The Chloroplast Genome of the Lichen Photobiont Trebouxiophyceae sp. DW1 and Its Phylogenetic Implications
Abstract
1. Introduction
2. Materials and Methods
2.1. Phycobiont Isolation and Culture Conditions
2.2. DNA Extraction, Sequencing and Raw Data Preprocessing
2.3. Genome Assembly
2.4. Gene Prediction
2.5. Codon Usage
2.6. Identification of Repeat and Simple Sequence Repeat
2.7. Phylogenetic Analysis
3. Results
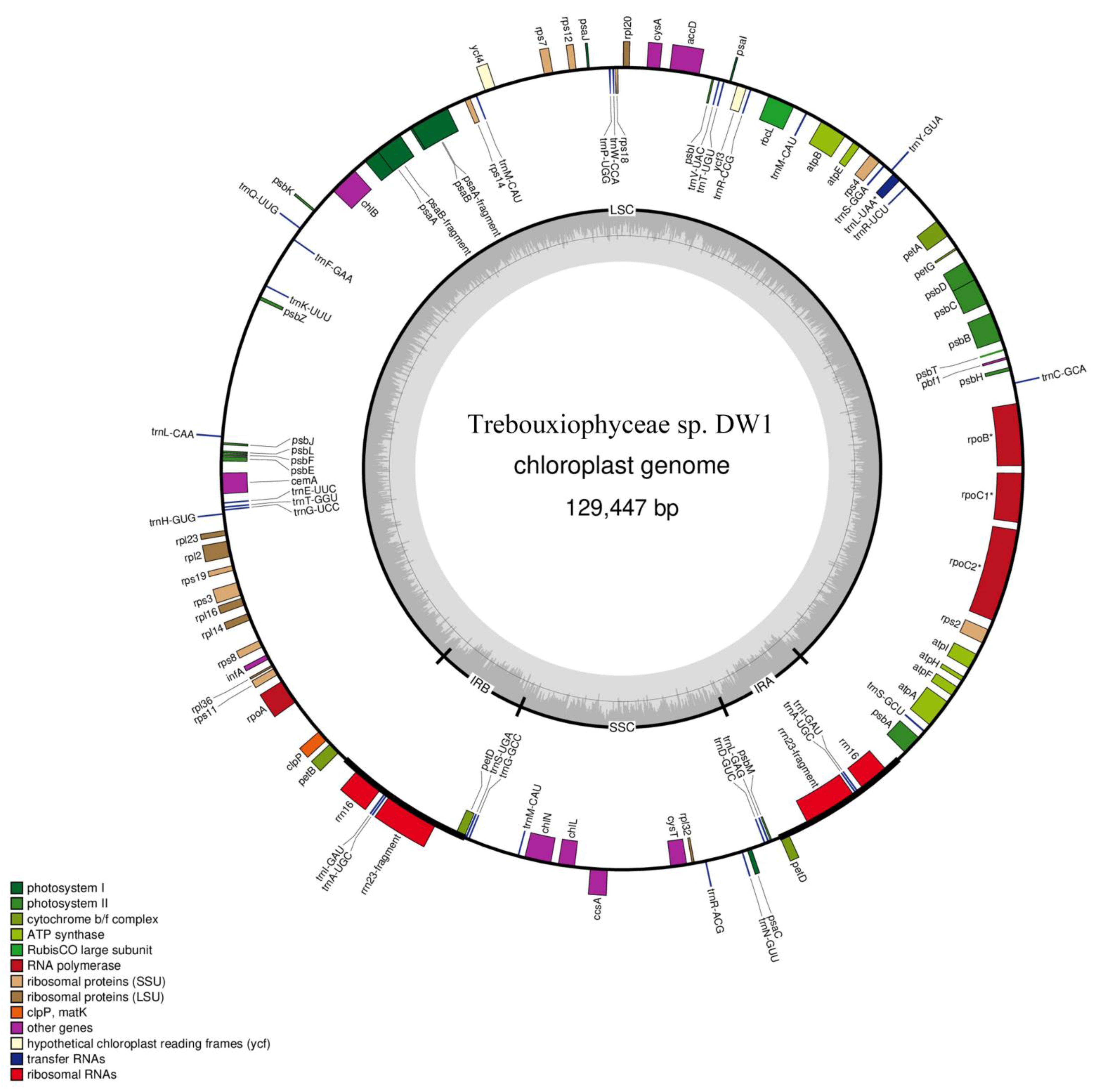
3.1. Structural Characteristics of Chloroplast Genome
3.2. Repeat Elements in the Trebouxiophyceae sp. DW1 Chloroplast Genome
3.3. Codon Usage Analysis
3.4. Phylogenetic Analysis
4. Discussion
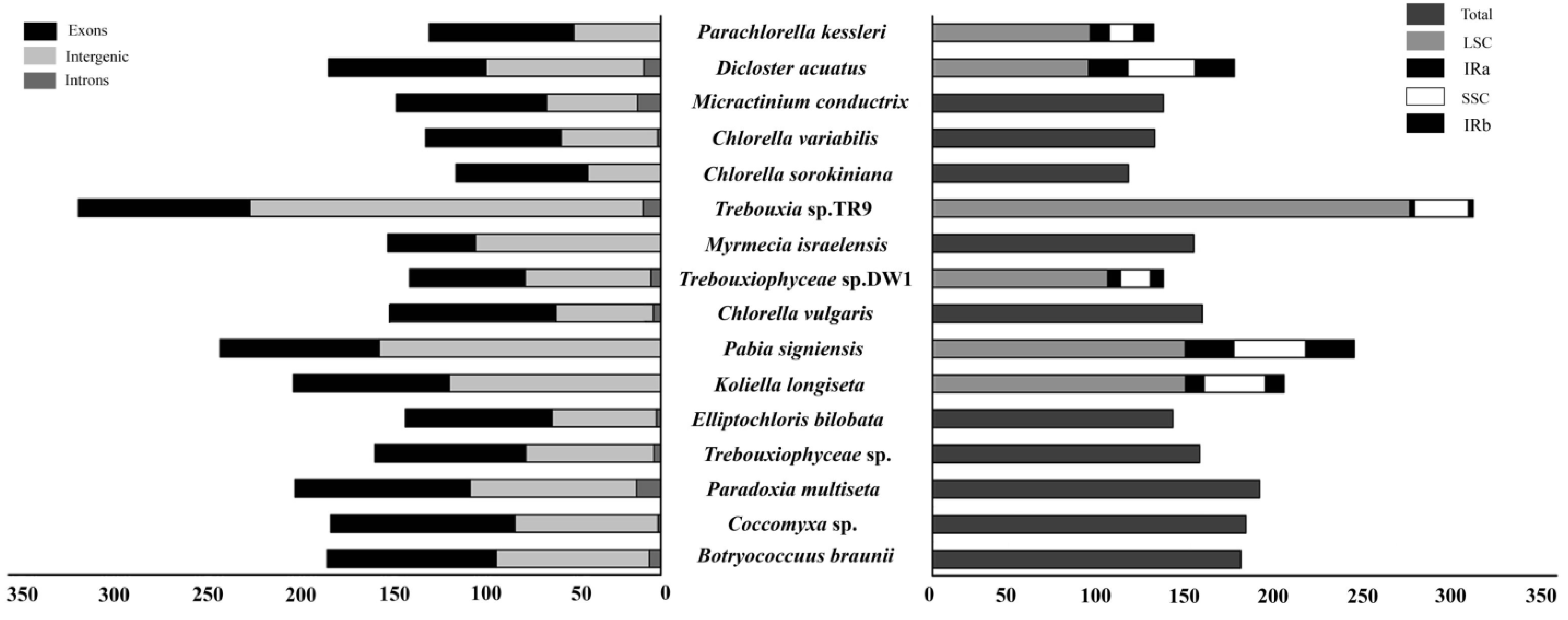
4.1. Features That Affect the Size of the Chloroplast Genome
4.2. Variations in Chloroplast Genome Structure
4.3. Repeat Elements: Significant Mononucleotide Repeats in SSRs
4.4. Codon Usage
4.5. Chloroplast Phylogenomics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Leliaert, F.; Smith, D.R.; Moreau, H.; Herron, M.D.; Verbruggen, H.; Delwiche, C.F.; De Clerck, O. Phylogeny and Molecular Evolution of the Green Algae. Crit. Rev. Plant. Sci. 2012, 31, 1–46. [Google Scholar] [CrossRef]
- Brodie, J.; Chan, C.X.; Clerck, O.D.; Cock, J.M.; Bhattacharya, D. The algal revolution. Trends Plant Sci. 2017, 22, 726–738. [Google Scholar] [CrossRef] [PubMed]
- Muscatine, L.; Trench, R.R.P.K. Symbiosis of algae and invertebrates: Aspects of the symbiont surface and the host-symbiont interface. Trans. Am. Microsc. Soc. 1975, 94, 450–469. [Google Scholar] [CrossRef]
- Wooldridge, S.A. Is the coral-algae symbiosis really ’mutually beneficial’ for the partners? Bioessays 2010, 32, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Kouzuma, A.; Watanabe, K. Exploring the potential of algae/bacteria interactions. Curr. Opin. Biotechnol. 2015, 33, 125–129. [Google Scholar] [CrossRef]
- Lewis, L.A.; McCourt, R.M. Green algae and the origin of land plants. Am. J. Bot. 2004, 91, 1535–1556. [Google Scholar] [CrossRef]
- Leliaert, F.; Verbruggen, H.; Zechman, F.W. Into the deep: New discoveries at the base of the green plant phylogeny. Bioessays 2011, 33, 683–692. [Google Scholar] [CrossRef]
- Moczydlowska, M.; Landing, E.D.; Zang, W.; Palacios, T. Proterozoic phytoplankton and timing of chlorophyte algae origins. Palaeontology 2011, 54, 721–733. [Google Scholar] [CrossRef]
- Boedeker, C.; O’ Kelly, C.J.; Star, W.; Leliaert, F. Molecular phylogeny and taxonomy of the Aegagropila clade (Cladophorales, Ulvophyceae), including the description of Aegagropilopsis gen. nov. and Pseudocladophora gen. nov. J. Phycol. 2012, 48, 808–825. [Google Scholar] [CrossRef]
- Carlile, A.; O’Kelly, C.J.; Sherwood, A.R. The green algal genus Cloniophora represents a novel lineage in the Ulvales: A proposal for Cloniophoraceae fam. nov. J. Phycol. 2011, 47, 1379–1387. [Google Scholar] [CrossRef]
