Abstract
B-box (BBX) is a zinc finger transcription factor, which is involved in regulating the growth and development of plants and resisting various stresses. In this study, 43 NtBBX genes were identified and divided into five subgroups in tobacco. The members in each subgroup had similar characteristics. The promoter region of NtBBX genes had cis-acting elements related to light response, hormone regulation and stress response. Transcriptome analysis showed that NtBBX30 was significantly up-regulated, and NtBBX12, NtBBX13, NtBBX16 and NtBBX17 were significantly down-regulated under abiotic stresses. The NtBBX genes also responded to the infection of Ralstonia solanacearum. NtBBX9, NtBBX1, NtBBX15 and NtBBX17 showed the greatest response under stresses. The NtBBX genes are expressed in various degrees under different tissues. This research will provide a solid foundation for further study of the biological function of NtBBX genes in tobacco.
1. Introduction
The B-box (BBX) protein, a zinc finger transcription factor with a B-box domain, has attracted much attention due to its multiple biological functions [1]. The BBX proteins have one to two B-box conservative domains at the N-terminal, and some BBX proteins also contain CCT (CONSTANS, CO-like, and TOC1) domains at the C-terminal [2]. The B-box domain is related to the interaction between specific proteins, and the CCT domain plays a regulatory role in gene transcription [3].
The BBX gene has been confirmed to be related to stress tolerance and photomorphogenesis in plants. The BBX genes can regulate abiotic stress to adapt plants to adverse external environment. In Arabidopsis, AtBBX18 negatively regulated heat tolerance [4]. CmBBX24 of chrysanthemum was confirmed to be tolerant to low temperature and drought stress [5,6]. SlBBX17 of tomato was related to heat stress resistance [7], and SlBBX7, SlBBX9 and SlBBX20 are involved in cold tolerance [8]. Molecular biological methods have demonstrated that the BBX genes could enhance the tolerance to salt and drought in transgenic Arabidopsis [9,10,11]. In addition, BBX genes were also related to biotic stress. The grapevine BBX genes can respond to powdery mildew infection [12]. The many functions of BBX genes were intrinsically linked to their ability to defend against abiotic stress through different pathways. MdBBX7, a target of MdMIEL1 E3 ligase, could improve drought tolerance in apple [13]. Sweet potato IbBBX24 could enhance abiotic stress tolerance by activating peroxidase IbPRX17 transcription to scavenge reactive oxygen species [14]. Some BBX genes have been shown to act in hormone signaling pathways [15]. AtBBX5 was upregulated by ABA under salt, osmotic, and dehydration stresses [16]. AtBBX18 antagonized hypocotyl elongation inhibition mediated by blue light by increasing the biological activity GA level [17]. CmBBX24 regulated the flowering time and abiotic stress tolerance by regulating GA biosynthesis [5,6]. MdBBX37 positively regulated JA-mediated cold stress resistance in apple [18]. BBX genes can be induced to express by light and play an important role in photomorphogenesis [19]. AtBBX30 and AtBBX31, acting downstream of HY5, negatively regulated photomorphogenesis in Arabidopsis [20]. Under blue light conditions, OsBBX14 positively regulated rice photomorphogenesis by activating OsHY5L1 expression [21]. The AtBBX22IR and AtBBX24IR, Arabidopsis BBX alternative splice variants produced by intron retention inhibited hypocotyl elongation by light mediation in Arabidopsis [22].
Tobacco is an important economic crop and one of the perfect model plants in scientific research. In recent years, due to global warming and natural disasters, tobacco is suffering from omnipresent threats. As a transcription factor, the BBX gene can participate in plant development, hormone signal transduction and the regulation of biotic and abiotic stresses. Therefore, it will be of great significance to study the function of the BBX gene family in stress tolerance and development in tobacco. The BBX gene family had been identified and analyzed at the whole genome level in many species, including rice [23], wheat [24], tomato [8], petunia [25], apple [26], pear [27] and grape [28]. With the complete sequencing of the whole genome of tobacco, the systematic identification and analysis of the BBX gene family become possible. In this study, the physicochemical properties, conservative domains, gene structure, cis-acting elements and gene expression pattern of the tobacco BBX gene family was comprehensively analyzed. Phylogenetic analysis can understand the evolutionary relationship of gene families among different species [29,30,31]. Transcriptome analysis has been used to identify potential genes that play an important role in plant development [32,33,34]. This study will provide valuable information for further functional research of BBX genes in tobacco and also provide a reference for subsequent molecular mechanism research.
2. Materials and Methods
2.1. Identification of BBX Genes in the Nicotiana Tabacum L. genome
The tobacco TN90 genome was obtained from the NCBI (https://www.ncbi.nlm.nih.gov/assembly/GCF_000715135.1/ (accessed on 8 May 2021)) [35]. The hidden Markov model (HMM) profile of the B-box domain (Pfam00643) downloaded from the Pfam database (https://pfam.xfam.org/ (accessed on 18 November 2021)) [36] was used as a query to identify BBX genes in the proteins sequences file of the tobacco genome using HMMER3.0. When there are multiple transcripts of the same gene, the longest transcript is selected as the BBX gene. Proteins sequences and the sequences ID of tobacco BBX genes were obtained after deduplication and identification for the B-box conserved domain using the Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (accessed on 2 December 2021)) [37]. The number of amino acids (AA), molecular weight (MW), isoelectric point (pI), and grand average of hydropathicity of the corresponding amino acid sequence (GRAVY) were calculated by the ExPASy website (http://web.expasy.org/protparam/ (accessed on 4 December 2021)) using proteins sequences of tobacco BBX genes [38]. The subcellular localization (Loc) of tobacco BBX proteins was predicted through WoLF PSORT (https://www.genscript.com/wolf-psort.html (accessed on 4 December 2021)) [39].
