Genome-Wide Identification of Membrane-Bound Fatty Acid Desaturase Genes in Three Peanut Species and Their Expression in Arachis hypogaea during Drought Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence Retrieval
2.2. Genome-Wide Identification and Phylogenetic Construction of FAD Genes
2.3. Characterization of FAD Genes
2.4. Gene Structure and Conserved Motif Analysis
2.5. Duplication and Synteny Analysis of FAD Genes
2.6. Cis-Element Prediction in Promoter Regions and Expression Analysis of Ah|FAD Genes
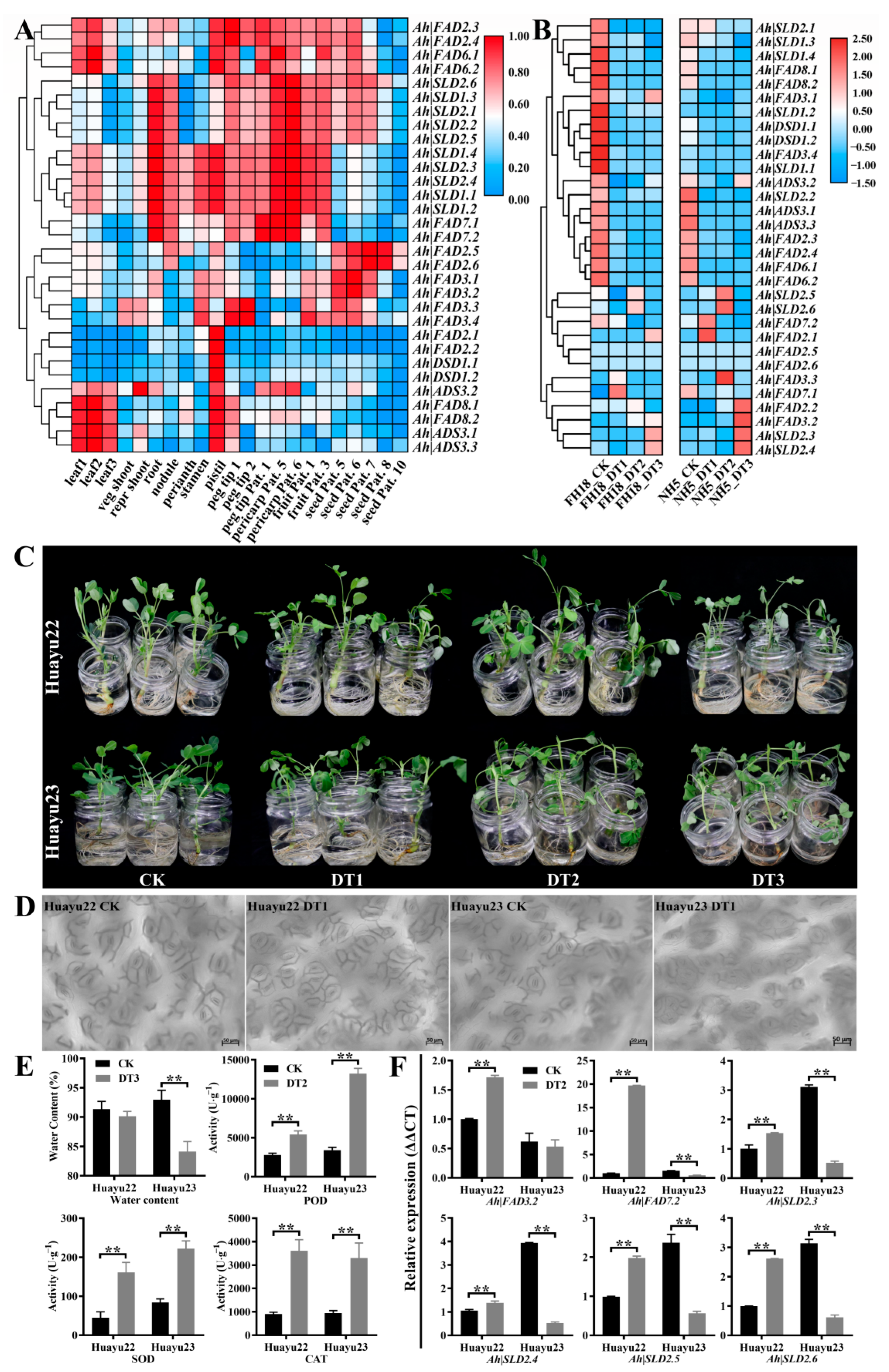
2.7. Assays for Water Content and Antioxidative Enzymes Activity
2.8. Prediction of miRNA Targeting Ah|FAD Genes
3. Results
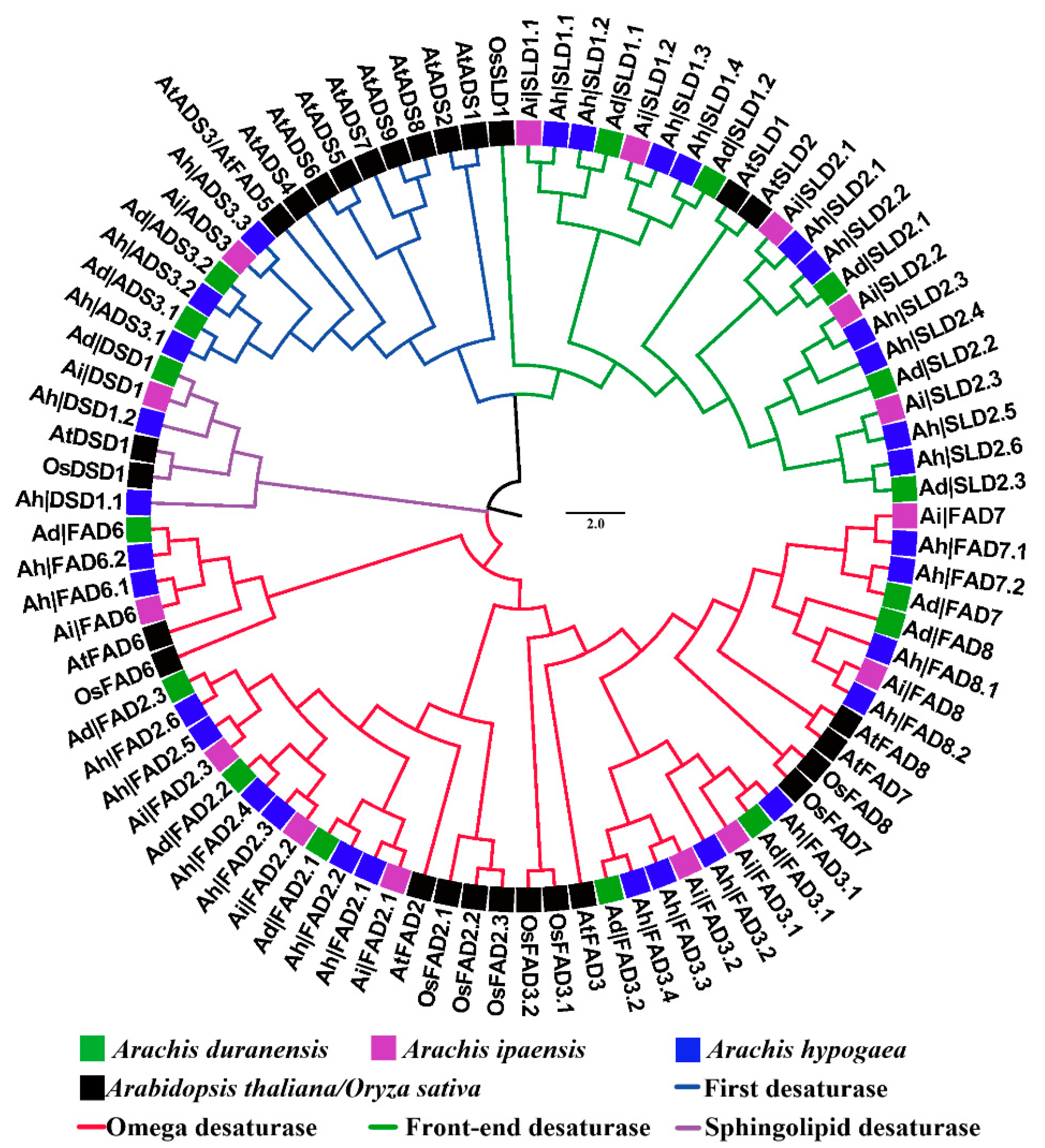
3.1. Identification and Phylogenetic Analysis of FAD Genes in Three Peanut Species
3.2. Characterization Analysis of FAD Genes in Three Peanut Species
3.3. Analysis of Gene Structure of FAD Genes and Domains and Motifs of Their Encoding Proteins
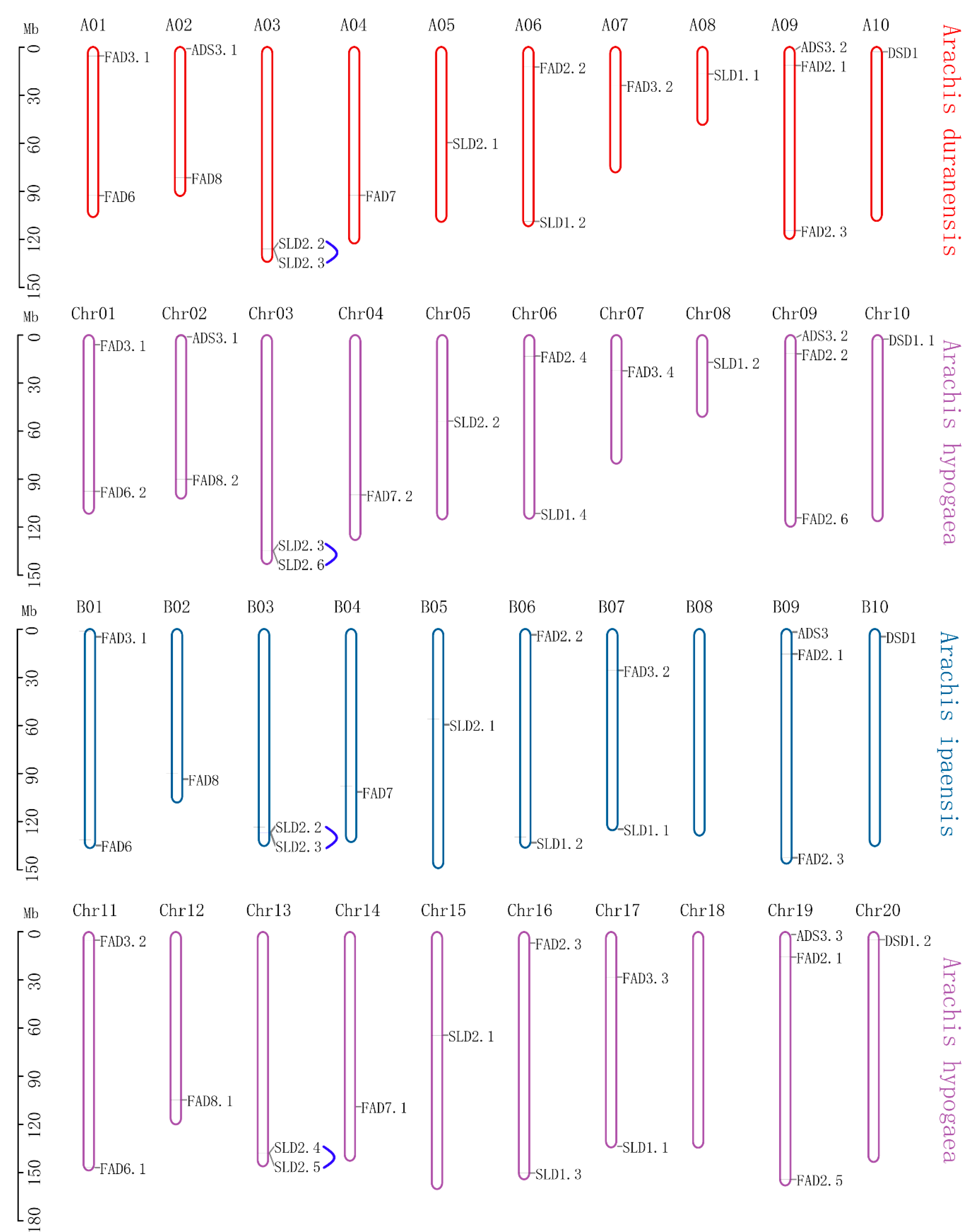
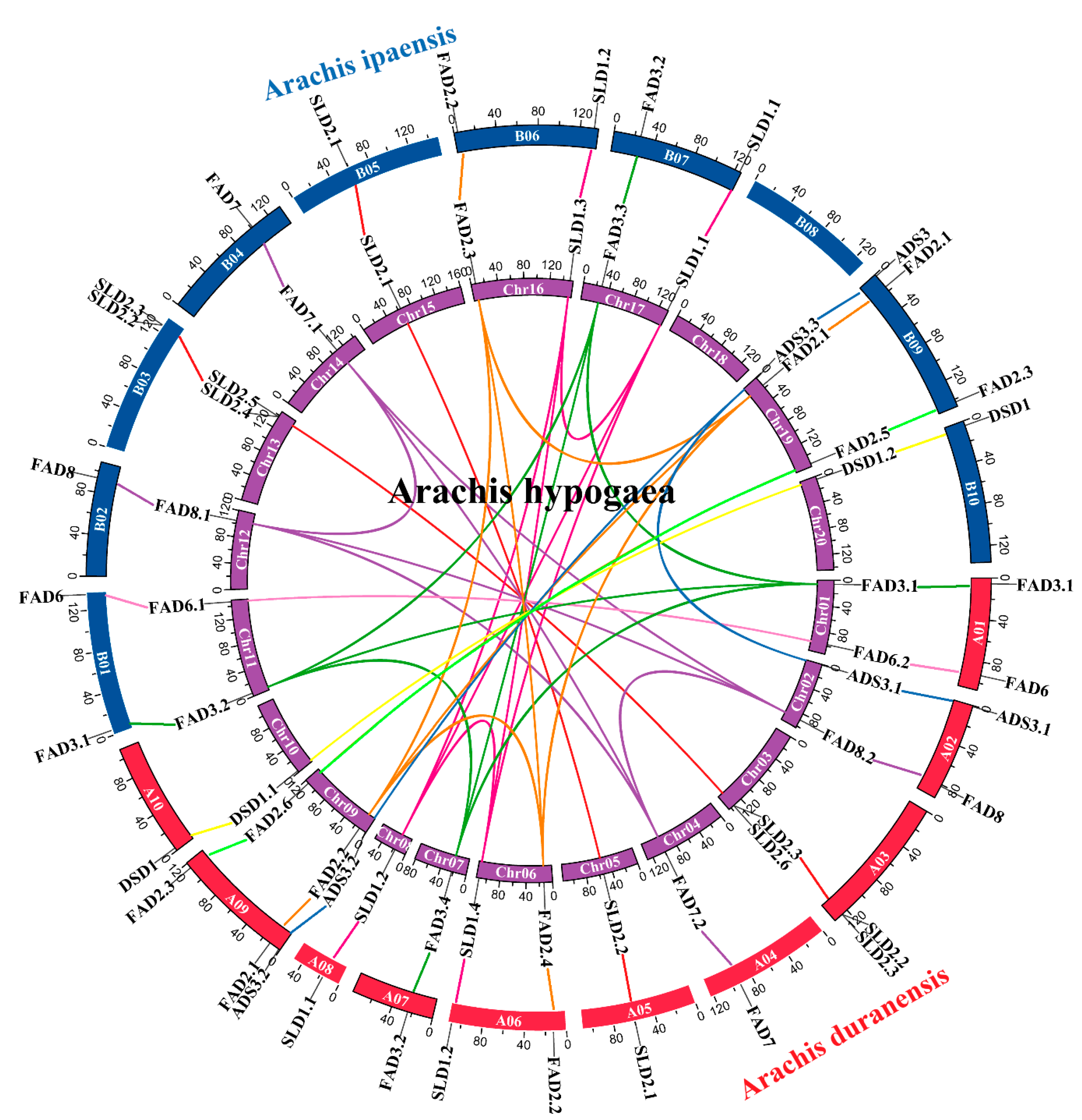
3.4. Chromosome Localization and Synteny Analysis of FAD Genes
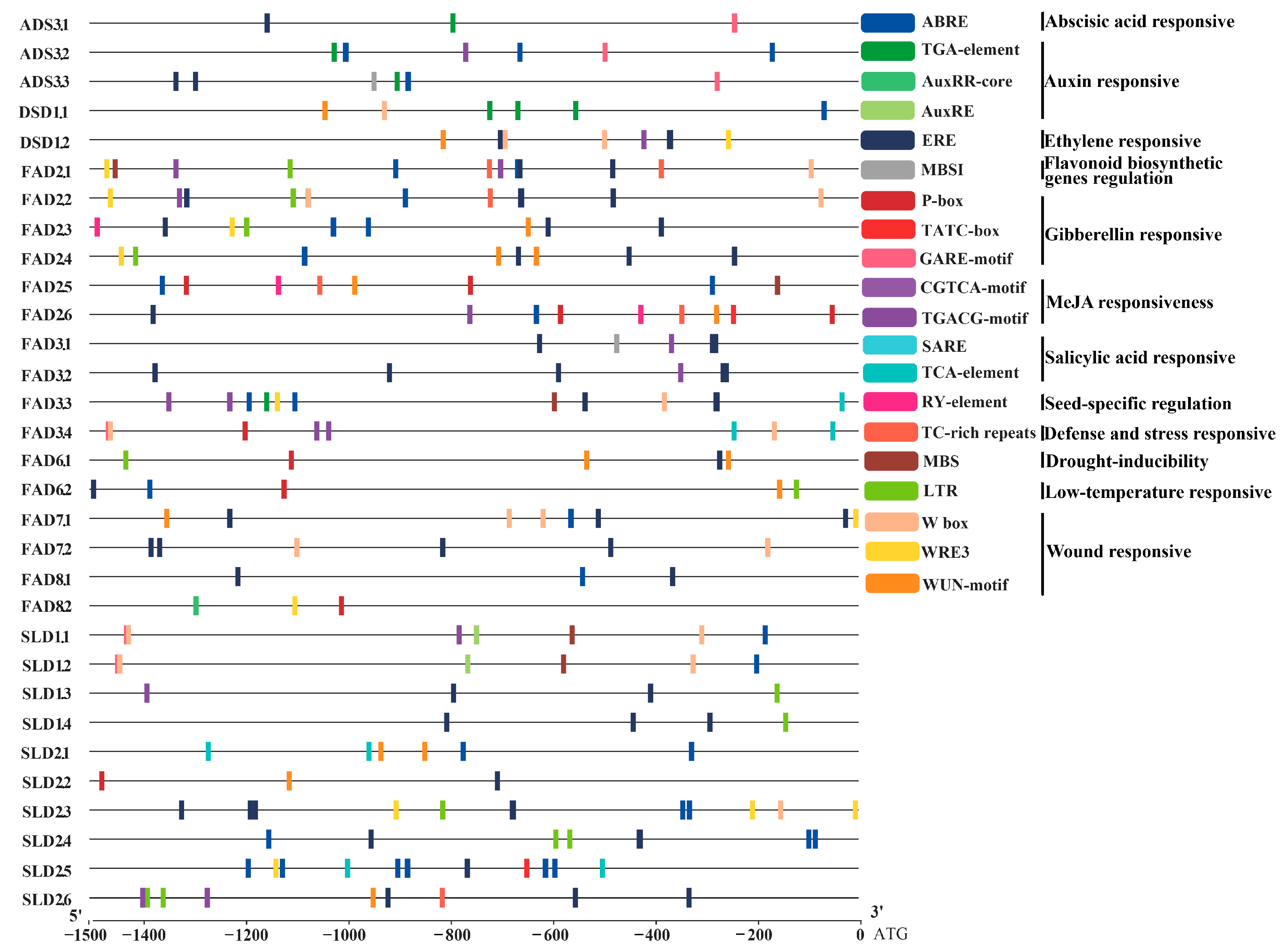
3.5. Stress-Related Cis-Elements in the Promoters of Ah|FAD Genes
3.6. Expression Profile Analysis of Ah|FAD Genes
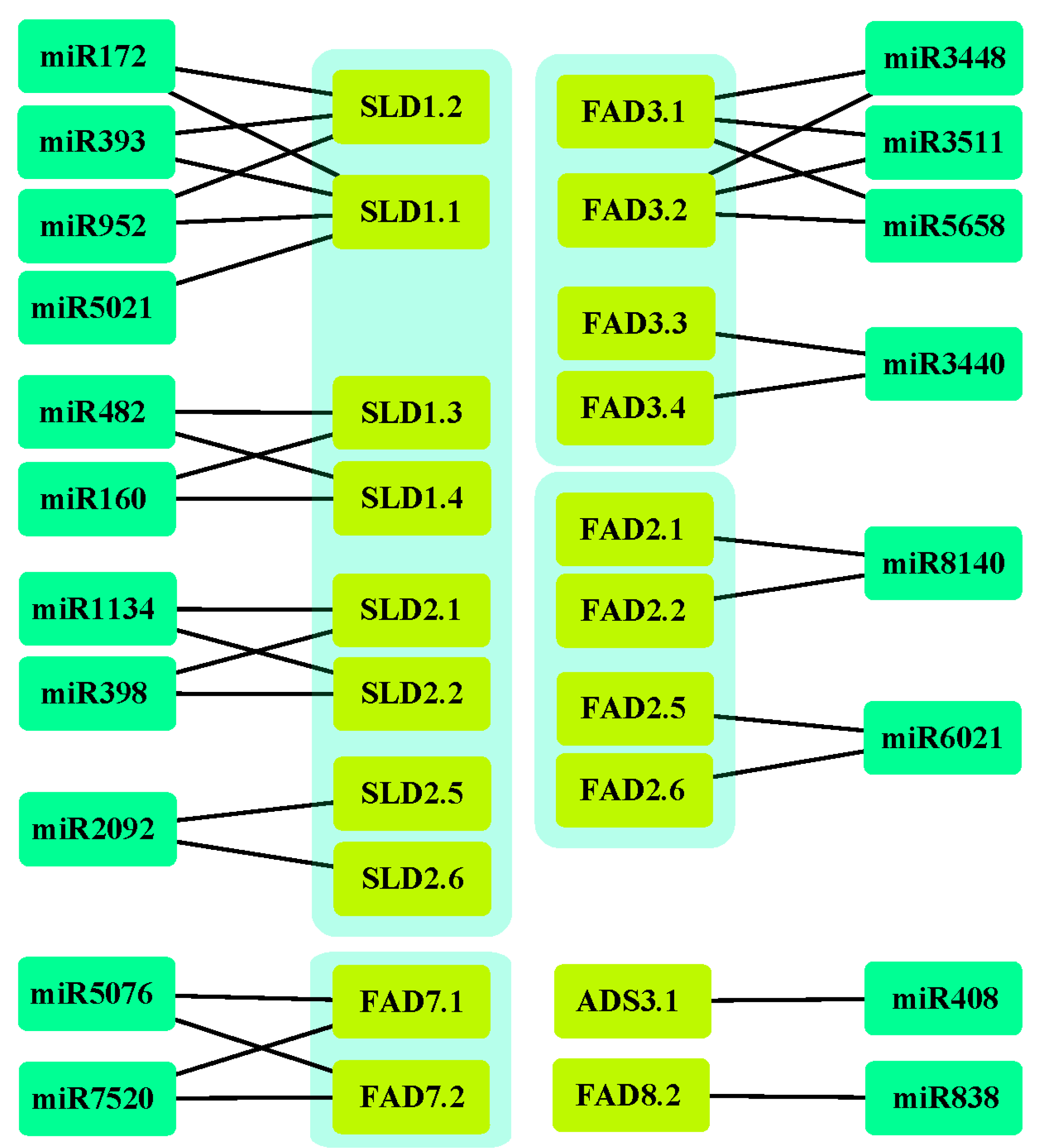
3.7. The miRNA Targeting Ah|FAD Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ramu, V.S.; Swetha, T.N.; Sheela, S.H.; Babitha, C.K.; Rohini, S.; Reddy, M.K.; Tuteja, N.; Reddy, C.P.; Prasad, T.G.; Udayakumar, M. Simultaneous expression of regulatory genes associated with specific drought-adaptive traits improves drought adaptation in peanut. Plant Biotechnol. J. 2016, 14, 1008–1020. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.B.; Chu, L.Y.; Jaleel, C.A.; Manivannan, P.; Panneerselvam, R.; Shao, M.A. Understanding water deficit stress-induced changes in the basic metabolism of higher plants—Biotechnologically and sustainably improving agriculture and the ecoenvironment in arid regions of the globe. Crit. Rev. Biotechnol. 2009, 29, 131–151. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.L.; Li, C.J.; Lu, X.D.; Zhao, X.B.; Yan, C.X.; Wang, J.; Sun, Q.X.; Shan, S.H. Comprehensive genomic characterization of NAC transcription factor family and their response to salt and drought stress in peanut. BMC Plant Biol. 2020, 20, 454. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, T.; Thankappan, R.; Kumar, A.; Mishra, G.P.; Dobaria, J.R. Stress inducible expression of AtDREB1A transcription factor in transgenic Peanut (Arachis hypogaea L.) conferred tolerance to soil-moisture deficit stress. Front. Plant Sci. 2016, 7, 935. [Google Scholar] [CrossRef]
- Dai, A.G. Increasing drought under global warming in observations and models. Nat. Clim. Change 2013, 3, 52–58. [Google Scholar] [CrossRef]
- Jiang, C.; Li, X.; Zou, J.; Ren, J.; Jin, C.; Zhang, H.; Yu, H.; Jin, H. Comparative transcriptome analysis of genes involved in the drought stress response of two peanut (Arachis hypogaea L.) varieties. BMC Plant Biol. 2021, 21, 64. [Google Scholar] [CrossRef] [PubMed]
- Sui, N.; Wang, Y.; Liu, S.S.; Yang, Z.; Wang, F.; Wan, S.B. Transcriptomic and physiological evidence for the relationship between unsaturated fatty acid and salt stress in peanut. Front. Plant Sci. 2018, 9, 7. [Google Scholar] [CrossRef]
- Singh, S.C.; Sinha, R.P.; Hader, D.P. Role of lipids and fatty acids in stress tolerance in cyanobacteria. Acta Protozool. 2002, 41, 297–308. [Google Scholar]
- Sui, N.; Tian, S.; Wang, W.; Wang, M.; Fan, H. Overexpression of glycerol-3-phosphate acyltransferase from Suaeda salsa improves salt tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 1337. [Google Scholar] [CrossRef]
- Zhang, H.; Dong, J.L.; Zhao, X.H.; Zhang, Y.M.; Ren, J.Y.; Xing, L.T.; Jiang, C.J.; Wang, X.G.; Wang, J.; Zhao, S.L.; et al. Research progress in membrane lipid metabolism and molecular mechanism in peanut cold tolerance. Front. Plant Sci. 2019, 10, 838. [Google Scholar] [CrossRef]
- Feng, J.; Dong, Y.; Liu, W.; He, Q.; Daud, M.K.; Chen, J.; Zhu, S. Genome-wide identification of membrane-bound fatty acid desaturase genes in Gossypium hirsutum and their expressions during abiotic stress. Sci Rep.-UK 2017, 7, 45711. [Google Scholar] [CrossRef]