- Fucikova, K.; Leliaert, F.; Cooper, E.D.; Skaloud, P.; Verbruggen, H. New phylogenetic hypotheses for the core Chlorophyta based on chloroplast sequence data. Front. Ecol. Evol. 2014, 2, 63. [Google Scholar]
- Mattox, K.R.; Stewart, K.D. Classifification of the green algae: A concept based on comparative cytology. In Systematics of the Green Algae; Academic Press: London, UK, 1984; Volume 1, pp. 29–72. [Google Scholar]
- Sluiman, H.J. The green algal class Ulvophyceae an ultrastructural survey and classifification. Crypt. Bot. 1989, 1, 83–94. [Google Scholar]
- Arora, M.; Anil, A.C.; Leliaert, F.; Delany, J.; Mesbahi, E. Tetraselmis indica (Chlorodendrophyceae, Chlorophyta), a new species isolated from salt pans in Goa, India. Eur. J. Phycol. 2013, 48, 61–78. [Google Scholar] [CrossRef]
- Norris, R.E.; Hori, T.; Chihara, M. Revision of the genus Tetraselmis (Class Prasinophyceae). J. Plant. Res. 1980, 93, 317–339. [Google Scholar]
- Marin, B. Nested in the Chlorellales or independent class? Phylogeny and classifification of the Pedinophyceae (Viridiplantae) revealed by molecular phylogenetic analyses of complete nuclear and plastid-encoded rRNA operons. Protist 2012, 163, 778–805. [Google Scholar] [CrossRef] [PubMed]
- Fucikova, K.; Lewis, P.O.; Lewis, L.A. Putting incertae sedis taxa in their place: A proposal for ten new families and three new genera in Sphaeropleales (Chlorophyceae, Chlorophyta). J. Phycol. 2014, 50, 14–25. [Google Scholar] [CrossRef]
- Fucikova, K.; Lewis, P.O.; Lewis, L.A. Widespread desert affiliations of Trebouxiophycean algae (Trebouxiophyceae, Chlorophyta) including discovery of three new desert genera. Phycol. Res. 2014, 62, 294–305. [Google Scholar] [CrossRef]
- Gaysina, L.; Nemcova, Y.; Skaloud, P.; Sevcikova, T.; Elias, M. Chloropyrula uraliensis gen. et sp. nov. (Trebouxiophyceae, Chlorophyta), a new green coccoid alga with a unique ultra structure, isolated from soil in South Urals. J. Syst. Evol. 2013, 51, 476–484. [Google Scholar] [CrossRef]
- Neustupa, J.; Elias, M.; Skaloud, P.; Nemcova, Y.; Sejnohova, L. Xylochloris irregularis gen. et sp. nov. (Trebouxiophyceae, Chlorophyta), a novel subaerial coccoid green alga. Phycologia 2011, 50, 57–66. [Google Scholar] [CrossRef][Green Version]
- Rindi, F.; Mcivor, L.; Sherwood, A.R.; Friedl, T.; Guiry, M.D.; Sheath, R.G. Molecular phylogeny of the green algal order Prasiolales (Trebouxiophyceae, Chlorophyta)1. J. Phycol. 2010, 43, 811–822. [Google Scholar] [CrossRef]
- Lucia, M.; Steven, L.; Eva, B. The hidden diversity of lichenised Trebouxiophyceae (Chlorophyta). Phycologia 2018, 57, 503–524. [Google Scholar]