2.2. Phylogenetic and Conserved Domain Alignments Analysis
The Arabidopsis BBX proteins sequences were attained from the TAIR database (http://www.arabidopsis.org (accessed on 17 September 2021)) [40]. The ClustalW module within MEGA-X was used to align the BBX proteins of tobacco and Arabidopsis thaliana, and the phylogenetic tree was constructed using the Neighbor-Joining (NJ) method with 1000 bootstrap-replications [41]. The iTOL website (https://itol.embl.de/ (accessed on 11 June 2022)) was used to beautify the phylogenetic tree [42]. Amino acid sequences of the B-box and CCT conserved domains arranged through CDD results were shown by TBtools [43]. These conserved domains sequences were aligned with ClustalW and GeneDoc software, and sequence logos were generated using Weblogo (http://weblogo.berkeley.edu/logo.cgi (accessed on 6 October 2022)) [44].
2.3. Analysis of Gene Structure and Conserved Motifs
The tobacco BBX gene structure information was extracted from a genome annotation GFF file. The conserved motif of the tobacco BBX proteins was predicted by the MEME website (https://meme-suite.org/meme/tools/meme (accessed on 6 October 2022)) with ten motifs; the minimum width was set to 6, and the maximum width was set to 50 [45]. The gene structure information was extracted from GFF gene annotation files. The conserved motif and exon–intron structure were drawn using TBtools.
2.4. Cis-Acting Elements Analysis in the BBX Genes Promoter
The promoters sequences were considered to be 2000 bp upstream of the first CDS of the tobacco BBX gene and was gained from the genome file and genome annotation file by TBtools. The cis-acting element was predicted using the PlantCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 21 November 2021)) [46] and was visualized by TBtools.
2.5. Expression Analysis of BBX Genes in Different Stress, Hormone and Tissue
RNA-seq data were used to analyze the expression patterns of BBX genes in tobacco under different conditions. RNA-seq data including NaCl (SRP193166), NaHCO3 (SRP193166), Cold (SRP097876), dehydration (SRP301492), Ralstonia solanacearum (R. solanacearum) (SRP336664), Melatonin (SRP301492), and different plant tissues (SRP101432) were obtained from the Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra/ (accessed on 10 May 2022)) [47]. The data of stems, stem apexes and roots were downloaded to analyze the difference of gene expression between different plant tissues. In addition, transcriptome data under abscisic acid (ABA) treatment came from laboratory data [48].
Trimmomatic [49] was used to remove the adapter and cut off the first 12 bases of reads (except ABA). Hisat2 [50] was exploited to establish the genome index and map reads. The Samtools [51] command was applied to convert the sam file to a bam file. Stringtie [52] was employed to calculate FPKM. The counts value was obtained by the prepDE.py3 program provided by stringtie. The differently expressed genes were analyzed by using DEseq2 [53]. The differently expressed genes screening standard was |log2FoldChange|≥ 1 and padj ≤ 0.05. The results were displayed by TBtools with a heatmap.
3. Results
3.1. Identification of BBX Genes in the Tobacco Genome
To identify BBX genes in the tobacco genome, the hidden Markov model (HMM) profile of the B-box domain (Pfam00643) was employed to search protein sequence files. In total, 43 tobacco BBX proteins sequences were obtained after deleting repeated sequences and identifying conserved domains. For the consistency of naming, tobacco BBX genes were named NtBBX1 to NtBBX43 (Table 1). In addition, multiple transcripts were found for four NtBBX genes (Table S1). The length of the NtBBX proteins sequences ranged from 176 amino acids to 473 amino acids. The maximum molecular weight and the minimum molecular weight were 51,997.34 and 19,539.31, respectively. Their pI ranged from 4.31 to 8.42 and grand average of hydropathicity (GRAVY) scores ranged from −0.938 to −0.282. Subcellular localization showed that most (79%) NtBBX proteins existed in the nucleus, such as NtBBX1, NtBBX2, and NtBBX8.

Table 1.
The fundamental information of NtBBX genes.
3.2. Conserved Domain and Phylogenetic Analysis of NtBBX Gene Family
In order to study the phylogenetic relationships of the NtBBX family, a phylogenetic tree was constructed using 43 tobacco BBX proteins and 32 Arabidopsis BBX proteins (Figure 1). According to the phylogenetic analysis, 43 NtBBX genes were divided into five subgroups. The number of members in the five subgroups was 7, 4, 6, 18, and 8, respectively. Each member of subgroup I and subgroup II had both contained two B-box domains and one CCT domain (B1+B2+CCT) (Figure 2), but they had differences in amino acid sequence (Figure 3A). Each member of subgroup III contained a B-box domain at the amino terminus and a CCT domain at the carboxy terminus. Each member of subgroup IV and subgroup V had only the B-box domain. The difference was that each member of subgroup IV had two B-box domains, and each member of subgroup V had one B-box domain.
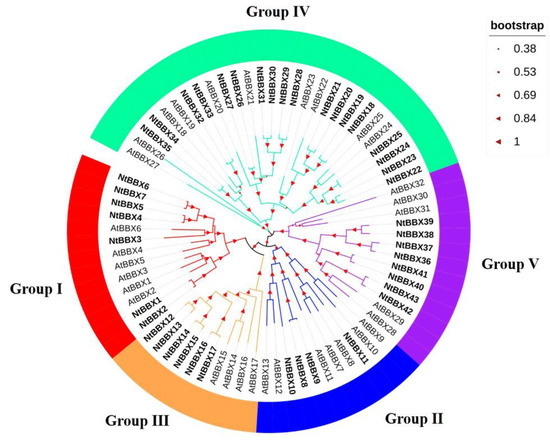
Figure 1.
Phylogenetic tree of BBX proteins sequences from tobacco and Arabidopsis. The phylogenetic tree was constructed using MEGA-X with the Neighbor-Joining (NJ) method with 1000 bootstrap replications. The tree was divided into five groups, and the bootstrap values were indicated by the size of asterisks.
