Development of Genomic Resources in Mexican Bursera (Section: Bullockia: Burseraceae): Genome Assembly, Annotation, and Marker Discovery for Three Copal Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Sequencing
2.2. Draft Genome Assembly and Annotation
2.3. Marker Discovery
3. Results
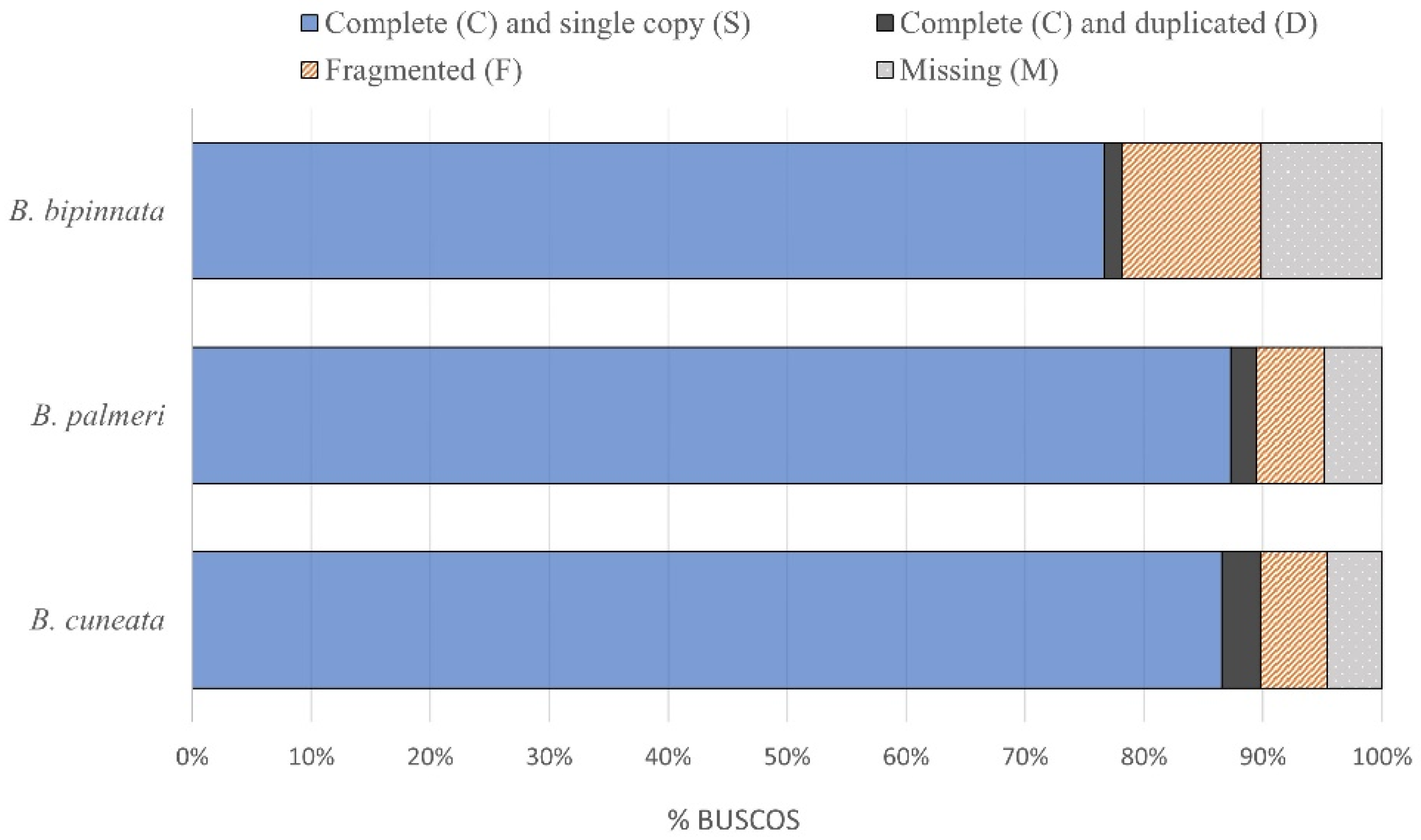
3.1. Genome Assembly and Annotation
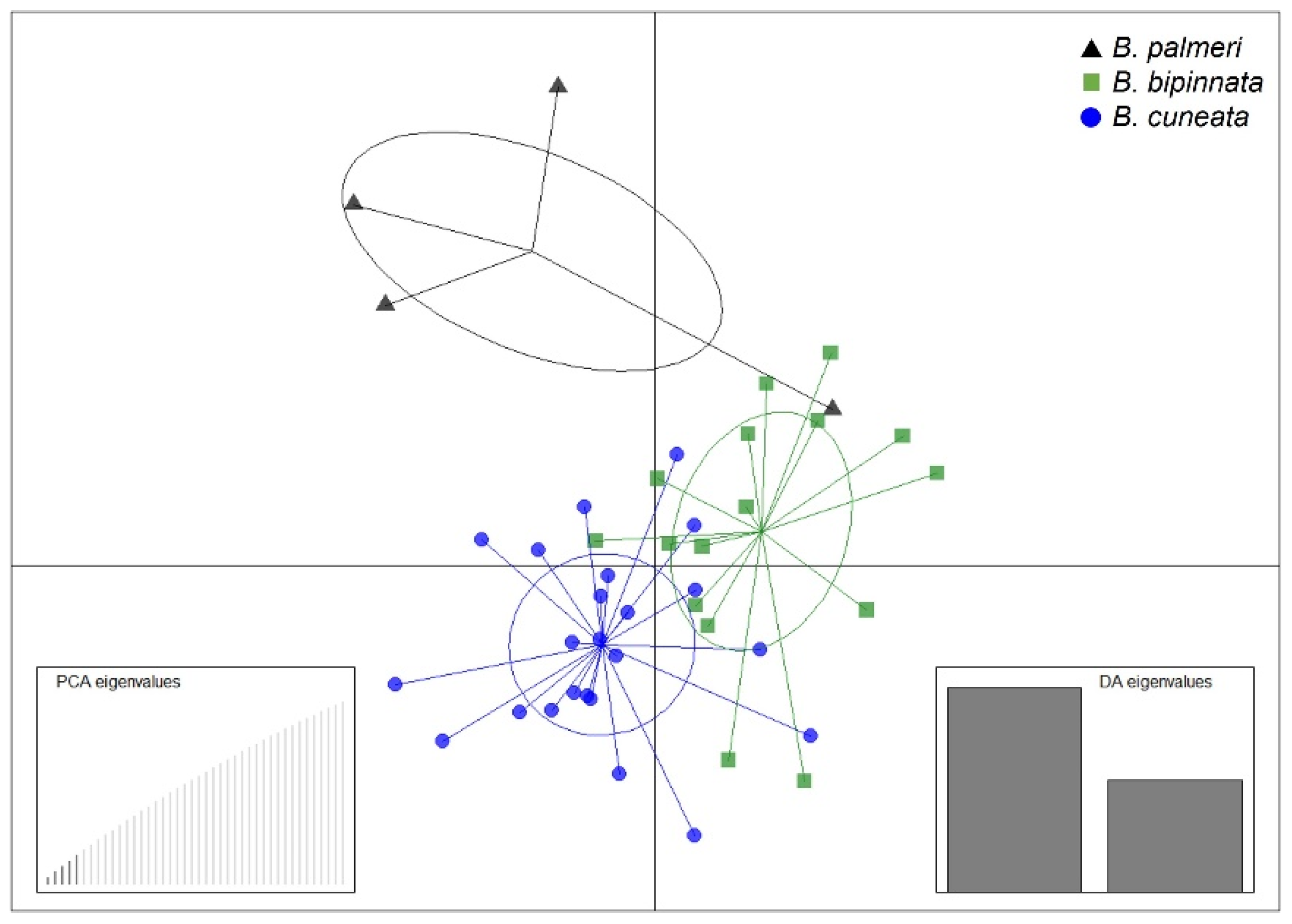
3.2. SSR and SNP Discovery
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sampling Permit
References
- Ekblom, R.; Wolf, J.B.W. A Field Guide to Whole-Genome Sequencing, Assembly and Annotation. Evol. Appl. 2014, 7, 1026–1042. [Google Scholar] [CrossRef] [PubMed]
- Bellin, D.; Ferrarini, A.; Chimento, A.; Kaiser, O.; Levenkova, N.; Bouffard, P.; Delledonne, M. Combining Next-Generation Pyrosequencing with Microarray for Large Scale Expression Analysis in Non-Model Species. BMC Genom. 