- Spatafora, J.W.; Bushley, K.E. Phylogenomics and evolution of secondary metabolism in plant-associated fungi. Curr. Opin. Plant Biol. 2015, 26, 37–44. [Google Scholar] [CrossRef]
- Monique, T.; Cambiaire, J.D.; Otis, C.; Lemieux, C. Distinctive architecture of the chloroplast genome in the Chlorodendrophycean green algae Scherffelia dubia and Tetraselmis sp. CCMP 881. PLoS ONE 2016, 11, e0148934. [Google Scholar]
- Parks, M.; Cronn, R.; Liston, A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 2009, 7, 84. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Ling, F.; Zhang, Z.; Xin, C.; Penny, D.; Zhong, B. Chloroplast phylogenomic inference of green algae relationships. Sci. Rep. 2016, 6, 20528. [Google Scholar] [CrossRef]
- Ruhfel, B.R.; Gitzendanner, M.A.; Soltis, P.S.; Soltis, D.E.; Burleigh, J.G. From algae to angiosperms-inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol. Biol. 2014, 14, 23. [Google Scholar] [CrossRef]
- Cambiaire, J.; Otis, C.; Turmel, M.; Lemieux, C. The chloroplast genome sequence of the green alga Leptosira terrestris: Multiple losses of the inverted repeat and extensive genome rearrangements within the Trebouxiophyceae. BMC Genom. 2007, 8, 213. [Google Scholar] [CrossRef]
- Lemieux, C.; Otis, C.; Turmel, M. Chloroplast phylogenomic analysis resolves deep-level relationships within the green algal class Trebouxiophyceae. BMC Evol. Biol. 2014, 14, 211. [Google Scholar] [CrossRef]
- Fernando, M.A.; Eva, B.; Leonardo, M.C.; Francisco, G.; Arantzazu, M.; Patricia, M.; Maria, G.H.; Eva, M.C. The chloroplast genome of the lichen-symbiont microalga Trebouxia sp. Tr9 (Trebouxiophyceae, Chlorophyta) shows short inverted repeats with a single gene and loss of the rps4 gene, which is encoded by the nucleus. J. Phycol. 2020, 56, 170–184. [Google Scholar]
- Rippka, R. Photoheterotrophy and chemoheterotrophy among unicellular blue-green algae. Arch. Microbiol. 1972, 87, 93–98. [Google Scholar] [CrossRef]
- Cota-Sánchez, J.H.; Remarchuk, K.; Ubayasena, K. Ready-to-use DNA extracted with a CTAB method adapted for herbarium specimens and mucilaginous plant tissue. Plant Mol. Biol. Rep. 2006, 24, 161–167. [Google Scholar] [CrossRef]
- Khan, M.A.; Bhatia, P.; Sadiq, M. Bbtool: A tool to generate the test cases. IJRTE 2012, 1, 192–197. [Google Scholar]
- Xu, M.; Guo, L.; Gu, S.; Wang, O.; Zhang, Y. Tgs-Gapcloser: A fast and accurate gap closer for large genomes with low coverage of error-prone long reads. GigaScience 2020, 9, gia094. [Google Scholar] [CrossRef] [PubMed]
- Boetzer, M.; Pirovano, W. Toward almost closed genomes with GapFiller. Genom. Biol. 2012, 13, R56. [Google Scholar] [CrossRef] [PubMed]
- Michael, T.; Pascal, L.; Tommaso, P.; Ulbricht-Jones, E.S.; Axel, F.; Ralph, B.; Stephan, G. Geseq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 5, W6–W11. [Google Scholar]