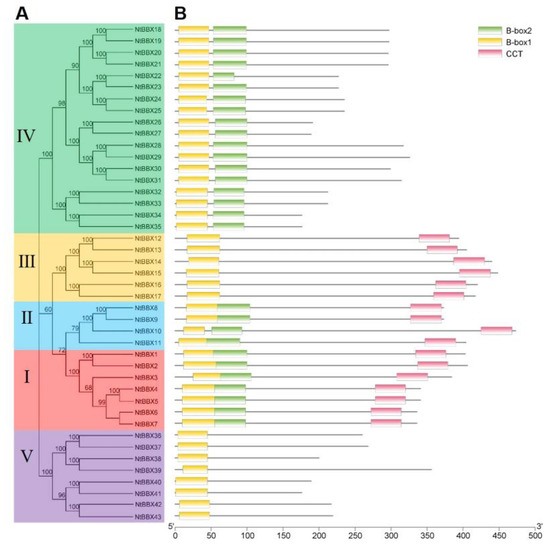
Figure 2.
Conserved domains of NtBBX proteins. (A) Phylogenetic tree of NtBBX proteins sequences from tobacco. The phylogenetic tree was constructed using MEGA-X with the Neighbor-Joining (NJ) method with 1000 bootstrap replications; (B) Conserved domains of NtBBX. B-box1 domains are indicated by yellow boxes, B-box2 domains are indicated by green boxes, and CCT domains are indicated by pink boxes.
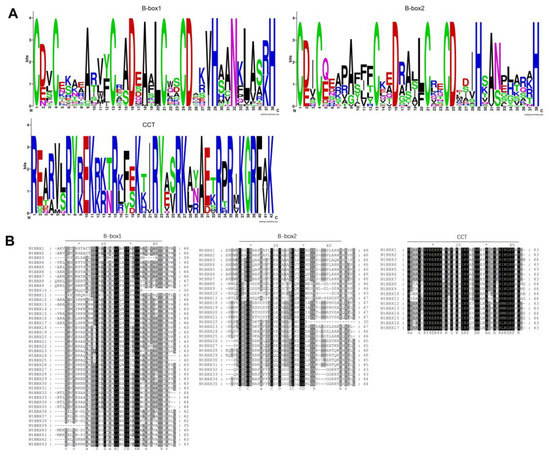
Figure 3.
Sequence characteristics of domains of NtBBX proteins. (A) Sequence logos alignment of the B-box1, B-box2, and CCT domains. Each colored letter represents an amino acid residue. The height of amino acids is directly proportional to the conservation of amino acids in this position; (B) Multi-sequence alignment of the B-box1, B-box2 and CCT domains of the NtBBX proteins. The black background means that the amino acids here are absolutely conservative.
3.3. Domain Alignments and Sequence Logos
In order to explore the amino acid sequence composition of each characteristic domain and its conservation, sequence logos (Figure 3A), and multi-sequence alignment (Figure 3B) of domains were performed. The results showed that B-box1 and B-box2 had high sequence similarity. In contrast to the B-box2 domain, the B-box1 domain was more conservative for the Alanine (A) of 9, 15, 18, and 35 of B-box1. In addition, the CCT domain was also highly conservative, such as Arginine (R), Lysine (K), and Tyrosine (Y).
3.4. Gene Structure and Conserved Motifs
The number of exons of NtBBX genes ranged from 2 to 6. In the same subgroup, NtBBX genes had similar numbers of exons (Figure 4B). Except for NtBBX1, NtBBX2 and NtBBX6 in subgroup I, had three exons, and the rest had two exons.
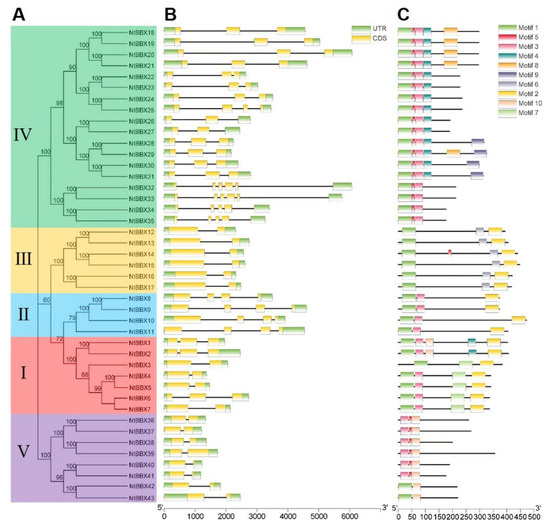
Figure 4.
Gene structure and motif of NtBBX gene family. (A) Phylogenetic tree of NtBBX proteins sequences from tobacco; (B) Gene structure of NtBBX genes. UTR are represented by green boxes, CDS are represented by yellow boxes, and black lines represent intron; (C) Motif of NtBBX proteins. Ten different categories of motif are represented by boxes of ten colors. Detailed sequence features of motifs are shown in Supplementary Figure S1.
All members of the subgroups II, III, and V had four, two, and two exons, respectively. Most members of the subgroup IV had three exons. Gene members of the same subgroup had similar motif types (Figure 4C).
3.5. Cis-Acting Elements of NtBBX Genes
The cis-acting elements of the promoter region of NtBBX genes were analyzed to reveal the potential function of the NtBBX family (Figure 5). Three main types of cis-acting elements in the NtBBX genes were light-responsive elements, hormone-responsive elements and stress-responsive elements. The light-responsive elements were widely distributed in each NtBBX gene. The MeJA responsiveness element, gibberellin responsiveness element, auxin responsiveness element, abscisic acid responsiveness element, and salicylic acid responsiveness element belonged to hormone-responsive elements. The anaerobic induction element, low-temperature responsiveness element, drought inducibility element and defense and stress responsiveness element belonged to abiotic stress-responsive elements. In addition to the three kinds of elements mentioned above, NtBBX genes contained elements related to circadian control, meristem expression, flavonoid biosynthetic, and zein metabolism regulation.
