A Novel Framework for Analysis of the Shared Genetic Background of Correlated Traits
Abstract
:1. Introduction
2. Materials and Methods
2.1. Shared Heredity Model
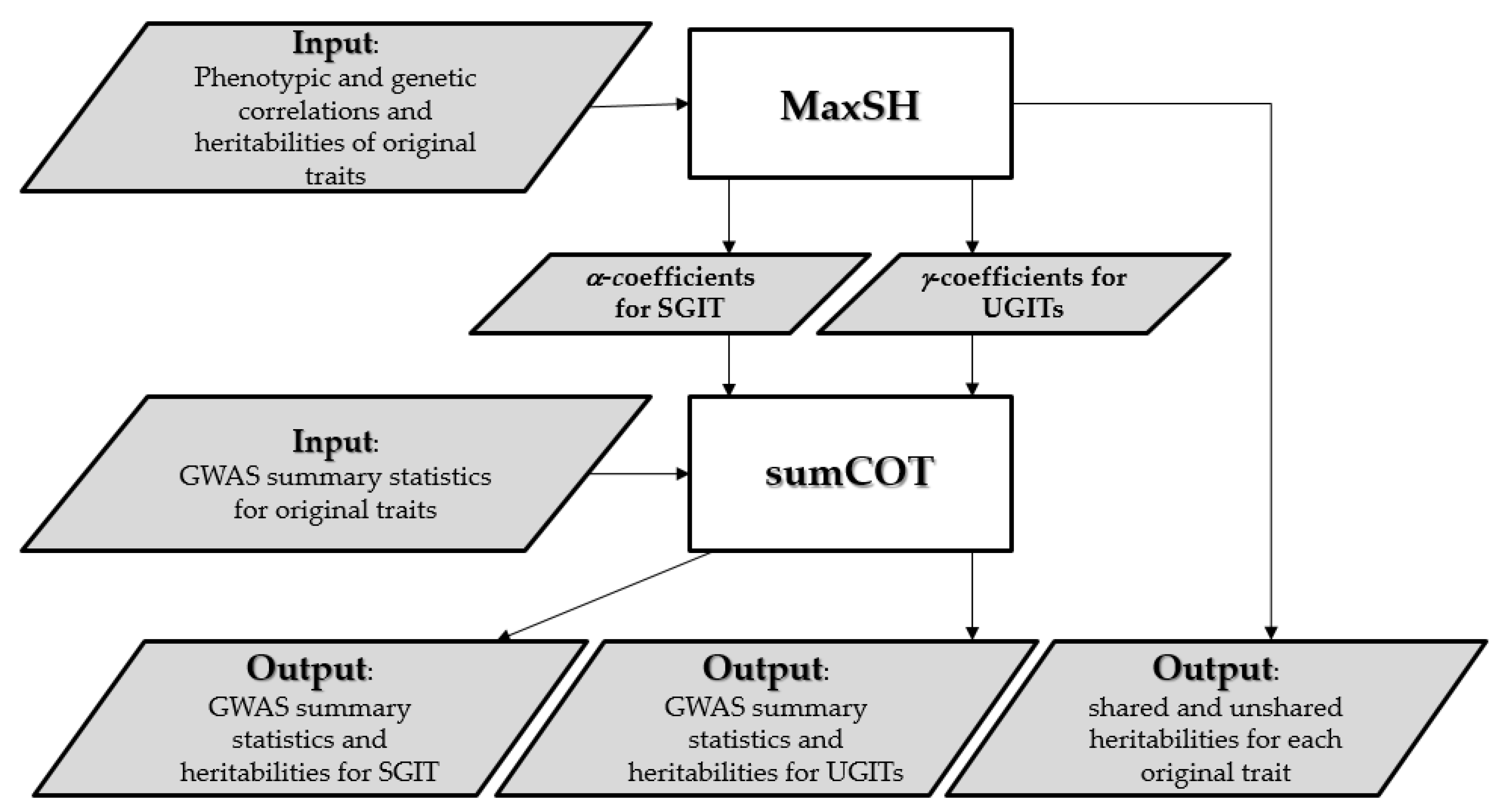
2.2. Overview of the SHAHER Framework
2.3. Simulation Study
2.4. Application to Real Data
2.4.1. Data Sets
2.4.2. Genetic Analysis
2.4.3. Gene set and Tissue/Cell Type Enrichment Analyses
2.4.4. The Number of Original Traits Associated with SGIT Loci
3. Results
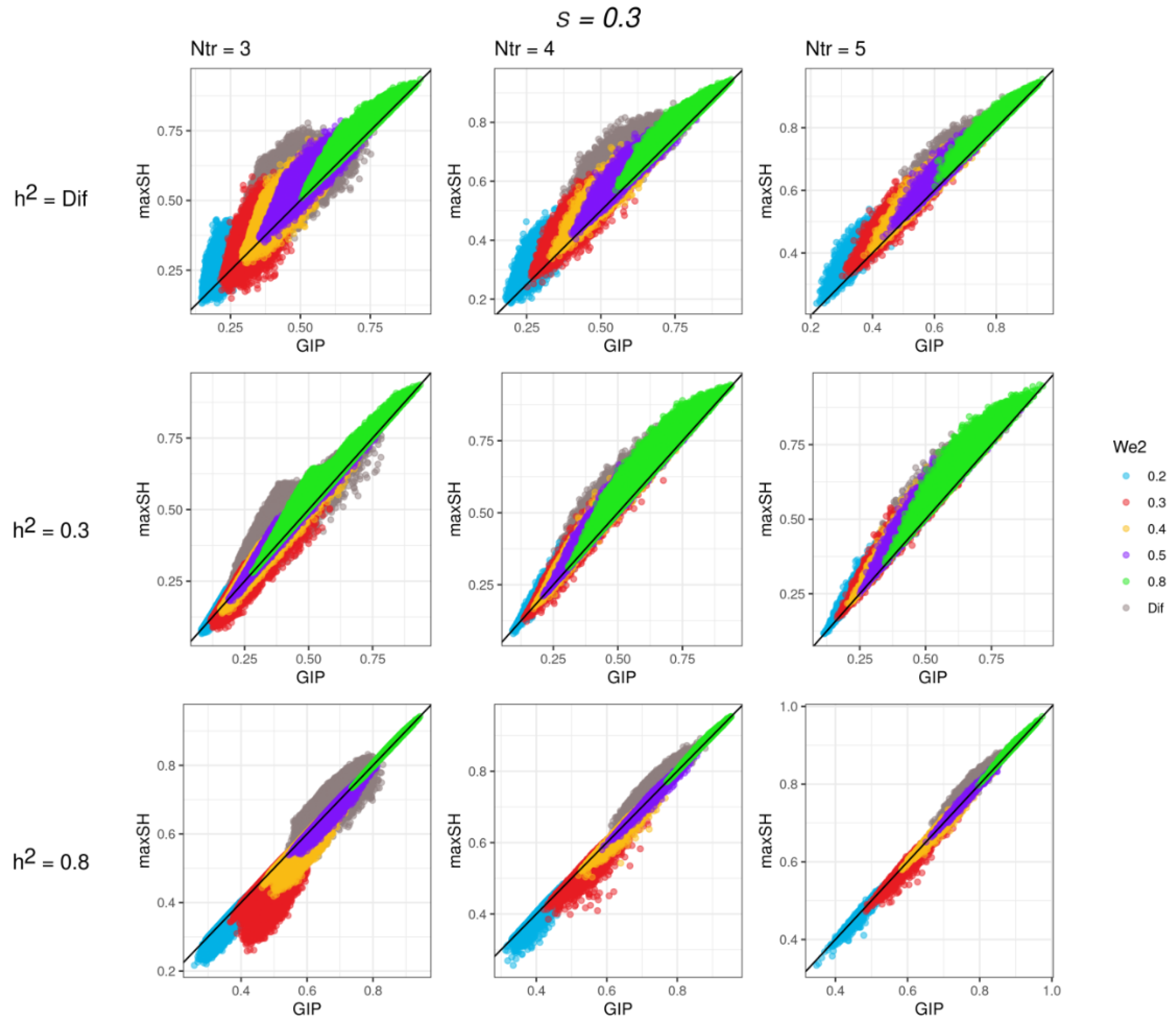
3.1. Simulation Study
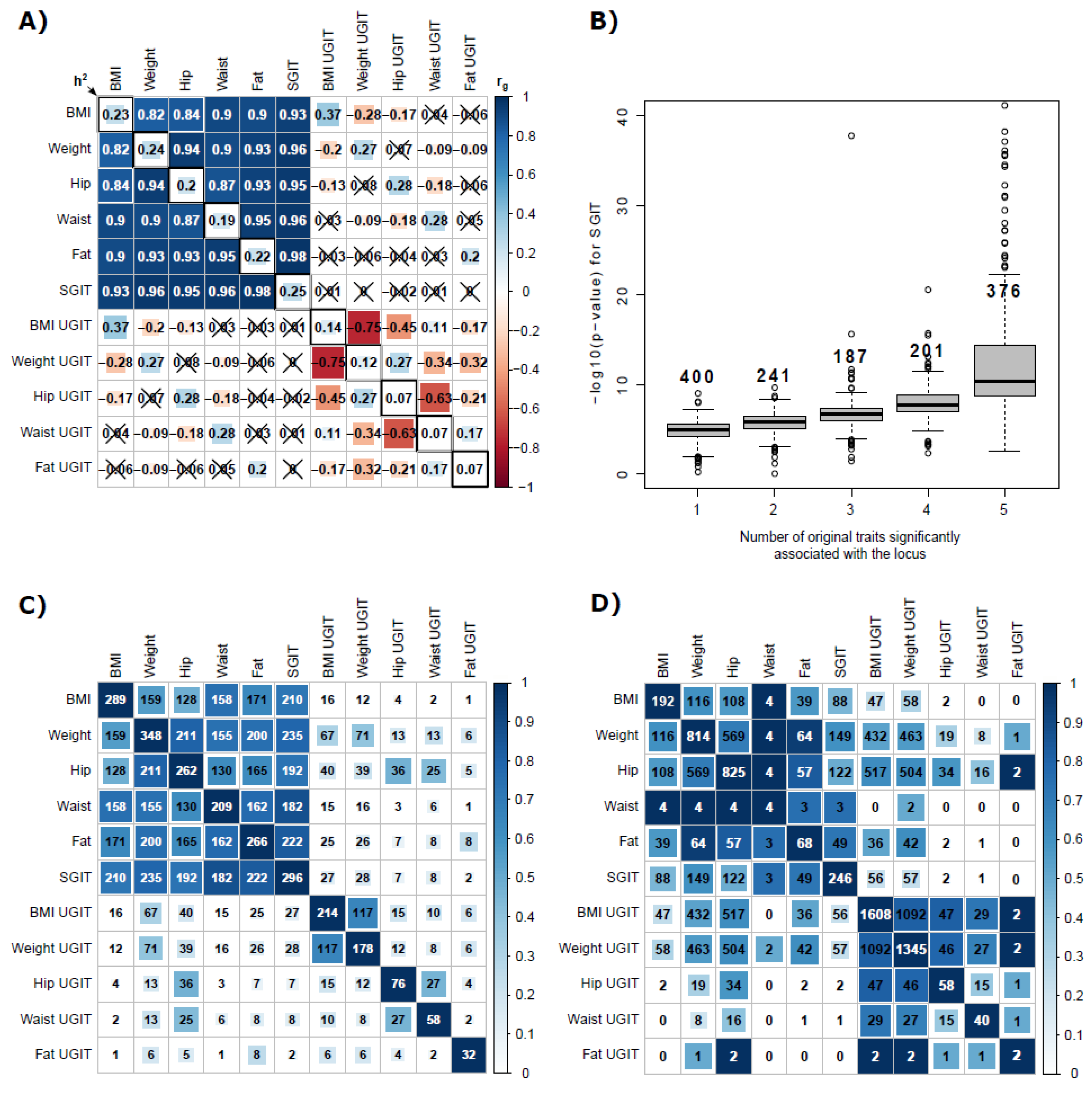
3.2. Real Data Assessment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Code Availability
References
- Jiang, X.; Finucane, H.K.; Schumacher, F.R.; Schmit, S.L.; Tyrer, J.P.; Han, Y.; Michailidou, K.; Lesseur, C.; Kuchenbaecker, K.B.; Dennis, J. Shared heritability and functional enrichment across six solid cancers. Nat. Commun. 2019, 10, 431. [Google Scholar] [CrossRef] [PubMed]
- Tsepilov, Y.A.; Freidin, M.B.; Shadrina, A.S.; Sharapov, S.Z.; Elgaeva, E.E.; van Zundert, J.; Karssen, L.C.; Suri, P.; Williams, F.M.; Aulchenko, Y.S. Analysis of genetically independent phenotypes identifies shared genetic factors associated with chronic musculoskeletal pain conditions. Commun. Biol. 2020, 3, 329. [Google Scholar] [CrossRef]
- Sampson, J.N.; Wheeler, W.A.; Yeager, M.; Panagiotou, O.; Wang, Z.; Berndt, S.I.; Lan, Q.; Abnet, C.C.; Amundadottir, L.T.; Figueroa, J.D. Analysis of heritability and shared heritability based on genome-wide association studies for 13 cancer types. JNCI J. Natl. Cancer Inst. 2015, 107, djv279. [Google Scholar] [CrossRef] [PubMed]