- Qin Cai, Z.Q.; Xia, G.M.; Wang, M.C. Synonymous codon usage bias is correlative to intron number and shows disequilibrium among exons in plants. BMC Genom. 2013, 14, 56. [Google Scholar] [CrossRef]
- Sudhir, K.; Glen, S.; Koichiro, T. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 7, 1870. [Google Scholar]
- Stefan, K.; Choudhuri, J.V.; Enno, O.; Chris, S.; Jens, S.; Robert, G. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 22, 4633–4642. [Google Scholar]
- Gary, B. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 2, 573–580. [Google Scholar]
- Sebastian, B.; Thomas, T.; Thomas, M.; Uwe, S.; Martin, M. Misa-web: A web server for microsatellite prediction. Bioinformatics 2017, 16, 2583–2585. [Google Scholar]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2017, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Subha, K.; Bui, Q.M.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Met. 2017, 14, 587–589. [Google Scholar]
- Bauman, N.; Akella, A.S.; Hann, A.E.; Morey, A.R.; Schwartz, A.A.S. Next-Generation Sequencing of Haematococcus lacustris Reveals an Extremely Large 1.35-Megabase Chloroplast Genome. Genome Announc. 2018, 6, e00181-18. [Google Scholar] [CrossRef]
- Hideto, S.; Rui, K.; Takaaki, I.; Masanobu, O.; Koichi, M.; Atsuhiko, H.; Hiroshi, K. In vitro susceptibility of Prototheca zopfii genotypes 1 and 2. Med. Mycol. 2011, 49, 222–224. [Google Scholar]
- Brouard, J.S.; Otis, C.; Lemieux, C.; Turmel, M. The exceptionally large chloroplast genome of the green alga Floydiella terrestris illuminates the evolutionary history of the Chlorophyceae. Gยืฟหenome Biol. Evol. 2010, 2, 240–256. [Google Scholar] [CrossRef]
- Cremen, M.; Frederik, L.; Marcelino, V.R.; Heroen, V. Large diversity of nonstandard genes and dynamic evolution of chloroplast genomes in siphonous green algae (Bryopsidales, Chlorophyta). Genome Biol. Evol. 2018, 10, 1048–1061. [Google Scholar] [CrossRef] [PubMed]
- Turmel, M.; Otis, C.; Lemieux, C. The complete chloroplast DNA sequences of the charophycean green algae Staurastrum and Zygnema reveal that the chloroplast genome underwent extensive changes during the evolution of the Zygnematales. BMC Biol. 2005, 3, 22. [Google Scholar] [CrossRef]
- Ohno, S. So much "junk" DNA in our genome. Brookhaven Symp. Biol. 1972, 23, 366–370. [Google Scholar]
- Petrov, D.A. Evolution of genome size: New approaches to an old problem. Trends Genet. 2001, 17, 23–28. [Google Scholar] [CrossRef]
- Schubert, I.; Vu, G. Genome stability and evolution: Attempting a holistic view. Trends Plant Sci. 2016, 9, 749–757. [Google Scholar] [CrossRef]
- Doolittle, W.F.; Sapienza, C. Selfish genes, the phenotype paradigm and genome evolution. Nature 1980, 284, 601–603. [Google Scholar] [CrossRef] [PubMed]