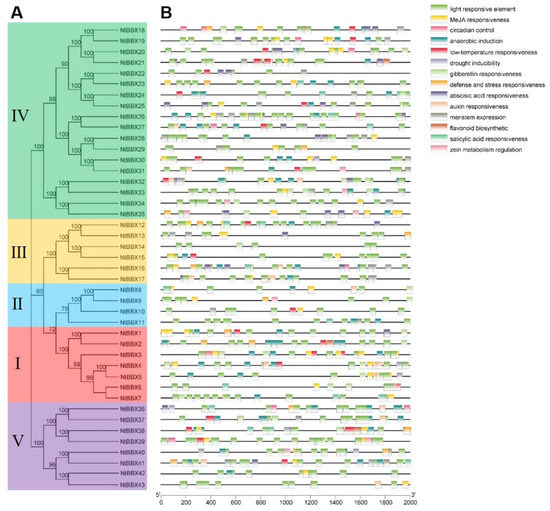
Figure 5.
Cis-acting elements in promoter region of NtBBX genes. (A) Phylogenetic tree of NtBBX proteins sequences from tobacco; (B) Cis-acting elements. Detailed information of cis-acting elements is provided in Supplementary Table S2.
3.6. Expression Patterns of NtBBX Genes under Abiotic Stress
Under salt stress, 13 NtBBX genes were differentially expressed (Figure 6A). The expression levels of NtBBX9 and NtBBX30 were significantly up-regulated, while the other NtBBX genes were significantly down-regulated. When tobacco was exposed to alkali stress, the expression levels of 18 NtBBX genes were significantly changed (Figure 6B). Five NtBBX genes were significantly up-regulated, which were NtBBX14, NtBBX17, NtBBX22, NtBBX27, and NtBBX30. Under dehydration stress, the expression levels of 28 NtBBX genes were changed significantly (Figure 6C). Twenty NtBBX genes were significantly down-regulated, and eight NtBBX genes were significantly up-regulated. Interestingly, all NtBBX genes of subgroup I and subgroup III were significantly down-regulated. The expression of 25 NtBBX genes was significantly different between cold stress and normal condition (Figure 6D). The majority (11/18) of the up-regulated NtBBX genes belonged to subgroup IV, and the majority (4/7) of the down-regulated NtBBX genes belonged to subgroup III.
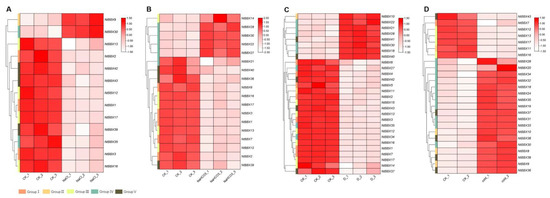
Figure 6.
Expression patterns of NtBBX genes under abiotic stress. (A) Expression pattern of NtBBX genes under NaCl stress; (B) Expression pattern of NtBBX genes under NaHCO3 stress; (C) Expression pattern of NtBBX genes under dehydration stress; (D) Expression pattern of NtBBX genes under cold stress. Subgroups I–V are represented by different colored boxes.
3.7. Expression Patterns of NtBBX Genes under Biotic Stress
After tobacco was infected by R. solanacearum, different NtBBX genes showed different expression patterns (Figure 7). The expression levels of some NtBBX genes were significantly increased after 10 days of infection with R. solanacearum, including NtBBX8, NtBBX14-15 and NtBBX28. The expression levels of some NtBBX genes were significantly decreased after 10 days, including NtBBX17, NtBBX21, NtBBX35 and NtBBX40-41. The expression level of NtBBX9 was significantly increased after 17 days of infection with R. solanacearum. The expression levels of some NtBBX genes were significantly decreased after 17 days, including NtBBX2, NtBBX16 and NtBBX22-23. After 10 and 17 days, the expression levels of some NtBBX genes were significantly decreased, including NtBBX1, NtBBX24-25 and NtBBX34.
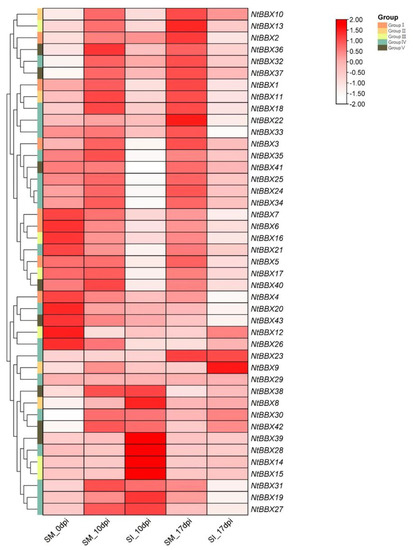
Figure 7.
Expression patterns of NtBBX genes under biotic stress. SI, no infection. SM, infection with R. solanacearum. Dpi, days after infection. Subgroups I–V are represented by different colored boxes.
3.8. Expression Patterns of NtBBX Genes under Hormone Treatment
Eleven NtBBX genes were differentially expressed under ABA treatment (Figure 8A). Six NtBBX genes were significantly up-regulated, which were NtBBX11, NtBBX30, NtBBX36–37 and NtBBX40–41. Five NtBBX genes were significantly down-regulated, which were NtBBX12–15 and NtBBX28.
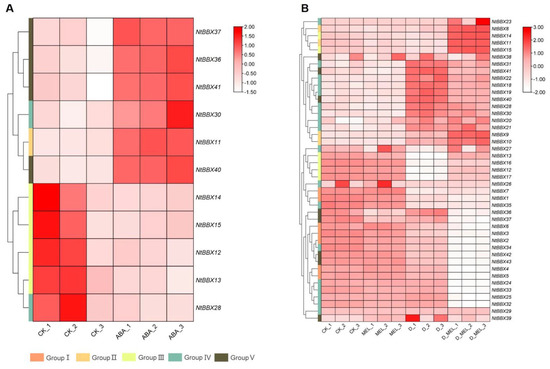
Figure 8.