- Brainstorm, C.; Anttila, V.; Bulik-Sullivan, B.; Finucane, H.K.; Walters, R.K.; Bras, J.; Duncan, L.; Escott-Price, V.; Falcone, G.J.; Gormley, P.; et al. Analysis of shared heritability in common disorders of the brain. Science 2018, 360, aap8757. [Google Scholar] [CrossRef]
- Yang, Y.; Zhao, H.; Heath, A.C.; Madden, P.A.; Martin, N.G.; Nyholt, D.R. Shared genetic factors underlie migraine and depression. Twin Res. Hum. Genet. 2016, 19, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. Correlation and Causation. J. Agric. Res. 1921, XX, 557–585. [Google Scholar]
- Rijsdijk, F.V.; Sham, P.C. Analytic approaches to twin data using structural equation models. Brief. Bioinform. 2002, 3, 119–133. [Google Scholar] [CrossRef]
- Galesloot, T.E.; Van Steen, K.; Kiemeney, L.A.; Janss, L.L.; Vermeulen, S.H. A comparison of multivariate genome-wide association methods. PLoS ONE 2014, 9, e95923. [Google Scholar] [CrossRef] [PubMed]
- Stephens, M. A unified framework for association analysis with multiple related phenotypes. PLoS ONE 2013, 8, e65245. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, S.; Sha, Q. Joint analysis of multiple phenotypes in association studies based on cross-validation prediction error. Sci. Rep. 2019, 9, 1073. [Google Scholar] [CrossRef]
- Turley, P.; Walters, R.K.; Maghzian, O.; Okbay, A.; Lee, J.J.; Fontana, M.A.; Nguyen-Viet, T.A.; Wedow, R.; Zacher, M.; Furlotte, N.A. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet. 2018, 50, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Fatumo, S.; Carstensen, T.; Nashiru, O.; Gurdasani, D.; Sandhu, M.; Kaleebu, P. Complimentary methods for multivariate genome-wide association study identify new susceptibility genes for blood cell traits. Front. Genet. 2019, 10, 334. [Google Scholar] [CrossRef]
