A Tremendous Reorganization Journey for the 3D Chromatin Structure from Gametes to Embryos
Abstract
1. Introduction
2. The Chromatin Organization in Male and Female Gametes
2.1. The Chromatin Organization in Sperm
2.2. The Chromatin Organization in Oocytes
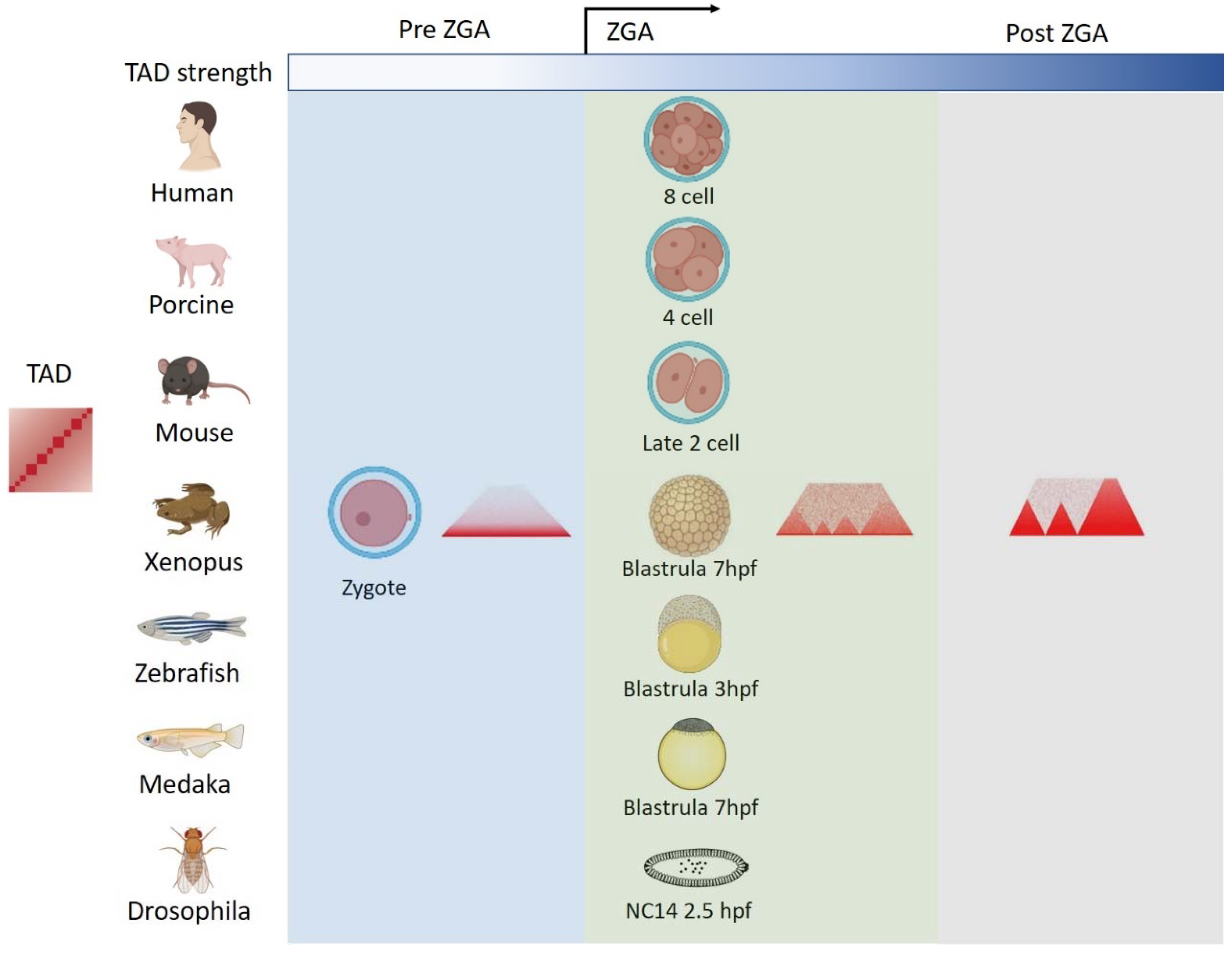
3. The Chromatin Structure Reorganization during Embryogenesis
4. Mechanisms of 3D Chromatin Structure Reorganization in Embryos
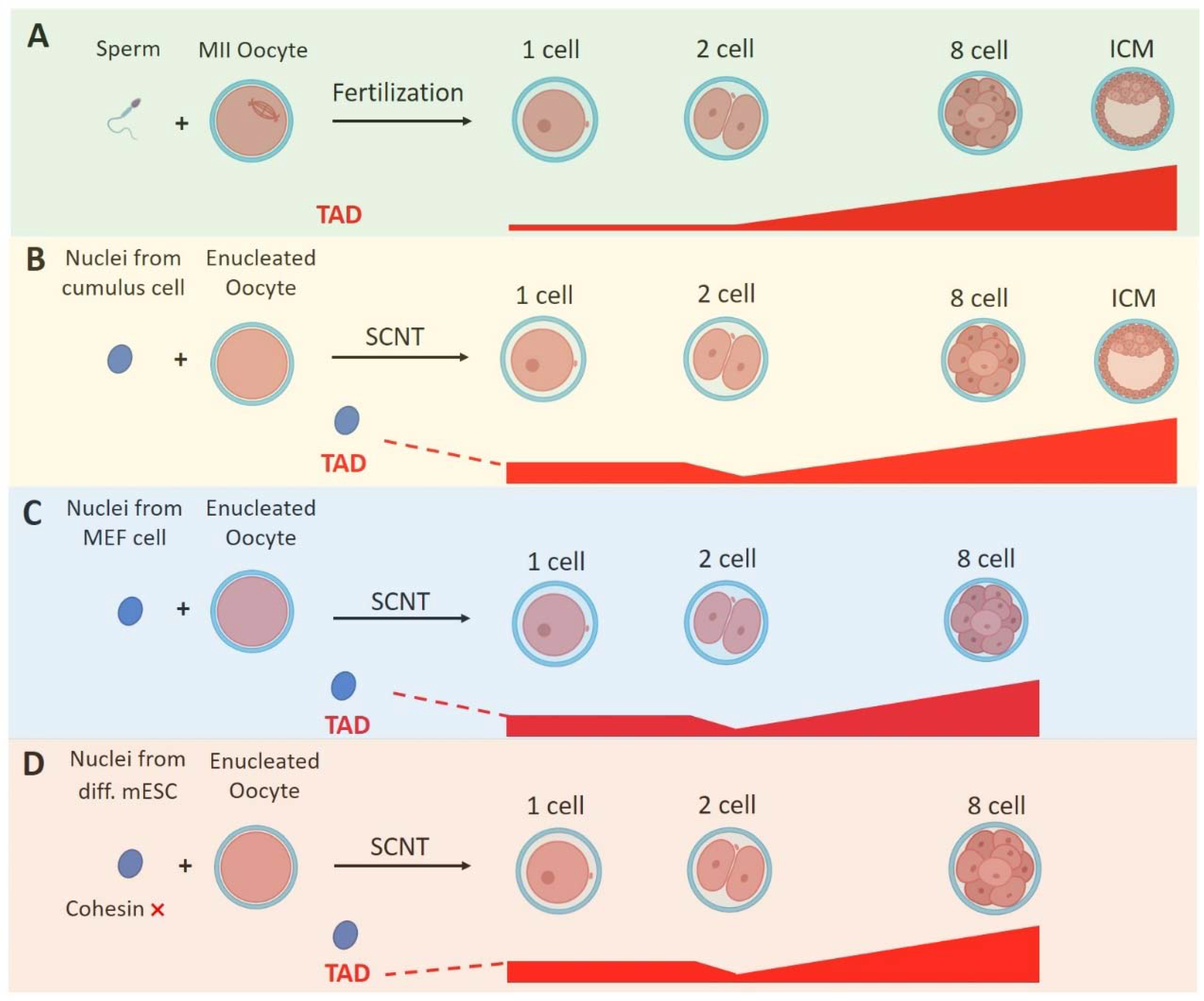
4.1. Zygotic Genome Activation
4.2. DNA Replication
4.3. Insulator Proteins
4.4. Non-Insulator Proteins
4.5. Histone Modifications
4.6. Phase Separation
5. The Role of 3D Chromatin Structures in the Cellular Totipotency
6. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Misteli, T. The Self-Organizing Genome: Principles of Genome Architecture and Function. Cell 2020, 183, 28–45. [Google Scholar] [CrossRef] [PubMed]
- Cremer, T.; Cremer, M. Chromosome territories. Cold Spring Harb. Perspect. Biol. 2010, 2, a003889. [Google Scholar] [CrossRef] [PubMed]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Denker, A.; de Laat, W. The second decade of 3C technologies: Detailed insights into nuclear organization. Genes Dev. 2016, 30, 1357–1382. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Nora, E.P.; Lajoie, B.R.; Schulz, E.G.; Giorgetti, L.; Okamoto, I.; Servant, N.; Piolot, T.; van Berkum, N.L.; Meisig, J.; Sedat, J.; et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 2012, 485, 381–385. [Google Scholar] [CrossRef]
- Andrey, G.; Mundlos, S. The three-dimensional genome: Regulating gene expression during pluripotency and development. Development 2017, 144, 3646–3658. [Google Scholar] [CrossRef]
- Flyamer, I.M.; Gassler, J.; Imakaev, M.; Brandao, H.B.; Ulianov, S.V.; Abdennur, N.; Razin, S.V.; Mirny, L.A.; Tachibana-Konwalski, K. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 2017, 544, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Sauerwald, N.; Kingsford, C. Quantifying the similarity of topological domains across normal and cancer human cell types. Bioinformatics 2018, 34, i475–i483. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Xie, W. The role of 3D genome organization in development and cell differentiation. Nat. Rev. Mol. Cell Biol. 2019, 20, 535–550. [Google Scholar] [CrossRef]
- Cremer, T.; Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001, 2, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Dekker, J.; Heard, E. Structural and functional diversity of Topologically Associating Domains. FEBS Lett. 2015, 589 Pt A, 2877–2884. [Google Scholar] [CrossRef]
- Sexton, T.; Cavalli, G. The role of chromosome domains in shaping the functional genome. Cell 2015, 160, 1049–1059. [Google Scholar] [CrossRef] [PubMed]
- Braude, P.; Bolton, V.; Moore, S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 1988, 332, 459–461. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, D.K.; Schubiger, G. Activation of transcription in Drosophila embryos is a gradual process mediated by the nucleocytoplasmic ratio. Genes Dev. 1996, 10, 1131–1142. [Google Scholar] [CrossRef] [PubMed]
- Erickson, J.W.; Cline, T.W. A bZIP protein, sisterless-a, collaborates with bHLH transcription factors early in Drosophila development to determine sex. Genes Dev. 1993, 7, 1688–1702. [Google Scholar] [CrossRef] [PubMed]