Expression of NtBBX genes under hormone induction. (A) Expression pattern of NtBBX genes under ABA treatment; (B) Expression pattern of NtBBX genes under dehydrate stress and melatonin treatment. D, dehydrate. MEL, melatonin. Subgroups I–V are represented by different colored boxes.
The NtBBX genes did not respond strongly under a single melatonin treatment, and the differentially expressed genes had only NtBBX15, NtBBX36, and NtBBX37 (Figure 8B). Under dehydration stress and melatonin treatment, 21 NtBBX genes were significantly down-regulated, including NtBBX1–7, NtBBX12–13, NtBBX16–17, NtBBX24–25, NtBBX32–37 and NtBBX42–43. Nine NtBBX genes were significantly up-regulated, including NtBBX9–11, NtBBX14–15, NtBBX21–22, NtBBX30 and NtBBX40.
3.9. Expression Patterns of NtBBX Genes in Different Tissues
NtBBX genes were expressed in all three tissues (Figure 9). NtBBX genes were mainly expressed in stems and stem apexes, but less in roots, such as NtBBX4, NtBBX5, NtBBX6, NtBBX7, NtBBX16 and NtBBX17. NtBBX genes with high expression in stems mainly belonged to subgroup I and subgroup IV. Some NtBBX genes were expressed more in stem apexes than in stems, for example, NtBBX6, NtBBX7 and NtBBX35.
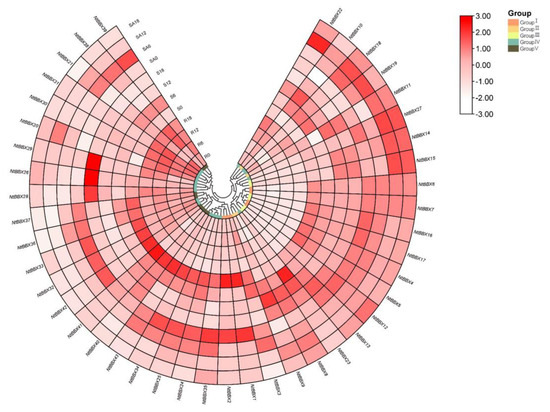
Figure 9.
Expression patterns of NtBBX genes in different tissues. R, roots. S, stems. SA, stem apexes. Subgroups I–V are represented by different colored boxes.
4. Discussion
BBX is a transcription factor of zinc finger protein, which plays a prominent role in plant growth and development, hormone response, and stress tolerance [54], but the function of BBX genes in tobacco has not been studied. In this study, the phylogeny, gene structure, cis-acting element, and expression pattern under diverse conditions of the NtBBX genes were comprehensively analyzed in tobacco. Genome-wide analysis of the NtBBX genes will establish a solid foundation for NtBBX genes function research in tobacco.
4.1. The Number and Classification of Tobacco NtBBX Genes
In this study, 43 NtBBX genes were obtained from the tobacco genome data. Even though both tobacco and tomato belong to Solanaceae, the number of BBX genes in tobacco is more than that in tomato (29) [8]. It might be that tobacco is an allotetraploid which is a combination of two diploid wild species and has more homologous genes [55]. Whole genome duplication (WGD) and segmental duplication may be important reasons for NtBBX gene family expansion [56,57]. The evolutionary tree was constructed using the BBX genes of Arabidopsis thaliana and tobacco, and finally, the NtBBX genes were divided into five subgroups. This was consistent with the grouping of Arabidopsis [2]. Coincidentally, the type and number of conserved domains in the AtBBX genes and the NtBBX genes were identical in each subgroup. For example, the AtBBX genes and the NtBBX genes in subgroup III both contain a B-box domain and a CCT domain. The BBX genes of tobacco and Arabidopsis in the same branch may have similar biological functions.
4.2. Tobacco NtBBX Genes and Photomorphogenesis
BBX genes are related to the development of light morphology in plants [58]. Cis-acting element analysis showed that a large number of photoresponsive elements were in the promoter region of NtBBX genes, for example, G-box, GT1-motif, and TCT-motif. The type and number of cis-acting elements in the promoter region are closely related to the gene’s function, leading to differential expression of the gene [59]. Studies on Arabidopsis have shown that AtBBX21 and AtBBX22 promote photomorphogenesis, while AtBBX24 and AtBBX25 inhibit photomorphogenesis [1,15,19]. From the evolutionary tree, NtBBX18–NtBBX35 were grouped into the same group as the four Arabidopsis BBX genes mentioned above, suggesting that they might be involved in the photomorphogenesis of tobacco. The NtBBX genes function in the photomorphogenesis will require further study in the future, which is very significant for tobacco improvement.
4.3. Tobacco NtBBX Genes and External Stresses
Stress cis-acting elements were widely present in the promoter region of tobacco NtBBX genes, suggesting that tobacco NtBBX genes may take part in tobacco response to stress. Therefore, the expression patterns of tobacco NtBBX genes were analyzed under NaCl, NaHCO3, cold, dehydration, and R. solanacearum stresses. The results showed that most tobacco NtBBX genes were differentially expressed under five stresses, and some genes distributed in the same subgroup had similar expression patterns (Figure 10). All NtBBX genes in subgroup I negatively regulated one or more stresses. All NtBBX genes in subgroup III had negative regulatory effects on dehydration stress. In four abiotic stresses, the number of NtBBX genes up-regulated by cold stress was the largest, which was followed by that by dehydration and NaHCO3 stress, and the number of NtBBX genes up-regulated by NaCl stress was the smallest. This may suggest that NtBBX genes play a more positive role in regulating temperature stress than osmotic stress [60]. Notably, most of the genes significantly up-regulated under these abiotic stresses conditions belong to subgroup IV. A study showed that the tomato subgroup IV SIBBX20 gene could induce the expression of cold-responsive (COR) genes to adapt to cold stress [8]; the study on Ginkgo biloba showed that the salt tolerance of transgenic poplar overexpressing GbBBX25 was improved [61]. These facts indicate that the fourth subfamily genes have an important effect in plants response to abiotic stress. NtBBX30 was significantly up-regulated, and NtBBX12, NtBBX13, NtBBX16 and NtBBX17 were significantly down-regulated under the four abiotic stresses. These genes are widely involved in the stress resistance process of tobacco and have an important influence on the growth and development of tobacco, which is worthy of further study on their functions. NtBBX9 and NtBBX1 showed the greatest response under abiotic stress. Tobacco NtBBX genes can respond to R. solanacearum infection in different degrees. The expression levels of NtBBX genes significantly differentially expressed under abiotic stress also changed under pathogen infection. NtBBX15 and NtBBX17 showed the greatest response under R. solanacearum stress. As a transcription factor, NtBBX genes were closely related to the abiotic and biotic stress tolerance of tobacco. These NtBBX genes could be used as the key genes of tobacco resistance to a variety of stresses, and it was expected to create tobacco multi-resistant germplasm by transgenic technology in the late stage. The NtBBX genes were highly expressed in stems and stem apexes, but they were relatively low in roots, which indicates that the expression of tobacco NtBBX genes was tissue-specific.