- Ning, Z.; Tsepilov, Y.A.; Sharapov, S.Z.; Wang, Z.; Grishenko, A.K.; Feng, X.; Shirali, M.; Joshi, P.K.; Wilson, J.F.; Pawitan, Y.; et al. Nontrivial Replication of Loci Detected by Multi-Trait Methods. Front. Genet. 2021, 12, 627989. [Google Scholar] [CrossRef] [PubMed]
- Pasaniuc, B.; Price, A.L. Dissecting the genetics of complex traits using summary association statistics. Nat. Rev. Genet. 2017, 18, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Timmers, P.R.H.J.; Tiys, E.S.; Sakaue, S.; Akiyama, M.; Kiiskinen, T.T.J.; Zhou, W.; Hwang, S.-J.; Yao, C.; Kamatani, Y.; Deelen, J.; et al. Mendelian randomization of genetically independent aging phenotypes identifies LPA and VCAM1 as biological targets for human aging. Nat Aging 2022, 2, 19–30. [Google Scholar] [CrossRef]
- Bulik-Sullivan, B.; Finucane, H.K.; Anttila, V.; Gusev, A.; Day, F.R.; Loh, P.R.; ReproGen Consortium; Psychiatric Genomics Consortium; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3; Duncan, L.; et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015, 47, 1236–1241. [Google Scholar] [CrossRef]
- Winkler, T.W.; Day, F.R.; Croteau-Chonka, D.C.; Wood, A.R.; Locke, A.E.; Mägi, R.; Ferreira, T.; Fall, T.; Graff, M.; Justice, A.E. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 2014, 9, 1192–1212. [Google Scholar] [CrossRef]
