 28, 49–57. [Google Scholar] [CrossRef]
- Wan, H.; Zhao, Z.; Qian, C.; Sui, Y.; Malik, A.A.; Chen, J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem. 2010, 399, 257–261. [Google Scholar] [CrossRef]
- Wang, H.L.; Chen, J.; Tian, Q.; Wang, S.; Xia, X.; Yin, W. Identification and validation of reference genes for Populus euphratica gene expression analysis during abiotic stresses by quantitative real-time PCR. Physiol. Plant. 2014, 152, 529–545. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Liu, Q.; Tong, H.; Zhang, T.; Gu, C.; Liu, L.; Huang, S.; Yuan, H. Selection and validation of appropriate reference genes for RT-qPCR analysis of flowering stages and different genotypes of Iris germanica L. Sci. Rep. 2021, 11, 1–10. [Google Scholar]
- Hua, Y.J.; Yue, Y.Z.; Yang, X.L.; He, Q. Selection of Reference Genes for Real-time Quantitative PCR in Clerodendrum trichotomum Thunb. Leaves. For. Res. 2022, 35, 193–201. [Google Scholar]
- Liu, M.; Wang, Q.; Xie, H.; Liu, S.; Wang, S.; Zhang, H.; Zhao, Y. UDP and NTF2 are the most consistently expressed genes in Panax ginseng roots at different growth stages. Mol. Med. Rep. 2017, 15, 4382–4390. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhou, L.; Shi, Q.; Wang, Y.; Li, K.; Zheng, B.; Miao, K. Evaluation of candidate reference genes for quantitative gene expression studies in tree peony. J. Am. Soc. Hortic. Sci. 2016, 141, 99–111. [Google Scholar] [CrossRef]
- Hossain, M.; Ahmed, R.; Haque, M.; Alam, M.; Islam, M. Identification and validation of reference genes for real-time quantitative RT-PCR analysis in jute. BMC Mol. Biol. 2019, 20, 1–13. [Google Scholar] [CrossRef]
- You, S.; Cao, K.; Chen, C.; Li, Y.; Wu, J.; Zhu, G.; Fang, W.; Wang, X.; Wang, L. Selection and validation reference genes for qRT-PCR normalization in different cultivars during fruit ripening and softening of peach (Prunus persica). Sci. Rep. 2021, 11, 1–13. [Google Scholar] [CrossRef]
- Chen, C.; Wu, J.; Hua, Q.; Tel-Zur, N.; Xie, F.; Zhang, Z.; Chen, J.; Zhang, R.; Hu, G.; Zhao, J. Identification of reliable reference genes for quantitative real-time PCR normalization in pitaya. Plant Methods 2019, 15, 1–12. [Google Scholar] [CrossRef]
- Pinheiro, T.; Nishimura, D.; De Nadai, F.; Figueira, A.; Latado, R. Selection of reference genes for expression analyses of red-fleshed sweet orange (Citrus sinensis). Genet. Mol. Res. 2015, 14, 18440–18451. [Google Scholar] [CrossRef]
- Xu, M.; Zhang, B.; Su, X.; Zhang, S.; Huang, M. Reference gene selection for quantitative real-time polymerase chain reaction in Populus. Anal. Biochem. 2011, 408, 337–339. [Google Scholar] [CrossRef]
- Deng, Y.; Li, Y.; Sun, H. Selection of reference genes for RT-qPCR normalization in blueberry (Vaccinium corymbosum× angustifolium) under various abiotic stresses. FEBS Open Bio 2020, 10, 1418–1435. [Google Scholar] [CrossRef]
- Zhu, L.; Yang, C.; You, Y.; Liang, W.; Wang, N.; Fengwang, M.; Li, C. Validation of reference genes for qRT-PCR analysis in peel and flesh of six apple cultivars (Malus domestica) at diverse stages of fruit development. Sci. Hortic. 2019, 244, 165–171. [Google Scholar] [CrossRef]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Tashiro, R.M.; Philips, J.G.; Winefield, C.S. Identification of suitable grapevine reference genes for qRT-PCR derived from heterologous species. Mol. Genet. Genom. 