- Hashimoto, K.; Yoshizawa, A.C.; Okuda, S.; Kuma, K.; Goto, S.; Kanehisa, M. The repertoire of desaturases and elongases reveals fatty acid variations in 56 eukaryotic genomes. J. Lipid Res. 2008, 49, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Heilmann, I.; Pidkowich, M.S.; Girke, T.; Shanklin, J. Switching desaturase enzyme specificity by alternate subcellular targeting. Proc. Natl. Acad. Sci. USA 2004, 101, 10266–10271. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Murata, N. Regulation of the desaturation of fatty acids and its role in tolerance to cold and salt stress. Curr. Opin. Microbiol. 2002, 5, 208–210. [Google Scholar] [CrossRef]
- Chen, M.; Markham, J.E.; Cahoon, E.B. Sphingolipid Delta8 unsaturation is important for glucosylceramide biosynthesis and low-temperature performance in Arabidopsis. Plant J. 2012, 69, 769–781. [Google Scholar] [CrossRef]
- Ternes, P.; Franke, S.; Zahringer, U.; Sperling, P.; Heinz, E. Identification and characterization of a sphingolipid delta 4-desaturase family. J. Biol. Chem. 2002, 277, 25512–25518. [Google Scholar] [CrossRef]
- Iba, K. Acclimative response to temperature stress in higher plants: Approaches of gene engineering for temperature tolerance. Annu. Rev. Plant Biol. 2002, 53, 225–245. [Google Scholar] [CrossRef]
- Routaboul, J.M.; Fischer, S.F.; Browse, J. Trienoic fatty acids are required to maintain chloroplast function at low temperatures. Plant Physiol. 2000, 124, 1697–1705. [Google Scholar] [CrossRef]
- Chen, Y.; Cui, Q.; Xu, Y.; Yang, S.; Gao, M.; Wang, Y. Effects of tung oilseed FAD2 and DGAT2 genes on unsaturated fatty acid accumulation in Rhodotorula glutinis and Arabidopsis thaliana. Mol. Genet. Genom. 2015, 290, 1605–1613. [Google Scholar] [CrossRef]
- Dominguez, T.; Hernandez, M.L.; Pennycooke, J.C.; Jimenez, P.; Martinez-Rivas, J.M.; Sanz, C.; Stockinger, E.J.; Sanchez-Serrano, J.J.; Sanmartin, M. Increasing omega-3 desaturase expression in tomato results in altered aroma profile and enhanced resistance to cold stress. Plant Physiol. 2010, 153, 655–665. [Google Scholar] [CrossRef]
- Wang, H.S.; Yu, C.; Tang, X.F.; Zhu, Z.J.; Ma, N.N.; Meng, Q.W. A tomato endoplasmic reticulum (ER)-type omega-3 fatty acid desaturase (LeFAD3) functions in early seedling tolerance to salinity stress. Plant Cell Rep. 2014, 33, 131–142. [Google Scholar] [CrossRef]