- Stahl, E.A.; Breen, G.; Forstner, A.J.; McQuillin, A.; Ripke, S.; Trubetskoy, V.; Mattheisen, M.; Wang, Y.; Coleman, J.R.; Gaspar, H.A. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 2019, 51, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Wray, N.R.; Ripke, S.; Mattheisen, M.; Trzaskowski, M.; Byrne, E.M.; Abdellaoui, A.; Adams, M.J.; Agerbo, E.; Air, T.M.; Andlauer, T.M. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 2018, 50, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Gorev, D.; Shashkova, T.; Pakhomov, E.; Torgasheva, A.; Klaric, L.; Severinov, A.; Sharapov, S.; Alexeev, D.; Aulchenko, Y. GWAS-MAP: A platform for storage and analysis of the results of thousands of genome-wide association scans. In Proceedings of the Bioinformatics of Genome Regulation and Structure\Systems Biology (BGRS\SB-2018), Novosibirsk, Russia, 20–25 August 2018; p. 43. [Google Scholar]
- Wei, T.; Simko, V. R Package ‘Corrplot’: Visualization of a Correlation Matrix. (Version 0.92). 2021. Available online: https://github.com/taiyun/corrplot (accessed on 1 September 2022).
- Pers, T.H.; Karjalainen, J.M.; Chan, Y.; Westra, H.-J.; Wood, A.R.; Yang, J.; Lui, J.C.; Vedantam, S.; Gustafsson, S.; Esko, T. Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 2015, 6, 5890. [Google Scholar] [CrossRef] [PubMed]
- Howe, L.J.; Lawson, D.J.; Davies, N.M.; Pourcain, B.S.; Lewis, S.J.; Smith, G.D.; Hemani, G. Genetic evidence for assortative mating on alcohol consumption in the UK Biobank. Nat. Commun. 2019, 10, 5039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcellini, F.; Giuli, C.; Papa, R.; Tirabassi, G.; Faloia, E.; Boscaro, M.; Polito, A.; Ciarapica, D.; Zaccaria, M.; Mocchegiani, E. Obesity and body mass index (BMI) in relation to life-style and psycho-social aspects. Arch. Gerontol. Geriatr. 