- Minami, N.; Suzuki, T.; Tsukamoto, S. Zygotic gene activation and maternal factors in mammals. J. Reprod. Dev. 2007, 53, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Eckersley-Maslin, M.A.; Alda-Catalinas, C.; Reik, W. Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell Biol. 2018, 19, 436–450. [Google Scholar] [CrossRef]
- Wike, C.L.; Guo, Y.; Tan, M.; Nakamura, R.; Shaw, D.K.; Diaz, N.; Whittaker-Tademy, A.F.; Durand, N.C.; Aiden, E.L.; Vaquerizas, J.M.; et al. Chromatin architecture transitions from zebrafish sperm through early embryogenesis. Genome Res. 2021, 31, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, R.; Motai, Y.; Kumagai, M.; Wike, C.L.; Nishiyama, H.; Nakatani, Y.; Durand, N.C.; Kondo, K.; Kondo, T.; Tsukahara, T.; et al. CTCF looping is established during gastrulation in medaka embryos. Genome Res. 2021, 31, 968–980. [Google Scholar] [CrossRef] [PubMed]
- Niu, L.; Shen, W.; Shi, Z.; Tan, Y.; He, N.; Wan, J.; Sun, J.; Zhang, Y.; Huang, Y.; Wang, W.; et al. Three-dimensional folding dynamics of the Xenopus tropicalis genome. Nat. Genet. 2021, 53, 1075–1087. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ke, Y.; Wu, K.; Zhao, H.; Sun, Y.; Gao, L.; Liu, Z.; Zhang, J.; Tao, W.; Hou, Z.; et al. Key role for CTCF in establishing chromatin structure in human embryos. Nature 2019, 576, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zheng, H.; Huang, B.; Ma, R.; Wu, J.; Zhang, X.; He, J.; Xiang, Y.; Wang, Q.; Li, Y.; et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 2017, 547, 232–235. [Google Scholar] [CrossRef]
- Gassler, J.; Brandao, H.B.; Imakaev, M.; Flyamer, I.M.; Ladstatter, S.; Bickmore, W.A.; Peters, J.M.; Mirny, L.A.; Tachibana, K. A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. EMBO J. 2017, 36, 3600–3618. [Google Scholar] [CrossRef] [PubMed]
- Hug, C.B.; Grimaldi, A.G.; Kruse, K.; Vaquerizas, J.M. Chromatin Architecture Emerges during Zygotic Genome Activation Independent of Transcription. Cell 2017, 169, 216–228.e19. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.W.; Xu, Y.A.; Chen, X.P.; Feng, S.K.; Liu, Z.B.; Sun, Y.Y.; Yao, X.L.; Li, F.Z.; Zhu, W.; Gao, L.; et al. 3D Chromatin Structures of Mature Gametes and Structural Reprogramming during Mammalian Embryogenesis. Cell 2017, 170, 367–381. [Google Scholar] [CrossRef] [PubMed]
- Ostrup, O.; Andersen, I.S.; Collas, P. Chromatin-linked determinants of zygotic genome activation. Cell Mol. Life Sci. 2013, 70, 1425–1437. [Google Scholar] [CrossRef]
- Ausio, J.; Gonzalez-Romero, R.; Woodcock, C.L. Comparative structure of vertebrate sperm chromatin. J. Struct. Biol. 2014, 188, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Carrell, D.T. Epigenetic marks in zebrafish sperm: Insights into chromatin compaction, maintenance of pluripotency, and the role of the paternal genome after fertilization. Asian J. Androl. 2011, 13, 620–621. [Google Scholar] [CrossRef] [PubMed]
- Rathke, C.; Baarends, W.M.; Jayaramaiah-Raja, S.; Bartkuhn, M.; Renkawitz, R.; Renkawitz-Pohl, R. Transition from a nucleosome-based to a protamine-based chromatin configuration during spermiogenesis in Drosophila. J. Cell Sci. 2007, 120 Pt 9, 1689–1700. [Google Scholar] [CrossRef] [PubMed]