- Yu, C.; Wang, H.S.; Yang, S.; Tang, X.F.; Duan, M.; Meng, Q.W. Overexpression of endoplasmic reticulum omega-3 fatty acid desaturase gene improves chilling tolerance in tomato. Plant Physiol. Biochem. 2009, 47, 1102–1112. [Google Scholar] [CrossRef]
- Zhang, M.; Barg, R.; Yin, M.; Gueta-Dahan, Y.; Leikin-Frenkel, A.; Salts, Y.; Shabtai, S.; Ben-Hayyim, G. Modulated fatty acid desaturation via overexpression of two distinct omega-3 desaturases differentially alters tolerance to various abiotic stresses in transgenic tobacco cells and plants. Plant J. 2005, 44, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Roman, A.; Hernandez, M.L.; Soria-Garcia, A.; Lopez-Gomollon, S.; Lagunas, B.; Picorel, R.; Martinez-Rivas, J.M.; Alfonso, M. Non-redundant contribution of the plastidial FAD8 omega-3 desaturase to glycerolipid unsaturation at different temperatures in Arabidopsis. Mol. Plant 2015, 8, 1599–1611. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Y.; Yang, J.H.; Li, B.; Yang, X.M.; Meng, Q.W. Antisense-mediated depletion of tomato chloroplast omega-3 fatty acid desaturase enhances thermal tolerance. J. Integr. Plant Biol. 2006, 48, 1096–1107. [Google Scholar] [CrossRef]
- Zhang, J.T.; Zhu, J.Q.; Zhu, Q.; Liu, H.; Gao, X.S.; Zhang, H.X. Fatty acid desaturase-6 (Fad6) is required for salt tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2009, 390, 469–474. [Google Scholar] [CrossRef]
- Zhang, J.T.; Liu, H.; Sun, J.; Li, B.; Zhu, Q.; Chen, S.L.; Zhang, H.X. Arabidopsis fatty acid desaturase FAD2 is required for salt tolerance during seed germination and early seedling growth. PLoS ONE 2012, 7, e30355. [Google Scholar] [CrossRef]
- Chi, X.; Zhang, Z.; Chen, N.; Zhang, X.; Wang, M.; Chen, M.; Wang, T.; Pan, L.; Chen, J.; Yang, Z.; et al. Isolation and functional analysis of fatty acid desaturase genes from peanut (Arachis hypogaea L.). PLoS ONE 2017, 12, e0189759. [Google Scholar] [CrossRef]