2009, 49, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Pulit, S.L.; Stoneman, C.; Morris, A.P.; Wood, A.R.; Glastonbury, C.A.; Tyrrell, J.; Yengo, L.; Ferreira, T.; Marouli, E.; Ji, Y. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum. Mol. Genet. 2019, 28, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Howard, D.M.; Adams, M.J.; Clarke, T.-K.; Hafferty, J.D.; Gibson, J.; Shirali, M.; McIntosh, A. Genome-wide meta-analysis of depression in 807,553 individuals identifies 102 independent variants with replication in a further 1,507,153 individuals. BioRxiv 2018, 6288, 433367. [Google Scholar]
- Falconer, D.; Mackay, T. Introduction to Quantitative Genetics, 2nd ed.; Longman: New York, NY, USA, 1981; p. 315. [Google Scholar]
- Khodadadi, Z.; Tarami, B. Robust Empirical Bayes Estimation of the Elliptically Countoured Covariance Matrix. J. Math. Ext. 2011, 5, 31–46. [Google Scholar]
- Konno, Y. Estimation of Multivariate Complex Normal Covariance Matrices Under an Invariant Quadratic Loss. Commun. Stat. Theory Methods 2010, 39, 1490–1497. [Google Scholar] [CrossRef]
- Oualkacha, K.; Labbe, A.; Ciampi, A.; Roy, M.A.; Maziade, M. Principal components of heritability for high dimension quantitative traits and general pedigrees. Stat. Appl. Genet. Mol. Biol. 2012, 11. [Google Scholar] [CrossRef] [Green Version]
- Mardia, K.; Kent, J.; Bibby, J. Multivariate Analysis; Academic Press Inc.: London, UK, 1979; Volume 15, p. 518. [Google Scholar]

| Trait Name | Number of Significant Loci | ||
|---|---|---|---|
| Real Trait | SGIT * | UGIT * | |
| Anthropometric Traits | |||
| BMI | 289 | 296 (210) | 214 (16) |
| Weight | 348 | 296 (235) | 178 (71) |