2009, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Unamba, C.I.N.; Nag, A.; Sharma, R.K. Next Generation Sequencing Technologies: The Doorway to the Unexplored Genomics of Non-Model Plants. Front. Plant Sci. 2015, 6, 1074. [Google Scholar] [CrossRef] [PubMed]
- Hoban, S.; Campbell, C.D.; da Silva, J.M.; Ekblom, R.; Funk, W.C.; Garner, B.A.; Godoy, J.A.; Kershaw, F.; MacDonald, A.J.; Mergeay, J.; et al. Genetic Diversity Is Considered Important but Interpreted Narrowly in Country Reports to the Convention on Biological Diversity: Current Actions and Indicators Are Insufficient. Biol. Conserv. 2021, 261, 109233. [Google Scholar] [CrossRef]
- Formenti, G.; Theissinger, K.; Fernandes, C.; Bista, I.; Bombarely, A.; Bleidorn, C.; Ciofi, C.; Crottini, A.; Godoy, J.A.; Höglund, J.; et al. The Era of Reference Genomes in Conservation Genomics. Trends Ecol. Evol. 2022, 37, 197–202. [Google Scholar] [CrossRef]
- McVaugh, R.; Rzedowski, J. Synopsis of the Genus bursera L. in Western Mexico, with Notes on the Material of Bursera Collected by Sesse & Mocino. Kew Bull. 1965, 18, 317. [Google Scholar] [CrossRef]
- Bonfil-Sanders, C.; Cajero-Lázaro, I.; Evans, R.Y. Germinación de Semillas de Seis Especies de Bursera Del Centro de México. Agrociencia 2008, 42, 827–834. [Google Scholar]
- Aurora Montufar, L. Identidad y Simbolismo Del Copal Prehispánico y Reciente. Arqueología 2004, 33, 60–71. [Google Scholar]
- Trejo, I.; Dirzo, R. Deforestation of Seasonally Dry Tropical Forest: A National and Local Analysis in Mexico. Biol. Conserv. 2000, 94, 133–142. [Google Scholar] [CrossRef]
- Hernández-Oria, J.G. Desaparición Del Bosque Seco En El Bajío Mexicano: Implicaciones Del Ensamblaje de Especies y Grupos Funcionales En La Dinámica de Una Vegetación Amenazada. Zonas Áridas 2007, 11, ág. 13–31. [Google Scholar] [CrossRef]
- Fuentes, A.C.D.; Samain, M.S.; Martínez-Salas, E. The IUCN Red List of Threatened Species 2019: E.T137371772A137376559. 2019. Available online: https://www.iucnredlist.org/es/species/137371772/137376559 (accessed on 2 May 2022).