- Vinogradov, A.E. Intron-genome size relationship on a large evolutionary scale. J. Mol. Evol. 1999, 49, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Voineagu, I.; Narayanan, V.; Lobachev, K.S.; Mirkin, S.M. Replication stalling at unstable inverted repeats: Interplay between DNA hairpins and fork stabilizing proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 9936–9941. [Google Scholar] [CrossRef] [PubMed]
- Lemoine, F.J.; Degtyareva, N.P.; Lobachev, K.; Petes, T.D. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell 2005, 120, 587–598. [Google Scholar] [CrossRef]
- Lobachev, K.S.; Shor, B.M.; Tran, H.T.; Taylor, W.; Keen, J.D.; Resnick, M.A.; Gordenin, D.A. Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics 1998, 148, 1507–1524. [Google Scholar] [CrossRef]
- Lobachev, K.S.; Gordenin, D.A.; Resnick, M.A. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rear rangements. Cell 2002, 108, 183–193. [Google Scholar] [CrossRef]
- Lobachev, K.S.; Stenger, J.E.; Kozyreva, O.G.; Jurka, J.; Resnick, M.A. Inverted Alu repeats unstable in yeast are excluded from the human genome. Embo J. 2000, 19, 3822–3830. [Google Scholar] [CrossRef]
- Nag, D.K.; Manisha, S.; Stenson, E.K. Both CAG repeats and inverted DNA repeats stimulate spontaneous unequal sister-chromatid exchange in Saccharomyces cerevisiae. Nucleic Acids Res. 2004, 32, 5677–5684. [Google Scholar] [CrossRef][Green Version]
- Lacomme, C.; Hrubikova, K.; Hein, I. Enhancement of virus-induced gene silencing through viral-based production of inverted-repeats. Plant J. 2003, 34, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Leach, D. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays 2010, 16, 893–900. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, K.; Miyabe, I.; Schalbetter, S.A.; Carr, A.M.; Murray, J.M. Recombination-restarted replication makes inverted chromosome fusions at inverted repeats. Nature 2013, 493, 246–249. [Google Scholar] [CrossRef] [PubMed]
- Smith, N.A.; Singh, S.P.; Wang, M.B.; Stoutjesdijk, P.A.; Green, A.G.; Waterhouse, P.M. Total silencing by intron-spliced hairpin RNAs. Nature 2000, 407, 319–320. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Ge, F.; Li, H.; Chen, Y.; Yang, L. PCIR: A database of plant chloroplast inverted repeats. Database 2019, 2019, baz127. [Google Scholar] [CrossRef] [PubMed]
- Wakasugi, T.; Nagai, T.; Kapoor, M.; Sugita, M.; Ito, M.; Ito, S.; Tsudzuki, J.; Nakashima, K.; Tsudzuki, T.; Suzuki, Y.; et al. Complete nucleotide sequence of the chloroplast genome from the green alga Chlorella vulgaris: The existence of genes possibly involved in chloroplast division. Proc. Natl. Acad. Sci. USA 1997, 94, 5967–5972. [Google Scholar] [CrossRef]