Figure 10.
Differential expression under multiple stresses and hormone treatment. The red boxes, blue boxes and gray boxes represent significant up-regulation, significant down-regulation and no significant change for NtBBX genes under corresponding conditions, respectively. The color depth of red boxes and blue boxes represents the size of log2 fold change.
4.4. Tobacco NtBBX Genes and Hormone Response
A large number of hormone-responsive elements were also present in tobacco NtBBX genes promoted such as ABA, GA and SA, suggesting that these genes may regulate many hormone signal transduction pathways and regulate the growth and development processes of tobacco. Transcriptome analysis showed that the tobacco NtBBX genes were regulated by ABA and melatonin (Figure 10).
Among the eleven differentially expressed genes under ABA treatment, more than half (4/6) of the up-regulated NtBBX genes were located in subgroup V, and more than half (4/5) of the down-regulated NtBBX genes were located in subgroup III. Subgroup preference suggests that two groups of NtBBX genes may defend against external stress through different signal transduction pathways. The CmBBX19 of chrysanthemum negatively regulated the drought tolerance through the ABA-dependent pathway [62]. The tobacco NtBBX genes may also regulate various abiotic stresses through the ABA pathway. NtBBX9, NtBBX11, NtBBX14 and NtBBX15 were significantly up-regulated under melatonin treatment and dehydration stress, which were contrary to the expression pattern under single dehydration stress, suggesting that these tobacco NtBBX genes participated in tobacco resistance to dehydration stress through the melatonin pathway.
ABA and melatonin regulate tobacco NtBBX gene to adapt to abiotic stress. The molecular mechanism of the hormone response of BBX genes in tobacco and the downstream pathway involved may be complex, which needs to be further explored using physiology and biochemistry combined with forward and reverse genetics.
5. Conclusions
In this study, the genome-wide analysis of the tobacco BBX gene family was performed, and we identified a total of 43 NtBBX genes. The physicochemical properties, phylogeny, conservative domain, gene structure, conservative motif, cis-acting element and expression patterns under various conditions of the NtBBX gene family were systematically analyzed in tobacco. A large number of light-responsive elements, hormone responsive elements, and stress-responsive elements existed in tobacco NtBBX genes. Transcriptome analysis showed that the NtBBX genes were responsive to salt, alkali, cold, dehydration and R. solanacearum infection. NtBBX9, NtBBX1, NtBBX15 and NtBBX17 showed the greatest response under stress. NtBBX30 expression was significantly up-regulated in all four abiotic stresses, and NtBBX12, NtBBX13, NtBBX16, and NtBBX17 were significantly down-regulated. The NtBBX genes of subgroup IV may have an essential impact on cold stress. The NtBBX genes also showed tissue specificity, with more expression in stems and stem apexes but less in roots. In conclusion, these studies will establish a solid foundation for further research on NtBBX gene function in tobacco.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13101841/s1, Figure S1: Detailed sequence features of ten motifs; Table S1: Multiple transcripts for four NtBBX genes. Table S2: Detailed information of cis-acting elements.
Author Contributions
Conceptualization, L.Y. and H.T.; methodology, L.Y.; software, B.L.; validation, K.S. and Y.L.; formal analysis, K.S.; investigation, K.S.; resources, K.S., L.Q. and X.C.; data curation, Y.S.; writing—original draft preparation, K.S. and B.L.; writing—review and editing, K.S. and H.W.; visualization, K.S.; supervision, H.T.; project administration, L.Y.; funding acquisition, L.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Foundation of Shandong Province Modern Agricultural Technology System Innovation Team (SDAIT-25-01) and Taishan brand cigarette high-quality core raw material development and application in Shandong (202102004).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article or Supplementary Material.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar] [CrossRef]
- Khanna, R.; Kronmiller, B.; Maszle, D.R.; Coupland, G.; Holm, M.; Mizuno, T.; Wu, S.H. The Arabidopsis B-box zinc finger family. Plant Cell 2009, 21, 3416–3420. [Google Scholar] [CrossRef]
- Gendron, J.M.; Pruneda-Paz, J.L.; Doherty, C.J.; Gross, A.M.; Kang, S.E.; Kay, S.A. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 2012, 109, 3167–3172. [Google Scholar] [CrossRef]
- Wang, Q.; Tu, X.; Zhang, J.; Chen, X.; Rao, L. Heat stress-induced BBX18 negatively regulates the thermotolerance in Arabidopsis. Mol. Biol. Rep. 2013, 40, 2679–2688. [Google Scholar] [CrossRef]