- Hammoud, S.S.; Nix, D.A.; Zhang, H.; Purwar, J.; Carrell, D.T.; Cairns, B.R. Distinctive chromatin in human sperm packages genes for embryo development. Nature 2009, 460, 473–478. [Google Scholar] [CrossRef]
- Brykczynska, U.; Hisano, M.; Erkek, S.; Ramos, L.; Oakeley, E.J.; Roloff, T.C.; Beisel, C.; Schubeler, D.; Stadler, M.B.; Peters, A.H. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat. Struct. Mol. Biol. 2010, 17, 679–687. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.F.; Zhang, H.; Cairns, B.R. Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome Res. 2011, 21, 578–589. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zheng, H.; Huang, B.; Li, W.; Xiang, Y.; Peng, X.; Ming, J.; Wu, X.; Zhang, Y.; Xu, Q.; et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 2016, 537, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wu, X.; Zhang, W.; Shen, W.; Sun, Q.; Liu, K.; Zhang, Y.; Wang, Q.; Li, Y.; Meng, A.; et al. Widespread Enhancer Dememorization and Promoter Priming during Parental-to-Zygotic Transition. Mol. Cell 2018, 72, 673–686.e6. [Google Scholar] [CrossRef]
- Patel, L.; Kang, R.; Rosenberg, S.C.; Qiu, Y.; Raviram, R.; Chee, S.; Hu, R.; Ren, B.; Cole, F.; Corbett, K.D. Dynamic reorganization of the genome shapes the recombination landscape in meiotic prophase. Nat. Struct. Mol. Biol. 2019, 26, 164–174. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Zhang, Y.; Du, Z.; Si, W.; Fan, S.; Qin, D.; Wang, M.; Duan, Y.; Li, L.; et al. Reprogramming of Meiotic Chromatin Architecture during Spermatogenesis. Mol. Cell 2019, 73, 547–561.e6. [Google Scholar] [CrossRef]
- Alavattam, K.G.; Maezawa, S.; Sakashita, A.; Khoury, H.; Barski, A.; Kaplan, N. and Namekawa, S.H. Attenuated chromatin compartmentalization in meiosis and its maturation in sperm development. Nat. Struct. Mol. Biol. 2019, 26, 175–184. [Google Scholar] [CrossRef]
- Rathke, C.; Baarends, W.M.; Awe, S.; Renkawitz-Pohl, R. Chromatin dynamics during spermiogenesis. Biochim. Biophys. Acta 2014, 1839, 155–168. [Google Scholar] [CrossRef]
- Naumova, N.; Imakaev, M.; Fudenberg, G.; Zhan, Y.; Lajoie, B.R.; Mirny, L.A.; Dekker, J. Organization of the mitotic chromosome. Science 2013, 342, 948–953. [Google Scholar] [CrossRef]
- Gibcus, J.H.; Samejima, K.; Goloborodko, A.; Samejima, I.; Naumova, N.; Nuebler, J.; Kanemaki, M.T.; Xie, L.; Paulson, J.R.; Earnshaw, W.C.; et al. A pathway for mitotic chromosome formation. Science 2018, 359, eaao6135. [Google Scholar] [CrossRef] [PubMed]
- Tadros, W.; Lipshitz, H.D. The maternal-to-zygotic transition: A play in two acts. Development 2009, 136, 3033–3042. [Google Scholar] [CrossRef] [PubMed]
- Blythe, S.A.; Wieschaus, E.F. Zygotic genome activation triggers the DNA replication checkpoint at the midblastula transition. Cell 2015, 160, 1169–1181. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, D.A.; Hong, J.W.; Zeitlinger, J.; Rokhsar, D.S.; Levine, M.S. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 2008, 105, 7762–7767. [Google Scholar] [CrossRef]
- Chen, K.; Johnston, J.; Shao, W.; Meier, S.; Staber, C.; Zeitlinger, J. A global change in RNA polymerase II pausing during the Drosophila midblastula transition. Elife 2013, 2, e00861. [Google Scholar] [CrossRef]