- Koning, A.; Keeling, P.J. The complete plastid genome sequence of the parasitic green alga Helicosporidium sp. is highly reduced and structured. BMC Biol. 2006, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Monique, T.; Christian, O.; Claude, L. Dynamic evolution of the chloroplast genome in the green algal classes Pedinophyceae and Trebouxiophyceae. Genome Biol. Evol. 2015, 7, 2062–2082. [Google Scholar]
- Sanderson, M.J.; Copetti, D.; Burquez, A.; Bustamante, E.; Charboneau, J.; Eguiarte, L.E.; Kumar, S.; Lee, H.O.; Lee, J.; Mcmahon, M. Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea): Loss of the ndh gene suite and inverted repeat. Am. J. Bot. 2015, 102, 1115–1127. [Google Scholar] [CrossRef]
- Tomonori, H.; Atsushi Manabu, K.; Teiji, K.; Katsuhiko, T. Complete nucleotide sequence of the Cryptomeria japonica D. Don. chloroplast genome and comparative chloroplast genomics: Diversified genomic structure of coniferous species. BMC Plant Biol. 2008, 8, 70. [Google Scholar]
- Wu, C.S.; Wang, Y.N.; Hsu, C.Y.; Lin, C.P.; Chaw, S.M. Loss of different inverted repeat copies from the chloroplast genomes of Pinaceae and Cupressophytes and influence of heterotachy on the evaluation of gymnosperm phylogeny. Genome Biol. Evol. 2011, 3, 1284–1295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ma, J.; Yang, B.; Li, R.; Zhu, W.; Sun, L.; Tian, J.; Zhang, L. The complete chloroplast genome sequence of Taxus chinensis var. mairei (Taxaceae): Loss of an inverted repeat region and comparative analysis with related species. Gene 2014, 540, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Hao, D.C.; Xiao, P.G.; Huang, B.; Ge, G.B.; Yang, L. Interspecific relationships and origins of Taxaceae and Cephalotaxaceae revealed by partitioned Bayesian analyses of chloroplast and nuclear DNA sequences. Plant Syst. Evol. 2008, 276, 89–104. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, X.; Liu, G.; Yin, Y.; Chen, K.; Yun, Q.; Zhao, D.; Al-Mssallem, I.S.; Yu, J. The Complete Chloroplast Genome Sequence of Date Palm (Phoenix dactylifera L.). PLoS ONE 2010, 5, e12762. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Mishra, R.K.; Singh, L. Genome-wide analysis of microsatellite repeats in humans: Their abundance and density in specific genomic regions. Genome Biol. 2003, 4, R13. [Google Scholar] [CrossRef]
- Legendre, M.; Pochet, N.; Pak, T.; Verstrepen, K.J. Sequence-based estimation of minisatellite and microsatellite repeat variability. Genome Res. 2007, 17, 1787–1796. [Google Scholar] [CrossRef]
- Kuntal, H.; Sharma, V.; Daniell, H. Microsatellite analysis in organelle genomes of Chlorophyta. Bioinformation 2012, 8, 255–259. [Google Scholar] [CrossRef]