2016, 291, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Sang, J.; Han, X.; Liu, M.; Qiao, G.; Jiang, J.; Zhuo, R. Selection and validation of reference genes for real-time quantitative PCR in hyperaccumulating ecotype of Sedum alfredii under different heavy metals stresses. PLoS ONE 2013, 8, e82927. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Han, S.; Yin, W.; Xia, X.; Liu, C. Comparison of reliable reference genes following different hormone treatments by various algorithms for qRT-PCR analysis of Metasequoia. Int. J. Mol. Sci. 2018, 20, 34. [Google Scholar] [CrossRef]
- Fei, X.; Shi, Q.; Yang, T.; Fei, Z.; Wei, A. Expression stabilities of ten candidate reference genes for RT-qPCR in Zanthoxylum bungeanum Maxim. Molecules 2018, 23, 802. [Google Scholar]
- Shen, Z.; Sun, J.; Yao, J.; Wang, S.; Ding, M.; Zhang, H.; Qian, Z.; Zhao, N.; Sa, G.; Zhao, R. High rates of virus-induced gene silencing by tobacco rattle virus in Populus. Tree Physiol. 2015, 35, 1016–1029. [Google Scholar] [CrossRef]
- Hochstrasser, M. Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol. 2000, 2, E153–E157. [Google Scholar] [CrossRef]
- Wu, Y.; Tian, Q.; Huang, W.; Liu, J.; Xia, X.; Yang, X.; Mou, H. Identification and evaluation of reference genes for quantitative real-time PCR analysis in Passiflora edulis under stem rot condition. Mol. Biol. Rep. 2020, 47, 2951–2962. [Google Scholar]
- Iskandar, H.M.; Simpson, R.S.; Casu, R.E.; Bonnett, G.D.; Maclean, D.J.; Manners, J.M. Comparison of reference genes for quantitative real-time polymerase chain reaction analysis of gene expression in sugarcane. Plant Mol. Biol. Rep. 2004, 22, 325–337. [Google Scholar] [CrossRef]
- Ye, Y.; Lu, Y.; Wang, G.; Liu, Y.; Zhang, Y.; Tang, H. Stable reference gene selection for qRT-PCR normalization in strawberry (Fragaria × ananassa) leaves under different stress and light-quality conditions. Horticulturae 2021, 7, 452. [Google Scholar] [CrossRef]
- Niu, K.; Shi, Y.; Ma, H. Selection of candidate reference genes for gene expression analysis in Kentucky bluegrass (Poa pratensis L.) under abiotic stress. Front. Plant Sci. 2017, 8, 193. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Niu, J.; Quan, S. Identification of appropriate reference genes for RT-qPCR analysis in Juglans regia L. PLoS ONE 2018, 13, e0209424. [Google Scholar] [CrossRef]
- De Jonge, H.J.; Fehrmann, R.S.; De Bont, E.S.; Hofstra, R.M.; Gerbens, F.; Kamps, W.A.; De Vries, E.G.; Van Der Zee, A.G.; Te Meerman, G.J.; Ter Elst, A. Evidence based selection of housekeeping genes. PLoS ONE 2007, 2, e898. [Google Scholar] [CrossRef] [PubMed]
| Reference Genes | Slope (K) | R2 | Amplification Efficiency (E) |
|---|---|---|---|
| UBQ | −2.8669 | 0.9987 | 123.13% |
| AP−2 | −2.9375 | 0.9875 | 118.99% |
| Gllα | −2.9570 | 0.9943 | 117.86% |
| UBC4 | −2.9876 | 0.9942 | 116.13% |
| elF−5α | −3.1023 | 0.9938 | 110.06% |
| TUB | −3.1214 | 0.9950 | 109.11% |
| GAPDH | −3.1213 | 0.9996 | 109.11% |
| RPL17 | −3.1270 | 0.9985 | 108.83% |
| UDP | −3.1845 | 0.9833 | 106.07% |
| EF1α | −3.1863 | 0.9998 | 105.99% |
| Actin | −3.4885 | 0.9953 | 93.49% |
| UBC20 | −3.5201 | 0.9930 | 92.35% |
| Rank | All Samples | Cold | Drought | Salt | Tissue | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SV | Gene | SV | Gene | SV | Gene | SV | Gene | SV | |
| 1 | AP-2 | 0.087 | UDP | 0.171 | AP-2 | 0.069 | AP-2 | 0.218 | UBQ | 0.066 |