- E, Z.; Chen, C.; Yang, J.; Tong, H.; Li, T.; Wang, L.; Chen, H. Genome-wide analysis of fatty acid desaturase genes in rice (Oryza sativa L.). Sci. Rep.-UK 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the expasy server. In Springer Protocols Handbooks; Humana Press: Totowa, NJ, USA, 2005; p. 571. [Google Scholar]
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Hwang, J.K. Prediction of protein subcellular localization. Proteins 2006, 64, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Rombauts, S.; Dehais, P.; Van Montagu, M.; Rouze, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef] [PubMed]
- Clevenger, J.; Chu, Y.; Scheffler, B.; Ozias-Akins, P. A developmental transcriptome map for allotetraploid Arachis hypogaea. Front. Plant Sci. 2016, 7, 1446. [Google Scholar] [CrossRef] [PubMed]
- Spurney, R.J.; Van den Broeck, L.; Clark, N.M.; Fisher, A.P.; de Luis Balaguer, M.A.; Sozzani, R. Tuxnet: A simple interface to process RNA sequencing data and infer gene regulatory networks. Plant J. 2020, 101, 716–730. [Google Scholar] [CrossRef]
- Zhang, Z.M.; Dai, L.X.; Ding, H.; Chen, D.X.; Yang, W.Q.; Song, W.W.; Wan, S.B. Identification and evaluation of drought resistance in different peanut varieties widely grown in Northern China. Acta Agron. Sin. 2012, 38, 495–504. [Google Scholar] [CrossRef]
- Ding, H.; Zhang, Z.; Dai, L.; Song, W.; Kang, T.; Ci, D. Responses of root morphology of peanut varieties differing in drought tolerance to water-deficient stress. Acta Ecol. Sin. 2013, 33, 5169–5176. [Google Scholar] [CrossRef]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Chen, Q.J.; Sun, G.Q.; Zheng, K.; Yao, Z.P.; Han, Y.H.; Wang, L.P.; Duan, Y.J.; Yu, D.Q.; Qu, Y.Y. Genome-wide identification of Cyclophilin gene family in cotton and expression analysis of the fibre development in Gossypium barbadense. Int. J. Mol. Sci. 2019, 20, 349. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cheng, Y.; Chen, D.; Liu, D.; Hu, M.; Dong, J.; Zhang, X.; Song, L.; Shen, F. The Catalase gene family in cotton: Genome-wide characterization and bioinformatics analysis. Cells 2019, 8, 86. [Google Scholar] [CrossRef]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef]
- Obayashi, T.; Aoki, Y.; Tadaka, S.; Kagaya, Y.; Kinoshita, K. ATTED-II in 2018: A plant coexpression database based on investigation of the statistical property of the mutual rank index. Plant Cell Physiol. 2018, 59, 440. [Google Scholar] [CrossRef]