| Hip | 262 | 296 (192) | 76 (36) |
| Waist | 209 | 296 (182) | 58 (6) |
| Fat | 266 | 296 (222) | 32 (8) |
| Psychometric Traits | |||
| BIP | 12 | 57 (8) | 2 (0) |
| MDD | 3 | 57 (0) | 2 (1) |
| SCZ | 92 | 57 (26) | 2 (0) |
| Happiness | 0 | 57 (0) | 1 (0) |
| Lipid Traits | |||
| LDL | 85 | 97 (69) | 43 (31) |
| Triglycerides | 71 | 97 (26) | 59 (30) |
| Cholesterol | 101 | 97 (84) | 51 (21) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Svishcheva, G.R.; Tiys, E.S.; Elgaeva, E.E.; Feoktistova, S.G.; Timmers, P.R.H.J.; Sharapov, S.Z.; Axenovich, T.I.; Tsepilov, Y.A. A Novel Framework for Analysis of the Shared Genetic Background of Correlated Traits. Genes 2022, 13, 1694. https://doi.org/10.3390/genes13101694
Svishcheva GR, Tiys ES, Elgaeva EE, Feoktistova SG, Timmers PRHJ, Sharapov SZ, Axenovich TI, Tsepilov YA. A Novel Framework for Analysis of the Shared Genetic Background of Correlated Traits. Genes. 2022; 13(10):1694. https://doi.org/10.3390/genes13101694
Chicago/Turabian StyleSvishcheva, Gulnara R., Evgeny S. Tiys, Elizaveta E. Elgaeva, Sofia G. Feoktistova, Paul R. H. J. Timmers, Sodbo Zh. Sharapov, Tatiana I. Axenovich, and Yakov A. Tsepilov. 2022. "A Novel Framework for Analysis of the Shared Genetic Background of Correlated Traits" Genes 13, no. 10: 1694. https://doi.org/10.3390/genes13101694
APA StyleSvishcheva, G. R., Tiys, E. S., Elgaeva, E. E., Feoktistova, S. G., Timmers, P. R. H. J., Sharapov, S. Z., Axenovich, T. I., & Tsepilov, Y. A. (2022). A Novel Framework for Analysis of the Shared Genetic Background of Correlated Traits. Genes, 13(10), 1694. https://doi.org/10.3390/genes13101694