- Blythe, S.A.; Wieschaus, E.F. Establishment and maintenance of heritable chromatin structure during early Drosophila embryogenesis. Elife 2016, 5, e20148. [Google Scholar] [CrossRef]
- Lu, F.; Liu, Y.; Inoue, A.; Suzuki, T.; Zhao, K.; Zhang, Y. Establishing Chromatin Regulatory Landscape during Mouse Preimplantation Development. Cell 2016, 165, 1375–1388. [Google Scholar] [CrossRef]
- Wu, J.; Huang, B.; Chen, H.; Yin, Q.; Liu, Y.; Xiang, Y.; Zhang, B.; Liu, B.; Wang, Q.; Xia, W.; et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 2016, 534, 652–657. [Google Scholar] [CrossRef]
- Li, X.Y.; Harrison, M.M.; Villalta, J.E.; Kaplan, T.; Eisen, M.B. Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. Elife 2014, 3, e03737. [Google Scholar] [CrossRef]
- Dahl, J.A.; Jung, I.; Aanes, H.; Greggains, G.D.; Manaf, A.; Lerdrup, M.; Li, G.; Kuan, S.; Li, B.; Lee, A.Y.; et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 2016, 537, 548–552. [Google Scholar] [CrossRef]
- Liu, X.; Wang, C.; Liu, W.; Li, J.; Li, C.; Kou, X.; Chen, J.; Zhao, Y.; Gao, H.; Wang, H.; et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 2016, 537, 558–562. [Google Scholar] [CrossRef]
- Collombet, S.; Ranisavljevic, N.; Nagano, T.; Varnai, C.; Shisode, T.; Leung, W.; Piolot, T.; Galupa, R.; Borensztein, M.; Servant, N.; et al. Parental-to-embryo switch of chromosome organization in early embryogenesis. Nature 2020, 580, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Ogiyama, Y.; Schuettengruber, B.; Papadopoulos, G.L.; Chang, J.M.; Cavalli, G. Polycomb-Dependent Chromatin Looping Contributes to Gene Silencing during Drosophila Development. Mol. Cell 2018, 71, 73–88.e5. [Google Scholar] [CrossRef]
- Li, F.; Wang, D.; Song, R.; Cao, C.; Zhang, Z.; Wang, Y.; Li, X.; Huang, J.; Liu, Q.; Hou, N.; et al. The asynchronous establishment of chromatin 3D architecture between in vitro fertilized and uniparental preimplantation pig embryos. Genome Biol. 2020, 21, 203. [Google Scholar] [CrossRef] [PubMed]
- Ing-Simmons, E.; Vaid, R.; Bing, X.Y.; Levine, M.; Mannervik, M.; Vaquerizas, J.M. Independence of chromatin conformation and gene regulation during Drosophila dorsoventral patterning. Nat. Genet. 2021, 53, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Espinola, S.M.; Gotz, M.; Bellec, M.; Messina, O.; Fiche, J.B.; Houbron, C.; Dejean, M.; Reim, I.; Cardozo Gizzi, A.M.; Lagha, M.; et al. Cis-regulatory chromatin loops arise before TADs and gene activation, and are independent of cell fate during early Drosophila development. Nat. Genet. 2021, 53, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Kaaij, L.J.T.; van der Weide, R.H.; Ketting, R.F.; de Wit, E. Systemic Loss and Gain of Chromatin Architecture throughout Zebrafish Development. Cell. Rep. 2018, 24, 1–10.e4. [Google Scholar] [CrossRef] [PubMed]
- Bonev, B.; Cavalli, G. Organization and function of the 3D genome. Nat. Rev. Genet. 2016, 17, 661–678. [Google Scholar] [CrossRef]
- Ryba, T.; Hiratani, I.; Lu, J.; Itoh, M.; Kulik, M.; Zhang, J.; Schulz, T.C.; Robins, A.J.; Dalton, S.; Gilbert, D.M. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010, 20, 761–770. [Google Scholar]