- Becerra, J.X.; Venable, D.L. Nuclear Ribosomal DNA Phylogeny and Its Implications for Evolutionary Trends in Mexican Bursera (Burseraceae). Am. J. Bot. 1999, 86, 1047–1057. [Google Scholar] [CrossRef] [PubMed]
- Becerra, J.X. Evolution of Mexican Bursera (Burseraceae) Inferred from ITS, ETS, and 5S Nuclear Ribosomal DNA Sequences. Mol. Phylogenet. Evol. 2003, 26, 300–309. [Google Scholar] [CrossRef]
- Weeks, A.; Simpson, B.B. Molecular Phylogenetic Analysis of Commiphora (Burseraceae) Yields Insight on the Evolution and Historical Biogeography of an “Impossible” Genus. Mol. Phylogenet. Evol. 2007, 42, 62–79. [Google Scholar] [CrossRef]
- Rosell, J.A.; Olson, M.E.; Weeks, A.; De-Nova, J.A.; Lemos, R.M.; Camacho, J.P.; Feria, T.P.; Gómez-Bermejo, R.; Montero, J.C.; Eguiarte, L.E. Diversification in Species Complexes: Tests of Species Origin and Delimitation in the Bursera Simaruba Clade of Tropical Trees (Burseraceae). Mol. Phylogenet. Evol. 2010, 57, 798–811. [Google Scholar] [CrossRef] [PubMed]
- Weeks, A.; Tye, A. Phylogeography of Palo Santo Trees (Bursera Graveolens and Bursera Malacophylla; Burseraceae) in the Galápagos Archipelago. Bot. J. Linn. Soc. 2009, 161, 396–410. [Google Scholar] [CrossRef][Green Version]
- Poelchau, M.F.; Hamrick, J.L. Comparative Phylogeography of Three Common Neotropical Tree Species. J. Biogeogr. 2013, 40, 618–631. [Google Scholar] [CrossRef]
- Dunphy, B.K.; Hamrick, J.L. Estimation of Gene Flow into Fragmented Populations of Bursera Simaruba (Burseraceae) in the Dry-Forest Life Zone of Puerto Rico. Am. J. Bot. 2007, 94, 1786–1794. [Google Scholar] [CrossRef]
- Weeks, A.; Simpson, B.B. Molecular Genetic Evidence for Interspecific Hybridization among Endemic Hispaniolan Bursera (Burseraceae). Am. J. Bot. 2017, 91, 976–984. [Google Scholar] [CrossRef]
- Melecio, E.Q.; Rico, Y.; Noriega, A.L.; Rodríguez, A.G. Molecular Evidence and Ecological Niche Modeling Reveal an Extensive Hybrid Zone among Three Bursera Species (Section Bullockia). PLoS ONE 2021, 16, e0260382. [Google Scholar] [CrossRef]
- Collins, E.S.; Gostel, M.R.; Weeks, A. An Expanded Nuclear Phylogenomic PCR Toolkit for Sapindales. Appl. Plant Sci. 2016, 4, 1600078. [Google Scholar] [CrossRef]
- Rzedowski, J.; Guevara-Féfer, F. Burseraceae. Flora Del Bajío Y De Reg. Adyac. 1992, 3, 46. [Google Scholar]
- Guimarães, R.; Forni-Martins, E.R. Chromosome Numbers and Their Evolutionary Meaning in the Sapindales Order: An Overview. Braz. J. Bot. 2021, 45, 77–91. [Google Scholar] [CrossRef]
- Khan, A.; Asaf, S.; Khan, A.L.; Al-Harrasi, A.; Al-Sudairy, O.; AbdulKareem, N.M.; Khan, A.; Shehzad, T.; Alsaady, N.; Al-Lawati, A.; et al. First Complete Chloroplast Genomics and Comparative Phylogenetic Analysis of Commiphora Gileadensis and C. Foliacea: Myrrh Producing Trees. PLoS ONE 2019, 14, e0208511. [Google Scholar] [CrossRef] [PubMed]
- Lai, R.-L.; Feng, X.; Chen, J.; Chen, Y.-T.; Wu, R.-J. The Complete Chloroplast Genome Characterization and Phylogenetic Analysis of Canarium Album. Mitochondrial. DNA B Resour. 2019, 4, 2948–2949. [Google Scholar] [CrossRef] [PubMed]
- Monpara, J.; Thaker, V. Phylogenic Position and Marker Studies Using CpDNA of C. Wightii: A Critically Endangered and Medicinally Important Plant in India. Vegetos 2021, 34, 300–308. [Google Scholar] [CrossRef]
- Healey, A.; Furtado, A.; Cooper, T.; Henry, R.J. Protocol: A Simple Method for Extracting next-Generation Sequencing Quality Genomic DNA from Recalcitrant Plant Species. Plant Methods 2014, 10, 21. [Google Scholar] [CrossRef] [PubMed]
- Yencho, G.C.; Olukolu, B.A. Compositions and Methods Related to Quantitative Reduced Representation Sequencing. 2022. Available online: https://patents.google.com/patent/US20220243267A1/ (accessed on 6 June 2022).