- Stackelberg, M.V.; Rensing, S.A.; Reski, R. Identification of genic moss SSR markers and a comparative analysis of twenty-four algal and plant gene indices reveal species-specific rather than group-specific characteristics of microsatellites. BMC Plant Biol. 2006, 6, 9. [Google Scholar] [CrossRef][Green Version]
- Huotari, T.; Korpelainen, H. Complete chloroplast genome sequence of Elodea canadensis and comparative analyses with other monocot plastid genomes. Gene 2012, 508, 96–105. [Google Scholar] [CrossRef]
- Gandhi, S.G.; Awasthi, P.; Bedi, Y.S. Analysis of SSR dynamics in chloroplast genomes of Brassicaceae family. Bioinformation 2010, 5, 16–20. [Google Scholar] [CrossRef]
- Lehmann, J.; Libchaber, A. Degeneracy of the genetic code and stability of the base pair at the second position of the anticodon. RNA 2008, 14, 1264–1269. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.-J.; Zhou, J.; Li, Z.-F.; Wang, L.; Gu, X.; Zhong, Y. Comparative analysis of codon usage patterns among mitochondrion, chloroplast and nuclear genes in Triticum aestivum L. J. Integr. Plant Biol. 2007, 49, 246–254. [Google Scholar] [CrossRef]
- Akashi, H. Synonymous codon usage in Drosophila melanogaster: Natural selection and translational accuracy. Genetics 1994, 136, 927–935. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Nilsson, L.; Kurland, C.G. Co-variation of tRNA abundance and codon usage in Escherichia coliat different growth rates. J. Mol. Biol. 1996, 260, 649–663. [Google Scholar] [CrossRef]
- Yu, C.-H.; Dang, Y.; Zhou, Z.; Wu, C.; Zhao, F.; Sachs, M.S.; Liu, Y. Codon Usage Influences the Local Rate of Translation Elongation to Regulate Co-translational Protein Folding. Mol. Cell 2015, 59, 744–754. [Google Scholar] [CrossRef]
- Chen, L.; Liu, T.; Yang, D.; Nong, X.; Xie, Y.; Fu, Y.; Wu, X.; Huang, X.; Gu, X.; Wang, S.; et al. Analysis of codon usage patterns in Taenia pisiformis through annotated transcriptome data. Biochem. Biophys. Res. Commun. 2013, 430, 1344–1348. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Hanmei, L.; Yong, G. Analysis of characteristic of codon usage in waxy gene of Zea mays. J. Maize Sci. 2008, 2, 432–436. [Google Scholar]
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; Depamphilis, C.W.; Leebens-Mack, J.; Müller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K.; et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef]
- Fang, L.; Leliaert, F.; Novis, P.M.; Zhang, Z.; Zhu, H.; Liu, G.; Penny, D.; Zhong, B. Improving phylogenetic inference of core Chlorophyta using chloroplast sequences with strong phylogenetic signals and heterogeneous models. Mol. Phylogenet. Evol. 2018, 127, 248–255. [Google Scholar] [CrossRef]
- Turmel, M.; Gagnon, M.-C.; O’Kelly, C.J.; Otis, C.; Lemieux, C. The Chloroplast Genomes of the Green Algae Pyramimonas, Monomastix, and Pycnococcus Shed New light on the Evolutionary History of Prasinophytes and the Origin of the Secondary Chloroplasts of Euglenids. Mol. Biol. Evol. 2008, 26, 631–648. [Google Scholar] [CrossRef]