- Yaffe, E.; Farkash-Amar, S.; Polten, A.; Yakhini, Z.; Tanay, A.; Simon, I. Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS Genet. 2010, 6, e1001011. [Google Scholar] [CrossRef]
- Marchal, C.; Sima, J.; Gilbert, D.M. Control of DNA replication timing in the 3D genome. Nat. Rev. Mol. Cell Biol. 2019, 20, 721–737. [Google Scholar] [CrossRef] [PubMed]
- Pope, B.D.; Ryba, T.; Dileep, V.; Yue, F.; Wu, W.; Denas, O.; Vera, D.L.; Wang, Y.; Hansen, R.S.; Canfield, T.K.; et al. Topologically associating domains are stable units of replication-timing regulation. Nature 2014, 515, 402–405. [Google Scholar] [CrossRef] [PubMed]
- Kurukuti, S.; Tiwari, V.K.; Tavoosidana, G.; Pugacheva, E.; Murrell, A.; Zhao, Z.; Lobanenkov, V.; Reik, W.; Ohlsson, R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl. Acad. Sci. USA 2006, 103, 10684–10689. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Mikkelsen, T.S.; Gnirke, A.; Lindblad-Toh, K.; Kellis, M.; Lander, E.S. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc. Natl. Acad. Sci. USA 2007, 104, 7145–7150. [Google Scholar] [CrossRef] [PubMed]
- Ong, C.T.; Corces, V.G. CTCF: An architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014, 15, 234–246. [Google Scholar] [CrossRef] [PubMed]
- Soshnikova, N.; Montavon, T.; Leleu, M.; Galjart, N.; Duboule, D. Functional analysis of CTCF during mammalian limb development. Dev. Cell 2010, 19, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Davidson, I.F.; Peters, J.M. Genome folding through loop extrusion by SMC complexes. Nat. Rev. Mol. Cell Biol. 2021, 22, 445–464. [Google Scholar] [CrossRef]
- Moore, J.M.; Rabaia, N.A.; Smith, L.E.; Fagerlie, S.; Gurley, K.; Loukinov, D.; Disteche, C.M.; Collins, S.J.; Kemp, C.J.; Lobanenkov, V.V.; et al. Loss of maternal CTCF is associated with peri-implantation lethality of Ctcf null embryos. PLoS ONE 2012, 7, e34915. [Google Scholar] [CrossRef] [PubMed]
- Andreu, M.J.; Alvarez-Franco, A.; Portela, M.; Gimenez-Llorente, D.; Cuadrado, A.; Badia-Careaga, C.; Tiana, M.; Losada, A.; Manzanares, M. Establishment of 3D chromatin structure after fertilization and the metabolic switch at the morula-to blastocyst transition require CTCF. BioRxiv 2021. [Google Scholar] [CrossRef]
- Franke, M.; De la Calle-Mustienes, E.; Neto, A.; Almuedo-Castillo, M.; Irastorza-Azcarate, I.; Acemel, R.D.; Tena, J.J.; Santos-Pereira, J.M.; Gomez-Skarmeta, J.L. CTCF knockout in zebrafish induces alterations in regulatory landscapes and developmental gene expression. Nat. Commun. 2021, 12, 5415. [Google Scholar] [CrossRef] [PubMed]
- Moretti, C.; Stevant, I.; Ghavi-Helm, Y. 3D genome organisation in Drosophila. Brief Funct. Genom. 2020, 19, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, A.; Mohana, G.; Dorier, J.; Ozdemir, I.; Omer, A.; Cousin, P.; Semenova, A.; Taschner, M.; Dergai, O.; Marzetta, F.; et al. CTCF loss has limited effects on global genome architecture in Drosophila despite critical regulatory functions. Nat. Commun. 2021, 12, 1011. [Google Scholar] [CrossRef] [PubMed]
- Strunnikov, A.V.; Larionov, V.L.; Koshland, D. SMC1: An essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J. Cell Biol. 1993, 123 Pt 2 Pt 2, 1635–1648. [Google Scholar] [CrossRef]