- Andrews, S. FASTQC. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 9 March 2022).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.M.; Birol, I. ABySS: A Parallel Assembler for Short Read Sequence Data. Genome Res. 2009, 19, 1117. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.T.; Durbin, R. Efficient de Novo Assembly of Large Genomes Using Compressed Data Structures. Genome Res. 2012, 22, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Kajitani, R.; Toshimoto, K.; Noguchi, H.; Toyoda, A.; Ogura, Y.; Okuno, M.; Yabana, M.; Harada, M.; Nagayasu, E.; Maruyama, H.; et al. Efficient de Novo Assembly of Highly Heterozygous Genomes from Whole-Genome Shotgun Short Reads. Genome Res. 2014, 24, 1384–1395. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Sahlin, K.; Vezzi, F.; Nystedt, B.; Lundeberg, J.; Arvestad, L. BESST—Efficient Scaffolding of Large Fragmented Assemblies. BMC Bioinform. 2014, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; An, H.; Li, L. Genome Survey Sequencing for the Characterization of the Genetic Background of Rosa Roxburghii Tratt and Leaf Ascorbate Metabolism Genes. PLoS ONE 2016, 11, e0147530. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.M.; Wang, Z.F.; Zhang, Y.; Wang, R.J. Chromosomal-Level Assembly of the Leptodermis Oblonga (Rubiaceae) Genome and Its Phylogenetic Implications. Genomics 2021, 113, 3072–3082. [Google Scholar] [CrossRef] [PubMed]
- Marçais, G.; Kingsford, C. A Fast, Lock-Free Approach for Efficient Parallel Counting of Occurrences of k-Mers. Bioinformatics 2011, 27, 764–770. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Humann, J.L.; Lee, T.; Ficklin, S.; Main, D. Structural and Functional Annotation of Eukaryotic Genomes with GenSAS. Methods Mol. Biol. 2019, 1962, 29–51. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Keller, O.; Gunduz, I.; Hayes, A.; Waack, S.; Morgenstern, B. AUGUSTUS: Ab Initio Prediction of Alternative Transcripts. Nucleic Acids Res. 2006, 34, W435. [Google Scholar] [CrossRef]
- Lomsadze, A.; Ter-Hovhannisyan, V.; Chernoff, Y.O.; Borodovsky, M. Gene Identification in Novel Eukaryotic Genomes by Self-Training Algorithm. Nucleic Acids Res. 2005, 33, 6494. [Google Scholar] [CrossRef]
- Li, H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Khan, A.L.; Al-Harrasi, A.; Wang, J.P.; Asaf, S.; Riethoven, J.J.M.; Shehzad, T.; Liew, C.S.; Song, X.M.; Schachtman, D.P.; Liu, C.; et al. Genome Structure and Evolutionary History of Frankincense Producing Boswellia Sacra. iScience 2022, 25, 104574. [Google Scholar] [CrossRef] [PubMed]
- Kuster, R.D.; Yencho, G.C.; Olukolu, B.A. NgsComposer: An Automated Pipeline for Empirically Based NGS Data Quality Filtering. Brief. Bioinform. 2021, 22, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T.; Devillard, S.; Dufour, A.-B.; Pontier, D. Revealing Cryptic Spatial Patterns in Genetic Variability by a New Multivariate Method. Heredity 2008, 101, 92–103. [Google Scholar] [CrossRef] [PubMed]
- De-Nova, J.A.; Medina, R.; Montero, J.C.; Weeks, A.; Rosell, J.A.; Olson, M.E.; Eguiarte, L.E.; Magallón, S. Insights into the Historical Construction of Species-Rich Mesoamerican Seasonally Dry Tropical Forests: The Diversification of Bursera (Burseraceae, Sapindales). New Phytol. 2012, 193, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; He, J.; Zhao, P.X.; Bouton, J.H.; Monteros, M.J. Genome-Wide Identification of Microsatellites in White Clover (Trifolium repens L.) Using FIASCO and PhpSSRMiner. Plant Methods 2008, 4, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Cardle, L.; Ramsay, L.; Milbourne, D.; Macaulay, M.; Marshall, D.; Waugh, R. Computational and Experimental Characterization of Physically Clustered Simple Sequence Repeats in Plants. Genetics 2000, 156, 847. [Google Scholar] [CrossRef]