| 2 | UDP | 0.251 | AP-2 | 0.183 | Actin | 0.086 | UDP | 0.228 | EF1α | 0.066 |
| 3 | UBC4 | 0.305 | eIF-5α | 0.221 | UBC4 | 0.176 | eIF-5α | 0.273 | UBC4 | 0.067 |
| 4 | Actin | 0.330 | EF1α | 0.309 | eIF-5α | 0.233 | UBC4 | 0.392 | Actin | 0.182 |
| 5 | UBQ | 0.368 | UBC4 | 0.323 | UDP | 0.292 | Actin | 0.421 | AP-2 | 0.237 |
| 6 | eIF-5α | 0.421 | GIIα | 0.398 | UBQ | 0.297 | UBQ | 0.513 | UDP | 0.271 |
| 7 | GIIα | 0.477 | UBQ | 0.399 | GIIα | 0.462 | GIIα | 0.522 | eIF-5α | 0.307 |
| 8 | EF1α | 0.551 | Actin | 0.466 | EF1α | 0.518 | EF1α | 0.612 | GIIα | 0.635 |
| 9 | RPL17 | 0.762 | GAPDH | 0.671 | UBC20 | 0.579 | RPL17 | 0.727 | RPL17 | 0.668 |
| 10 | UBC20 | 1.223 | RPL17 | 0.678 | GAPDH | 0.694 | UBC20 | 0.837 | UBC20 | 0.953 |
| 11 | GAPDH | 1.246 | UBC20 | 1.254 | RPL17 | 0.755 | GAPDH | 1.175 | GAPDH | 1.176 |
| 12 | TUB | 2.448 | TUB | 1.641 | TUB | 2.861 | TUB | 2.741 | TUB | 1.760 |
| Rank | All Samples | Cold | Drought | Salt | Tissue | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SD | CV | Gene | SD | CV | Gene | SD | CV | Gene | SD | CV | Gene | SD | CV | |
| 1 | UBQ | 0.31 | 1.20 | UBQ | 0.14 | 0.53 | UBQ | 0.16 | 0.65 | UBQ | 0.21 | 0.80 | RPL17 | 0.40 | 1.64 |
| 2 | EF1α | 0.45 | 1.70 | EF1α | 0.31 | 1.14 | UBC4 | 0.26 | 0.98 | eIF-5α | 0.27 | 1.17 | GIIα | 0.67 | 2.54 |
| 3 | UBC4 | 0.48 | 1.78 | eIF-5α | 0.42 | 1.94 | AP-2 | 0.34 | 1.19 | RPL17 | 0.33 | 1.38 | eIF-5α | 0.68 | 3.06 |
| 4 | Actin | 0.50 | 2.19 | UDP | 0.49 | 1.87 | eIF-5α | 0.34 | 1.56 | Actin | 0.35 | 1.54 | EF1α | 0.72 | 2.66 |
| 5 | AP-2 | 0.51 | 1.79 | UBC4 | 0.49 | 1.80 | Actin | 0.34 | 1.51 | AP-2 | 0.37 | 1.30 | GAPDH | 0.73 | 2.88 |
| 6 | GIIα | 0.53 | 1.99 | AP-2 | 0.51 | 1.80 | EF1α | 0.36 | 1.36 | GIIα | 0.40 | 1.46 | UBQ | 0.78 | 3.01 |
| 7 | RPL17 | 0.54 | 2.26 | Actin | 0.54 | 2.39 | GIIα | 0.39 | 1.43 | UBC4 | 0.42 | 1.54 | UBC4 | 0.78 | 2.90 |
| 8 | eIF-5α | 0.58 | 2.62 | GIIα | 0.58 | 2.18 | GAPDH | 0.44 | 1.72 | EF1α | 0.47 | 1.77 | Actin | 0.94 | 4.14 |
| 9 | UDP | 0.60 | 2.28 | RPL17 | 0.76 | 3.19 | UDP | 0.47 | 1.76 | UDP | 0.67 | 2.51 | AP-2 | 0.95 | 3.30 |
| 10 | GAPDH | 1.02 | 3.86 | GAPDH | 0.80 | 2.93 | RPL17 | 0.50 | 2.07 | GAPDH | 0.94 | 3.47 | UDP | 1.04 | 3.89 |
| 11 | UBC20 | 1.18 | 4.10 | TUB | 1.03 | 3.77 | UBC20 | 0.65 | 2.19 | UBC20 | 0.97 | 3.33 | UBC20 | 1.11 | 4.03 |
| 12 | TUB | 1.97 | 6.90 | UBC20 | 1.28 | 4.55 | TUB | 2.33 | 8.05 | TUB | 2.25 | 7.87 | TUB | 1.95 | 6.48 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, J.; Zhu, G.; Liang, D.; He, B.; Wang, Y.; Cai, Y.; Zhang, Q. Reference Gene Selection for qPCR Analysis in Schima superba under Abiotic Stress. Genes 2022, 13, 1887. https://doi.org/10.3390/genes13101887
Yao J, Zhu G, Liang D, He B, Wang Y, Cai Y, Zhang Q. Reference Gene Selection for qPCR Analysis in Schima superba under Abiotic Stress. Genes. 2022; 13(10):1887. https://doi.org/10.3390/genes13101887
Chicago/Turabian StyleYao, Jun, Gang Zhu, Dongcheng Liang, Boxiang He, Yingli Wang, Yanling Cai, and Qian Zhang. 2022. "Reference Gene Selection for qPCR Analysis in Schima superba under Abiotic Stress" Genes 13, no. 10: 1887. https://doi.org/10.3390/genes13101887
APA StyleYao, J., Zhu, G., Liang, D., He, B., Wang, Y., Cai, Y., & Zhang, Q. (2022). Reference Gene Selection for qPCR Analysis in Schima superba under Abiotic Stress. Genes, 13(10), 1887. https://doi.org/10.3390/genes13101887