- Guacci, V.; Koshland, D.; Strunnikov, A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 1997, 91, 47–57. [Google Scholar] [CrossRef]
- Wohlschlegel, J.A.; Dwyer, B.T.; Dhar, S.K.; Cvetic, C.; Walter, J.C.; Dutta, A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 2000, 290, 2309–2312. [Google Scholar] [CrossRef] [PubMed]
- Losada, A.; Hirano, M.; Hirano, T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998, 12, 1986–1997. [Google Scholar] [CrossRef]
- Haering, C.H.; Farcas, A.M.; Arumugam, P.; Metson, J.; Nasmyth, K. The cohesin ring concatenates sister DNA molecules. Nature 2008, 454, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Fudenberg, G.; Imakaev, M.; Lu, C.; Goloborodko, A.; Abdennur, N.; Mirny, L.A. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016, 15, 2038–2049. [Google Scholar] [CrossRef] [PubMed]
- Davidson, I.F.; Bauer, B.; Goetz, D.; Tang, W.; Wutz, G.; Peters, J.M. DNA loop extrusion by human cohesin. Science 2019, 366, 1338–1345. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, W.; Abdennur, N.; Goloborodko, A.; Pekowska, A.; Fudenberg, G.; Loe-Mie, Y.; Fonseca, N.A.; Huber, W.; Haering, C.H.; Mirny, L.; et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 2017, 551, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.S.P.; Huang, S.C.; Glenn St Hilaire, B.; Engreitz, J.M.; Perez, E.M.; Kieffer-Kwon, K.R.; Sanborn, A.L.; Johnstone, S.E.; Bascom, G.D.; Bochkov, I.D.; et al. Cohesin Loss Eliminates All Loop Domains. Cell 2017, 171, 305–320.e24. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, J.D.P.; Feldmann, A.; Hernandez-Rodriguez, B.; Diaz, N.; Brown, J.M.; Fursova, N.A.; Blackledge, N.P.; Prathapan, P.; Dobrinic, P.; Huseyin, M.K.; et al. Cohesin Disrupts Polycomb-Dependent Chromosome Interactions in Embryonic Stem Cells. Cell Rep. 2020, 30, 820–835.e10. [Google Scholar] [CrossRef] [PubMed]
- Vogelmann, J.; Le Gall, A.; Dejardin, S.; Allemand, F.; Gamot, A.; Labesse, G.; Cuvier, O.; Negre, N.; Cohen-Gonsaud, M.; Margeat, E.; et al. Chromatin insulator factors involved in long-range DNA interactions and their role in the folding of the Drosophila genome. PLoS Genet. 2014, 10, e1004544. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Lacroix, L.; Gamot, A.; Cuddapah, S.; Queille, S.; Lhoumaud, P.; Lepetit, P.; Martin, P.G.; Vogelmann, J.; Court, F.; et al. Chromatin immunoprecipitation indirect peaks highlight long-range interactions of insulator proteins and Pol II pausing. Mol. Cell 2014, 53, 672–681. [Google Scholar] [CrossRef] [PubMed]
- Rowley, M.J.; Lyu, X.; Rana, V.; Ando-Kuri, M.; Karns, R.; Bosco, G.; Corces, V.G. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 2008, 456, 400–403. [Google Scholar]
- Schulz, K.N.; Bondra, E.R.; Moshe, A.; Villalta, J.E.; Lieb, J.D.; Kaplan, T.; McKay, D.J.; Harrison, M.M. Zelda is differentially required for chromatin accessibility, transcription factor binding, and gene expression in the early Drosophila embryo. Genome Res. 2015, 25, 1715–1726. [Google Scholar] [CrossRef]