- Rossetto, M.; Rymer, P.D. Applications of Molecular Markers in Plant Conservation. In Molecular Markers in Plants, 1st ed.; Henry, R.J., Ed.; Johh Wiley & Sons, Inc.: Chichester, UK, 2013; pp. 81–98. [Google Scholar] [CrossRef]
| Species | Genome Assembly Size (Mb) | Contigs (n) | N50 (bp) | L50 (n) | Largest Contig (bp) | GC Content (%) |
|---|---|---|---|---|---|---|
| B. cuneata | 252.9 | 70,005 | 13,532 | 4324 | 203,078 | 34.97 |
| B. bipinnata | 229.2 | 93,723 | 5699 | 9652 | 104,624 | 34.14 |
| B. palmeri | 237.4 | 67,240 | 11,112 | 4918 | 230,450 | 34.11 |
| Species | Total Primary Reads (Million) | Mapped Reads (Million) | Properly Paired Reads (Million) |
|---|---|---|---|
| B. cuneata | 169.8 | 62.9 (37.0%) | 43.5 (25.6%) |
| B. bipinnata | 141.9 | 53.6 (37.8%) | 35.6 (25.1%) |
| B. palmeri | 145.2 | 62.9 (43.4%) | 48.6 (33.5%) |
| Species | Reads (M) | Boswellia sacra | Canarium album | Commiphora foliacea | Commiphora gileadensis | Commiphora wightii |
|---|---|---|---|---|---|---|
| B. cuneata | 169.8 | 7,481,910 (4.40%) | 7,253,921 (4.27%) | 7,304,511 (4.30%) | 7,364,709 (4.34%) | 7,514,759 (4.42%) |
| B. bipinnata | 141.9 | 4,080,000 (2.87%) | 3,908,183 (2.75%) | 3,948,175 (2.78%) | 3,970,894 (2.80%) | 4,122,751 (2.90%) |
| B. palmeri | 145.2 | 5,187,716 (3.57%) | 4,953,793 (3.41%) | 5,043,110 (3.47%) | 5,042,119 (3.47%) | 5,238,798 (3.61%) |
| Species | Total | Di | Tri | Tetra | Penta | Hexa |
|---|---|---|---|---|---|---|
| B. cuneata | 107,270 | 39,769 (37.1%) | 20,606 (19.2%) | 31,871 (29.7%) | 10,011 (9.3%) | 5013 (4.7%) |
| B. bipinnata | 76,766 | 27,915 (36.4%) | 15,049 (19.6%) | 23,067 (30%) | 6988 (9.1%) | 3747 (4.9%) |
| B. palmeri | 100,614 | 37,480 (37.3%) | 19,431 (19.3%) | 29,818 (29.6%) | 9212 (9.2%) | 4673 (4.6%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rico, Y.; Lorenzana, G.P.; Benítez-Pineda, C.A.; Olukolu, B.A. Development of Genomic Resources in Mexican Bursera (Section: Bullockia: Burseraceae): Genome Assembly, Annotation, and Marker Discovery for Three Copal Species. Genes 2022, 13, 1741. https://doi.org/10.3390/genes13101741
Rico Y, Lorenzana GP, Benítez-Pineda CA, Olukolu BA. Development of Genomic Resources in Mexican Bursera (Section: Bullockia: Burseraceae): Genome Assembly, Annotation, and Marker Discovery for Three Copal Species. Genes. 2022; 13(10):1741. https://doi.org/10.3390/genes13101741
Chicago/Turabian StyleRico, Yessica, Gustavo P. Lorenzana, Carlos A. Benítez-Pineda, and Bode A. Olukolu. 2022. "Development of Genomic Resources in Mexican Bursera (Section: Bullockia: Burseraceae): Genome Assembly, Annotation, and Marker Discovery for Three Copal Species" Genes 13, no. 10: 1741. https://doi.org/10.3390/genes13101741
APA StyleRico, Y., Lorenzana, G. P., Benítez-Pineda, C. A., & Olukolu, B. A. (2022). Development of Genomic Resources in Mexican Bursera (Section: Bullockia: Burseraceae): Genome Assembly, Annotation, and Marker Discovery for Three Copal Species. Genes, 13(10), 1741. https://doi.org/10.3390/genes13101741