- Imtiaz, M.; Yang, Y.; Liu, R.; Xu, Y.; Khan, M.A.; Wei, Q.; Gao, J.; Hong, B. Identification and functional characterization of the BBX24 promoter and gene from chrysanthemum in Arabidopsis. Plant Mol. Biol. 2015, 89, 1–19. [Google Scholar] [CrossRef]
- Yang, Y.; Ma, C.; Xu, Y.; Wei, Q.; Imtiaz, M.; Lan, H.; Gao, S.; Cheng, L.; Wang, M.; Fei, Z.; et al. A Zinc Finger Protein Regulates Flowering Time and Abiotic Stress Tolerance in Chrysanthemum by Modulating Gibberellin Biosynthesis. Plant Cell 2014, 26, 2038–2054. [Google Scholar] [CrossRef]
- Xu, X.; Wang, Q.; Li, W.; Hu, T.; Wang, Q.; Yin, Y.; Liu, X.; He, S.; Zhang, M.; Liang, Y.; et al. Overexpression of SlBBX17 affects plant growth and enhances heat tolerance in tomato. Int. J. Biol. Macromol. 2022, 206, 799–811. [Google Scholar] [CrossRef]
- Bu, X.; Wang, X.; Yan, J.; Zhang, Y.; Zhou, S.; Sun, X.; Yang, Y.; Ahammed, G.J.; Liu, Y.; Qi, M.; et al. Genome-Wide Characterization of B-Box Gene Family and Its Roles in Responses to Light Quality and Cold Stress in Tomato. Front. Plant Sci. 2021, 12, 698525. [Google Scholar] [CrossRef]
- Liu, X.; Li, R.; Dai, Y.; Yuan, L.; Sun, Q.; Zhang, S.; Wang, X. A B-box zinc finger protein, MdBBX10, enhanced salt and drought stresses tolerance in Arabidopsis. Plant Mol. Biol. 2019, 99, 437–447. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, H.; Ping, Q.; Zhang, Z.; Guan, Z.; Fang, W.; Chen, S.; Chen, F.; Jiang, J.; Zhang, F. The heterologous expression of CmBBX22 delays leaf senescence and improves drought tolerance in Arabidopsis. Plant Cell Rep. 2019, 38, 15–24. [Google Scholar] [CrossRef]
- Wu, H.; Wang, X.; Cao, Y.; Zhang, H.; Hua, R.; Liu, H.; Sui, S. CpBBX19, a B-Box Transcription Factor Gene of Chimonanthus praecox, Improves Salt and Drought Tolerance in Arabidopsis. Genes 2021, 12, 1456. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, L.; Ji, M.; Wu, Y.; Zhang, S.; Zhu, Y.; Yao, J.; Li, Z.; Gao, H.; Wang, X. Genome-wide identification and expression analysis of the B-box transcription factor gene family in grapevine (Vitis vinifera L.). BMC Genom. 2021, 22, 221. [Google Scholar] [CrossRef]
- Chen, P.; Zhi, F.; Li, X.; Shen, W.; Yan, M.; He, J.; Bao, C.; Fan, T.; Zhou, S.; Ma, F.; et al. Zinc-finger protein MdBBX7/MdCOL9, a target of MdMIEL1 E3 ligase, confers drought tolerance in apple. Plant Physiol. 2022, 188, 540–559. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Z.; Li, X.; Gao, X.; Dai, Z.; Cui, Y.; Zhi, Y.; Liu, Q.; Zhai, H.; Gao, S.; et al. The IbBBX24-IbTOE3-IbPRX17 module enhances abiotic stress tolerance by scavenging reactive oxygen species in sweet potato. New Phytol. 2022, 233, 1133–1152. [Google Scholar] [CrossRef]
- Vaishak, K.P.; Yadukrishnan, P.; Bakshi, S.; Kushwaha, A.K.; Ramachandran, H.; Job, N.; Babu, D.; Datta, S. The B-box bridge between light and hormones in plants. J. Photochem. Photobiol. B 2019, 191, 164–174. [Google Scholar] [CrossRef]
- Min, J.H.; Chung, J.S.; Lee, K.H.; Kim, C.S. The CONSTANS-like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis. J. Integr. Plant Biol. 2015, 57, 313–324. [Google Scholar] [CrossRef]
- Wang, Q.; Zeng, J.; Deng, K.; Tu, X.; Zhao, X.; Tang, D.; Liu, X. DBB1a, involved in gibberellin homeostasis, functions as a negative regulator of blue light-mediated hypocotyl elongation in Arabidopsis. Planta 2011, 233, 13–23. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Zhang, X.W.; You, C.X.; Hao, Y.J. Apple B-box protein BBX37 regulates jasmonic acid mediated cold tolerance through the JAZ-BBX37-ICE1-CBF pathway and undergoes MIEL1-mediated ubiquitination and degradation. New Phytol. 2020, 229, 2707–2729. [Google Scholar] [CrossRef]
- Yadav, A.; Ravindran, N.; Singh, D.; Rahul, P.V.; Datta, S. Role of Arabidopsis BBX proteins in light signaling. J. Plant Biochem. Biotechnol. 2020, 29, 623–635. [Google Scholar] [CrossRef]
- Heng, Y.; Lin, F.; Jiang, Y.; Ding, M.; Yan, T.; Lan, H.; Zhou, H.; Zhao, X.; Xu, D.; Deng, X.W. B-Box Containing Proteins BBX30 and BBX31, Acting Downstream of HY5, Negatively Regulate Photomorphogenesis in Arabidopsis. Plant Physiol. 2019, 180, 497–508. [Google Scholar] [CrossRef]
- Bai, B.; Lu, N.; Li, Y.; Guo, S.; Yin, H.; He, Y.; Sun, W.; Li, W.; Xie, X. OsBBX14 promotes photomorphogenesis in rice by activating OsHY5L1 expression under blue light conditions. Plant Sci. 2019, 284, 192–202. [Google Scholar] [CrossRef]
- Huang, C.K.; Lin, W.D.; Wu, S.H. An improved repertoire of splicing variants and their potential roles in Arabidopsis photomorphogenic development. Genome Biol. 2022, 23, 50. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhao, X.; Weng, X.; Wang, L.; Xie, W. The rice B-box zinc finger gene family: Genomic identification, characterization, expression profiling and diurnal analysis. PLoS ONE 2012, 7, e48242. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Jiang, W.; Yin, J.; Wang, S.; Fang, Z.; Ma, D.; Gao, D. Genome-wide mining of wheat B-BOX zinc finger (BBX) gene family provides new insights into light stress responses. Crop Pasture Sci. 2021, 72, 17–37. [Google Scholar] [CrossRef]