- Zenk, F.; Zhan, Y.; Kos, P.; Loser, E.; Atinbayeva, N.; Schachtle, M.; Tiana, G.; Giorgetti, L.; Iovino, N. HP1 drives de novo 3D genome reorganization in early Drosophila embryos. Nature 2021, 593, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.L.; Nien, C.Y.; Liu, H.Y.; Metzstein, M.M.; Kirov, N.; Rushlow, C. Mammalian ISWI and SWI/SNF selectively mediate binding of distinct transcription factors. Nature 2019, 569, 136–140. [Google Scholar]
- Barisic, D.; Stadler, M.B.; Iurlaro, M.; Schubeler, D. Condensin II Counteracts Cohesin and RNA Polymerase II in the Establishment of 3D Chromatin Organization. Cell Rep. 2019, 26, 2890–2903.e3. [Google Scholar]
- Li, E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 2002, 3, 662–673. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.S.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef]
- Yang, H.; Luan, Y.; Liu, T.; Lee, H.J.; Fang, L.; Wang, Y.; Wang, X.; Zhang, B.; Jin, Q.; Ang, K.C.; et al. A map of cis-regulatory elements and 3D genome structures in zebrafish. Nature 2020, 588, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Borsos, M.; Torres-Padilla, M.E. Building up the nucleus: Nuclear organization in the establishment of totipotency and pluripotency during mammalian development. Genes Dev. 2016, 30, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Fulka, H.; Aoki, F. Nucleolus Precursor Bodies and Ribosome Biogenesis in Early Mammalian Embryos: Old Theories and New Discoveries. Biol. Reprod. 2016, 94, 143. [Google Scholar] [CrossRef] [PubMed]
- Percharde, M.; Lin, C.J.; Yin, Y.; Guan, J.; Peixoto, G.A.; Bulut-Karslioglu, A.; Biechele, S.; Huang, B.; Shen, X.; Ramalho-Santos, M. A LINE1-Nucleolin Partnership Regulates Early Development and ESC Identity. Cell 2018, 174, 391–405.e19. [Google Scholar] [CrossRef]
- Yu, H.; Sun, Z.; Tan, T.; Pan, H.; Zhao, J.; Zhang, L.; Chen, J.; Lei, A.; Zhu, Y.; Chen, L.; et al. rRNA biogenesis regulates mouse 2C-like state by 3D structure reorganization of peri-nucleolar heterochromatin. Nat. Commun. 2021, 12, 6365. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yu, H.; Ma, Q.; Zeng, P.; Wu, D.; Hou, Y.; Liu, X.; Jia, L.; Sun, J.; Chen, Y.; et al. Phase separation of OCT4 controls TAD reorganization to promote cell fate transitions. Cell Stem Cell 2021, 28, 1868–1883.e11. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.K.; Bouchoux, C.; Liu, H.W.; Kim, E.; Minamino, M.; de Groot, R.; Katan, A.J.; Bonato, A.; Marenduzzo, D.; Michieletto, D.; et al. Bridging-induced phase separation induced by cohesin SMC protein complexes. Sci. Adv. 2021, 7, eabe5905. [Google Scholar] [CrossRef]
- Chen, M.; Zhu, Q.; Li, C.; Kou, X.; Zhao, Y.; Li, Y.; Xu, R.; Yang, L.; Yang, L.; Gu, L.; et al. Chromatin architecture reorganization in murine somatic cell nuclear transfer embryos. Nat. Commun. 2020, 11, 1813. [Google Scholar] [CrossRef]
- Zhang, K.; Wu, D.Y.; Zheng, H.; Wang, Y.; Sun, Q.R.; Liu, X.; Wang, L.Y.; Xiong, W.J.; Wang, Q.; Rhodes, J.D.P.; et al. Analysis of Genome Architecture during SCNT Reveals a Role of Cohesin in Impeding Minor ZGA. Mol. Cell 2020, 79, 234–250.e9. [Google Scholar] [CrossRef] [PubMed]
- Macfarlan, T.S.; Gifford, W.D.; Driscoll, S.; Lettieri, K.; Rowe, H.M.; Bonanomi, D.; Firth, A.; Singer, O.; Trono, D.; Pfaff, S.L. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 2012, 487, 57–63. [Google Scholar] [CrossRef]