- Zhang, Z.S.; Wei, X.Y.; Liu, W.X.; Min, X.Y.; Jin, X.Y.; Ndayambaza, B.; Wang, Y.R. Genome-wide identification and expression analysis of the fatty acid desaturase genes in Medicago truncatula. Biochem. Biophys. Res. Commun. 2018, 499, 361–367. [Google Scholar] [CrossRef]
- Xue, Y.; Chen, B.; Wang, R.; Win, A.N.; Li, J.; Chai, Y. Genome-wide survey and characterization of fatty acid desaturase gene family in Brassica napus and Its parental species. Appl. Biochem. Biotechnol. 2018, 184, 582–598. [Google Scholar] [CrossRef]
- Chen, X.; Lu, Q.; Liu, H.; Zhang, J.; Hong, Y.; Lan, H.; Li, H.; Wang, J.; Liu, H.; Li, S.; et al. Sequencing of cultivated peanut, Arachis hypogaea, yields insights into genome evolution and oil improvement. Mol. Plant 2019, 12, 920–934. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef]
- Peng, Z.; Ruan, J.; Tian, H.; Shan, L.; Meng, J.; Guo, F.; Zhang, Z.; Ding, H.; Wan, S.; Li, X. The family of peanut fatty acid desaturase genes and a functional analysis of four ω-3 AhFAD3 members. Plant Mol. Biol. Rep. 2020, 38, 209–221. [Google Scholar] [CrossRef]
- Nguyen, V.C.; Nakamura, Y.; Kanehara, K. Membrane lipid polyunsaturation mediated by FATTY ACID DESATURASE 2 (FAD2) is involved in endoplasmic reticulum stress tolerance in Arabidopsis thaliana. Plant J. 2019, 99, 478–493. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Li, M.; Jiang, Y.; Duan, X. Genome-wide identification, characterization and expression profile of glutaredoxin gene family in relation to fruit ripening and response to abiotic and biotic stresses in banana (Musa acuminata). Int. J. Biol. Macromol. 2020, 170, 636–651. [Google Scholar] [CrossRef] [PubMed]
- Ku, Y.S.; Sintaha, M.; Cheung, M.Y.; Lam, H.M. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef]
- Sunkar, R.; Li, Y.F.; Jagadeeswaran, G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012, 17, 196–203. [Google Scholar] [CrossRef]