- Huss, V.A.R.; Frank, C.; Hartmann, E.C.; Hirmer, M.; Kloboucek, A.; Seidel, B.M.; Wenzeler, P.; Kessler, E. Biochemical taxonomy and molecular phylogeny of the genus Chlorella sensu Lato (Chlorophyta). J. Phycol. 1999, 35, 587–598. [Google Scholar] [CrossRef]




| Species | GenBank No. |
|---|---|
| Botryococcuus braunii | NC_025545 |
| Coccomyxa sp. | NC_015084 |
| Paradoxia multiseta | NC_025540 |
| Trebouxiophyceae sp. | NC_018569 |
| Elliptochloris bilobata | NC_025548 |
| K. longiseta | NC_025531 |
| P. signiensis | NC_025529 |
| Chlorella vulgaris | NC_001565 |
| Trebouxiophyceae sp. DW1 | MW_255987 |
| Myrmecia israelensis | NC_025525 |
| Trebouxia sp. TR9 | MK_643158 |
| Chlorella sorokiniana | NC_023835 |
| Chlorella variabilis | NC_015359 |
| Micractinium conductrix | NC_036806 |
| Dicloster acuatus | NC_025546 |
| Parachlorella kessleri | NC_012978 |
| Pedinomonas minor | NC_016733 |
| Pedinomonas tuberculata | NC_025530 |
| Marsupiomonas sp. | KM_462870 |
| Nephroselmis astigmatica | NC_024829 |
| Nephroselmis olivacea | NC_000927 |
| Chara vulgaris | NC_008097 |
| Chlorokybus atmophyticus | NC_008822 |
| Number of Bases. | Type | Number |
|---|---|---|
| 30–35 | P | 20 |
| F | 14 | |
| 36–40 | P | 7 |
| F | 4 | |
| 41–45 | P | 2 |
| F | 1 | |
| 46–50 | P | 1 |
| F | 0 |
| Indices | Period | Copy | Consensus | Percent | Percent | Score | A | C | G | T | Entropy |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Size | Number | Size | Matches | Indels | (0–2) | ||||||
| 28,773–28,933 | 77 | 2.1 | 77 | 87 | 5 | 227 | 37 | 14 | 9 | 37 | 1.79 |
| 106,572–106,604 | 12 | 2.8 | 12 | 100 | 0 | 66 | 33 | 18 | 9 | 39 | 1.82 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Zhang, S.; Fang, J.; Jin, X.; Mamut, R.; Li, P. The Chloroplast Genome of the Lichen Photobiont Trebouxiophyceae sp. DW1 and Its Phylogenetic Implications. Genes 2022, 13, 1840. https://doi.org/10.3390/genes13101840
Wang L, Zhang S, Fang J, Jin X, Mamut R, Li P. The Chloroplast Genome of the Lichen Photobiont Trebouxiophyceae sp. DW1 and Its Phylogenetic Implications. Genes. 2022; 13(10):1840. https://doi.org/10.3390/genes13101840
Chicago/Turabian StyleWang, Lidan, Shenglu Zhang, Jinjin Fang, Xinjie Jin, Reyim Mamut, and Pan Li. 2022. "The Chloroplast Genome of the Lichen Photobiont Trebouxiophyceae sp. DW1 and Its Phylogenetic Implications" Genes 13, no. 10: 1840. https://doi.org/10.3390/genes13101840
APA StyleWang, L., Zhang, S., Fang, J., Jin, X., Mamut, R., & Li, P. (2022). The Chloroplast Genome of the Lichen Photobiont Trebouxiophyceae sp. DW1 and Its Phylogenetic Implications. Genes, 13(10), 1840. https://doi.org/10.3390/genes13101840