- Zhu, Y.; Yu, J.; Gu, J.; Xue, C.; Zhang, L.; Chen, J.; Shen, L. Relaxed 3D genome conformation facilitates the pluripotent to totipotent-like state transition in embryonic stem cells. Nucleic Acids Res. 2021, 49, 12167–12177. [Google Scholar] [CrossRef] [PubMed]
- Beagrie, R.A.; Scialdone, A.; Schueler, M.; Kraemer, D.C.; Chotalia, M.; Xie, S.Q.; Barbieri, M.; de Santiago, I.; Lavitas, L.M.; Branco, M.R.; et al. Complex multi-enhancer contacts captured by genome architecture mapping. Nature 2017, 543, 519–524. [Google Scholar] [CrossRef] [PubMed]
- Payne, A.C.; Chiang, Z.D.; Reginato, P.L.; Mangiameli, S.M.; Murray, E.M.; Yao, C.C.; Markoulaki, S.; Earl, A.S.; Labade, A.S.; Jaenisch, R.; et al. In situ genome sequencing resolves DNA sequence and structure in intact biological samples. Science 2021, 371, eaay3446. [Google Scholar] [CrossRef]
- Quinodoz, S.A.; Ollikainen, N.; Tabak, B.; Palla, A.; Schmidt, J.M.; Detmar, E.; Lai, M.M.; Shishkin, A.A.; Bhat, P.; Takei, Y.; et al. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell 2018, 174, 744–757.e24. [Google Scholar] [CrossRef] [PubMed]
- Quinodoz, S.A.; Jachowicz, J.W.; Bhat, P.; Ollikainen, N.; Banerjee, A.K.; Goronzy, I.N.; Blanco, M.R.; Chovanec, P.; Chow, A.; Markaki, Y.; et al. RNA promotes the formation of spatial compartments in the nucleus. Cell 2021, 184, 5775–5790.e30. [Google Scholar] [CrossRef]
- Conte, M.; Fiorillo, L.; Bianco, S.; Chiariello, A.M.; Esposito, A.; Nicodemi, M. Polymer physics indicates chromatin folding variability across single-cells results from state degeneracy in phase separation. Nat. Commun. 2020, 11, 3289. [Google Scholar] [CrossRef]


| Gene | Species | Functions | References |
|---|---|---|---|
| CTCF | Z,X,M,H | function as an insulator protein in TAD establishment | [21,22,69,70] |
| Cohesin | D,M,H | function as a driver to extrude a chromatin loop | [80,81,82,85] |
| BEAF-32 | D | BEAF-32 binds to specific DNA sequences, mediate long-range chromosomal contacts | [83,84] |
| Cp190 | D | bind to BEAF-32, mediate long-range chromosomal contacts | [83,84] |
| Chromator | D | bind to BEAF-32, mediate long-range chromosomal contacts | [83,84] |
| Zelda | D | may relax local chromatin environment to help TAD boundaries and multi-way interaction formation | [25] |
| GAF | D | associate with Zelda to open chromatin | [86] |
| HP1α | D | bind to H3K9me3 in constitutive heterochromatin to establish B compartments | [87] |
| Snf2h | X | mediate CTCF binding to DNA for TAD establishment | [21] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Chen, X. A Tremendous Reorganization Journey for the 3D Chromatin Structure from Gametes to Embryos. Genes 2022, 13, 1864. https://doi.org/10.3390/genes13101864
Chen Z, Chen X. A Tremendous Reorganization Journey for the 3D Chromatin Structure from Gametes to Embryos. Genes. 2022; 13(10):1864. https://doi.org/10.3390/genes13101864
Chicago/Turabian StyleChen, Zhenping, and Xuepeng Chen. 2022. "A Tremendous Reorganization Journey for the 3D Chromatin Structure from Gametes to Embryos" Genes 13, no. 10: 1864. https://doi.org/10.3390/genes13101864
APA StyleChen, Z., & Chen, X. (2022). A Tremendous Reorganization Journey for the 3D Chromatin Structure from Gametes to Embryos. Genes, 13(10), 1864. https://doi.org/10.3390/genes13101864