- Zhu, H.; Xia, R.; Zhao, B.; An, Y.Q.; Dardick, C.D.; Callahan, A.M.; Liu, Z. Unique expression, processing regulation, and regulatory network of peach (Prunus persica) miRNAs. BMC Plant Biol. 2012, 12, 149. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, S.; Yan, C.; Li, C.; Zhao, X.; Wan, S.; Shan, S. Mining, identification and function analysis of microRNAs and target genes in peanut (Arachis hypogaea L.). Plant Physiol. Biochem. 2017, 111, 85–96. [Google Scholar] [CrossRef]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef]
- Akdogan, G.; Tufekci, E.D.; Uranbey, S.; Unver, T. miRNA-based drought regulation in wheat. Funct. Integr. Genom. 2016, 16, 221–233. [Google Scholar] [CrossRef]
- Kantar, M.; Lucas, S.J.; Budak, H. miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta 2011, 233, 471–484. [Google Scholar] [CrossRef]
- Xia, K.; Wang, R.; Ou, X.; Fang, Z.; Tian, C.; Duan, J.; Wang, Y.; Zhang, M. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS ONE 2012, 7, e30039. [Google Scholar] [CrossRef] [PubMed]
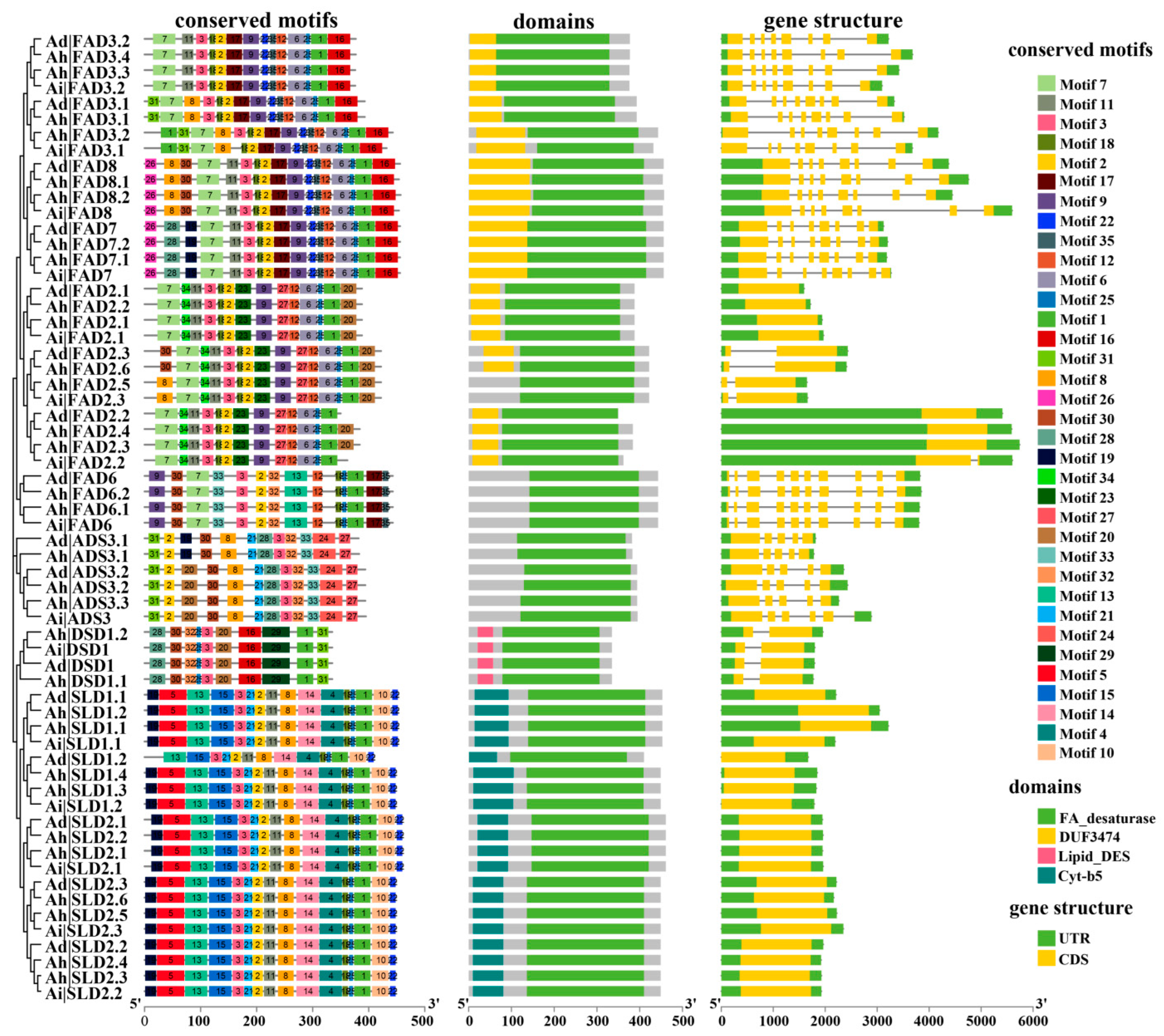

| Subfamily | Gene | Number of Genes | Conserved Motifs | Number of Motifs | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ω-3 desaturase | FAD3 | 4 | 7 | 11 | 3 | 18 | 2 | 17 | 9 | 22 | 35 | 12 | 6 | 25 | 1 | 16 | 14 | |||||
| FAD3 | 2 | 31 | 7 | 8 | 3 | 18 | 2 | 17 | 9 | 22 | 35 | 12 | 6 | 25 | 1 | 16 | 15 | |||||
| FAD3 | 2 | 1 | 31 | 7 | 8 | 3 | 18 | 2 | 17 | 9 | 22 | 35 | 12 | 6 | 25 | 1 | 16 | 16 | ||||
| FAD8 | 4 | 26 | 8 | 30 | 7 | 11 | 3 | 18 | 2 | 17 | 9 | 22 | 35 | 12 | 6 | 25 | 1 | 16 | 17 | |||
| FAD7 | 4 | 26 | 28 | 19 | 7 | 11 | 3 | 18 | 2 | 17 | 9 | 22 | 35 | 12 | 6 | 25 | 1 | 16 | 17 | |||
| Motif set 1, shared in ω-3 desaturases | 7 | 3 | 18 | 2 | 17 | 9 | 22 | 35 | 12 | 6 | 25 | 1 | 16 | |||||||||
| ω-6 desaturase | FAD2 | 4 | 7 | 34 | 11 | 3 | 18 | 2 | 23 | 9 | 27 | 12 | 6 | 25 | 1 | 20 | 14 | |||||
| FAD2 | 2 | 30 | 7 | 34 | 11 | 3 | 18 | 2 | 23 | 9 | 27 | 12 | 6 | 25 | 1 | 20 | 15 | |||||
| FAD2 | 2 | 8 | 7 | 34 | 11 | 3 | 18 | 2 | 23 | 9 | 27 | 12 | 6 | 25 | 1 | 20 | 15 | |||||
| FAD2 | 2 | 7 | 34 | 11 | 3 | 18 | 2 | 23 | 9 | 27 | 12 | 6 | 25 | 1 | 20 | 14 | ||||||
| FAD2 | 2 | 7 | 34 | 11 | 3 | 18 | 2 | 23 | 9 | 27 | 12 | 6 | 25 | 1 | 13 | |||||||
| FAD6 | 4 | 7 | 33 | 3 | 2 | 32 | 13 | 12 | 18 | 25 | 1 | 17 | 35 | 12 | ||||||||