- Wen, S.; Zhang, Y.; Deng, Y.; Chen, G.; Yu, Y.; Wei, Q. Genomic identification and expression analysis of the BBX transcription factor gene family in Petunia hybrida. Mol. Biol. Rep. 2020, 47, 6027–6041. [Google Scholar] [CrossRef]
- Liu, X.; Li, R.; Dai, Y.; Chen, X.; Wang, X. Genome-wide identification and expression analysis of the B-box gene family in the Apple (Malus domestica Borkh.) genome. Mol. Genet. Genom. 2018, 293, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Han, Y.; Meng, D.; Li, D.; Jiao, C.; Jin, Q.; Lin, Y.; Cai, Y. B-BOX genes: Genome-wide identification, evolution and their contribution to pollen growth in pear (Pyrus bretschneideri Rehd.). BMC Plant Biol. 2017, 17, 156. [Google Scholar] [CrossRef]
- Liu, W.; Tang, R.; Zhang, Y.; Liu, X.; Gao, Y.; Dai, Z.; Li, S.; Wu, B.; Wang, L. Genome-wide identification of B-box proteins and VvBBX44 involved in light-induced anthocyanin biosynthesis in grape (Vitis vinifera L.). Planta 2021, 253, 114. [Google Scholar] [CrossRef]
- Zhang, Z.; Quan, S.; Niu, J.; Guo, C.; Kang, C.; Liu, J.; Yuan, X. Genome-Wide Identification, Classification, Expression and Duplication Analysis of bZIP Family Genes in Juglans regia L. Int. J. Mol. Sci. 2022, 23, 5961. [Google Scholar] [CrossRef]
- Chen, C.; Xie, F.; Shah, K.; Hua, Q.; Chen, J.; Zhang, Z.; Zhao, J.; Hu, G.; Qin, Y. Genome-Wide Identification of WRKY Gene Family in Pitaya Reveals the Involvement of HmoWRKY42 in Betalain Biosynthesis. Int. J. Mol. Sci. 2022, 23, 10568. [Google Scholar] [CrossRef]
- Jin, M.; Gong, X.; Zhang, Q.; Chen, Y.; Ma, H.; Zhang, T.; Wu, C.; Zhang, R.; Zhang, Q.; Tao, S.; et al. Genome-wide analysis and expression pattern of the PIN gene family during Korla fragrant pear calyx development. Acta Physiol. Plant. 2022, 44, 55. [Google Scholar] [CrossRef]
- Groen, S.C.; Calic, I.; Joly-Lopez, Z.; Platts, A.E.; Choi, J.Y.; Natividad, M.; Dorph, K.; Mauck, W.M., 3rd; Bracken, B.; Cabral, C.L.U.; et al. The strength and pattern of natural selection on gene expression in rice. Nature 2020, 578, 572–576. [Google Scholar] [CrossRef]
- Sun, S.; Chen, H.; Yang, Z.; Lu, J.; Wu, D.; Luo, Q.; Jia, J.; Tan, J. Identification of WRKY transcription factor family genes in Pinus massoniana Lamb. and their expression patterns and functions in response to drought stress. BMC Plant Biol. 2022, 22, 424. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Li, B.; Rizwan, H.M.; Sun, K.; Zeng, J.; Shi, M.; Guo, T.; Chen, F. Genome-wide identification and comprehensive analyses of NAC transcription factor gene family and expression analysis under Fusarium kyushuense and drought stress conditions in Passiflora edulis. Front. Plant Sci. 2022, 13, 972734. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Katz, K.; Shutov, O.; Lapoint, R.; Kimelman, M.; Brister, J.R.; O’Sullivan, C. The Sequence Read Archive: A decade more of explosive growth. Nucleic Acids Res. 2022, 50, D387–D390. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Li, H.; Zhang, W.; Tang, H.; Yang, L. Transcriptional regulation and functional analysis of Nicotiana tabacum under salt and ABA stress. Biochem. Biophys. Res. Commun. 2021, 570, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zhang, X.; Jia, H.; Li, T.; Wu, J.; Nagarajan, R.; Lei, L.; Powers, C.; Kan, C.C.; Hua, W.; Liu, Z.; et al. TaCol-B5 modifies spike architecture and enhances grain yield in wheat. Science 2022, 376, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Sierro, N.; Battey, J.N.; Ouadi, S.; Bakaher, N.; Bovet, L.; Willig, A.; Goepfert, S.; Peitsch, M.C.; Ivanov, N.V. The tobacco genome sequence and its comparison with those of tomato and potato. Nat. Commun. 2014, 5, 3833. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef]
- Yu, L.; Lyu, Z.; Liu, H.; Zhang, G.; He, C.; Zhang, J. Insights into the evolutionary origin and expansion of the BBX gene family. Plant Biotechnol. Rep. 2022, 16, 205–214. [Google Scholar] [CrossRef]
- Bursch, K.; Toledo-Ortiz, G.; Pireyre, M.; Lohr, M.; Braatz, C.; Johansson, H. Identification of BBX proteins as rate-limiting cofactors of HY5. Nat. Plants 2020, 6, 921–928. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Huang, S.; Chen, C.; Xu, M.; Wang, G.; Xu, L.A.; Wu, Y. Overexpression of Ginkgo BBX25 enhances salt tolerance in Transgenic Populus. Plant Physiol. Biochem. 2021, 167, 946–954. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhao, X.; Aiwaili, P.; Mu, X.; Zhao, M.; Zhao, J.; Cheng, L.; Ma, C.; Gao, J.; Hong, B. A zinc finger protein BBX19 interacts with ABF3 to affect drought tolerance negatively in chrysanthemum. Plant J. 2020, 103, 1783–1795. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).