| Motif set 2, shared in ω-6 desaturases | 7 | 34 | 11 | 3 | 18 | 2 | 17 | 9 | 27 | 12 | 6 | 25 | 1 | 20 | ||||||||
| Motif set 3, shared in ω desaturases | 7 | 3 | 18 | 2 | 9 | 12 | 6 | 25 | 1 | |||||||||||||
| First desaturase | ADS | 2 | 31 | 2 | 19 | 30 | 8 | 21 | 28 | 3 | 32 | 33 | 24 | 27 | 12 | |||||||
| ADS | 4 | 31 | 2 | 20 | 30 | 8 | 21 | 28 | 3 | 32 | 33 | 24 | 27 | 12 | ||||||||
| Motif set 4, shared in ADSs | 30 | 8 | 21 | 28 | 3 | 32 | 33 | 24 | 27 | |||||||||||||
| Sphingolipid desaturase | DSD | 4 | 20 | 30 | 32 | 25 | 3 | 20 | 16 | 29 | 1 | 31 | 10 | |||||||||
| Frond-end desaturase | SLD | 19 | 19 | 5 | 13 | 15 | 3 | 21 | 2 | 11 | 8 | 14 | 4 | 18 | 25 | 1 | 10 | 22 | 16 | |||
| SLD | 1 | 13 | 15 | 3 | 21 | 2 | 11 | 8 | 14 | 4 | 18 | 25 | 1 | 10 | 22 | 14 | ||||||
| Motif set 5, shared in SLDs | 13 | 15 | 3 | 21 | 2 | 11 | 8 | 14 | 4 | 18 | 25 | 1 | 10 | 22 | ||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gai, W.; Sun, H.; Hu, Y.; Liu, C.; Zhang, Y.; Gai, S.; Yuan, Y. Genome-Wide Identification of Membrane-Bound Fatty Acid Desaturase Genes in Three Peanut Species and Their Expression in Arachis hypogaea during Drought Stress. Genes 2022, 13, 1718. https://doi.org/10.3390/genes13101718
Gai W, Sun H, Hu Y, Liu C, Zhang Y, Gai S, Yuan Y. Genome-Wide Identification of Membrane-Bound Fatty Acid Desaturase Genes in Three Peanut Species and Their Expression in Arachis hypogaea during Drought Stress. Genes. 2022; 13(10):1718. https://doi.org/10.3390/genes13101718
Chicago/Turabian StyleGai, Wenyu, Hua Sun, Ya Hu, Chunying Liu, Yuxi Zhang, Shupeng Gai, and Yanchao Yuan. 2022. "Genome-Wide Identification of Membrane-Bound Fatty Acid Desaturase Genes in Three Peanut Species and Their Expression in Arachis hypogaea during Drought Stress" Genes 13, no. 10: 1718. https://doi.org/10.3390/genes13101718
APA StyleGai, W., Sun, H., Hu, Y., Liu, C., Zhang, Y., Gai, S., & Yuan, Y. (2022). Genome-Wide Identification of Membrane-Bound Fatty Acid Desaturase Genes in Three Peanut Species and Their Expression in Arachis hypogaea during Drought Stress. Genes, 13(10), 1718. https://doi.org/10.3390/genes13101718





