Abstract
RNA silencing serves key roles in a multitude of cellular processes, including development, stress responses, metabolism, and maintenance of genome integrity. Dicer, Argonaute (AGO), double-stranded RNA binding (DRB) proteins, RNA-dependent RNA polymerase (RDR), and DNA-dependent RNA polymerases known as Pol IV and Pol V form core components to trigger RNA silencing. Common bean (Phaseolus vulgaris) is an important staple crop worldwide. In this study, we aimed to unravel the components of the RNA-guided silencing pathway in this non-model plant, taking advantage of the availability of two genome assemblies of Andean and Meso-American origin. We identified six PvDCLs, thirteen PvAGOs, 10 PvDRBs, 5 PvRDRs, in both genotypes, suggesting no recent gene amplification or deletion after the gene pool separation. In addition, we identified one PvNRPD1 and one PvNRPE1 encoding the largest subunits of Pol IV and Pol V, respectively. These genes were categorized into subgroups based on phylogenetic analyses. Comprehensive analyses of gene structure, genomic localization, and similarity among these genes were performed. Their expression patterns were investigated by means of expression models in different organs using online data and quantitative RT-PCR after pathogen infection. Several of the candidate genes were up-regulated after infection with the fungus Colletotrichum lindemuthianum.
1. Introduction
Small RNAs have regulatory roles in a multitude of biological processes, including stress responses, development, metabolism, and maintenance of genome integrity, in a sequence-specific manner [1]. Although heterogeneous in size, sequence, genomic distribution, biogenesis, and action, most of these small RNA molecules mediate repressive gene regulation through RNA silencing [2]. RNA silencing refers to a variety of mechanisms where a small RNA molecule interferes with a given nucleotide sequence. Plant RNA silencing operates via RNA-directed DNA-methylation (RdDM) to repress transcription or by targeting mRNAs via post-transcriptional gene silencing (PTGS) [3].
RNA silencing is triggered by double-stranded RNA (dsRNA), and the generation and function of the small RNAs depend on key protein families such as Dicer-like (DCLs), Argonautes (AGOs), and RNA-dependent RNA polymerases (RDRs) [4]. The RNA silencing pathways rely on distinct DCL proteins that cleave dsRNA precursors into small RNAs 21–26 nucleotides in length [5], small-interfering RNAs (siRNAs), or microRNAs (miRNAs) [6]. In Arabidopsis thaliana, dsRNAs are processed into specifically-sized sRNA duplexes by one of the four DCL (AtDCL1–4) proteins. dsRNA processing, called dicing, is facilitated by one of the six dsRNA-binding proteins (HYPONASTIC1 or AtHYL1, AtDRB2–5, and AtDRB7) that interact with specific DCLs [7,8]. dsRNA might derive directly from virus replication, inverted repeats, or convergent transcription. dsRNA formation may also be genetically programmed at endogenous loci that produce transcripts with internal stem-loop structures. Alternatively, in A. thaliana, dsRNA may be synthesized by one of the six RDRs (AtRDR1–6) that copy single-stranded RNA (ssRNA) to initiate a new round of RNA silencing. These small RNAs are then incorporated into AGO-containing RNA-induced silencing complexes (RISCs) that guide small RNAs to their targets by sequence complementarity resulting in target RNA degradation, translational inhibition, or heterochromatin formation [6]. The A. thaliana genome encodes 10 AGO proteins (AGO1-10), with various functions such as implication in the RdDM pathway (AGO4) or viral defense (AGO2).
RdDM requires specialized transcriptional machinery centered on two plant-specific RNA polymerase II (Pol II)-related enzymes called Pol IV and Pol V [9]. Pol II, Pol IV, and Pol V have each have 12 subunits. Half of these subunits are common in Pols II, IV, and V, but each Pol also has specialized subunits. Subunits are named nuclear RNA polymerase B (NRPB) for Pol II subunits, NRPD for Pol IV subunits, and NRPE for Pol V subunits. The largest specialized subunits in Pol IV and Pol V are NRPD1 and NRPE1, respectively, and they bind to a shared subunit NRPD2/NRPE2 to form the catalytic cores [9]. NRPD1 and NRPE1 differ from NRPB1 by many substitutions or deletions of conserved amino acids, which probably contribute to their specialized functions in RdDM. Pol IV and Pol V are essential for the biogenesis and function of heterochromatic (hc)-siRNAs, which mediate TGS by RdDM (or histone modification) [10].
The availability of an increasing number of plant genomes shows that there is a large variation in the number of gene members of the core families encoding key components of RNA silencing. For example, A. thaliana, rice, tomato, soybean, and Medicago truncatula present four, eight, seven, five, and six DCL genes, respectively [11,12,13,14,15]. Similarly, the AGO gene family has expanded from three members in green algae [16] to 6 in moss, 10 in Arabidopsis, 17 in maize, 19 in rice, 25 in tomato, 22 in soybean, 27 in Brassica napus, and 11 in potato and coffee [12,13,15,17,18,19,20,21,22]. Plant AGO proteins are grouped into three major clades: AGO1/5/10, AGO2/3/7, and AGO4/6/8/9 [17,23]. These phylogenetic analyses showed that the diversification of the AGO gene family is an ancient and probably continuous process. This could mirror a functional diversification of AGO and DCL proteins, presumably reflecting expanding small RNA-directed regulatory pathways [17]. Likewise, the RDR family has also been expanded in different plant species, for example, from 6 members in rice and tomato to 7 and 16 in soybean and B. napus, respectively [12,13,15,24].
Common bean (P. vulgaris) is the most important grain legume for direct human consumption in the world, particularly in developing countries where it constitutes an important source of protein and essential micronutrients [25]. Unfortunately, bean production can be drastically impaired by environmental conditions and particularly by fungal diseases. Anthracnose, caused by the hemibiotrophic fungal pathogen C. lindemuthianum, is one of the most widespread and economically important diseases [25,26]. Common bean is an autogamous diploid (2n = 2x = 22) species with a relatively small genome (∼630 Mb) [27]. P. vulgaris is not only a major pulse crop, but is also an ideal model for crop evolutionary studies because of its complex evolution, which led to two major gene pools known as the Andean and Meso-American gene pools [28]. The divergence between these two gene pools was estimated to have occurred ca. 110 000 to 165 000 years ago [29,30]. In that context, two genome assemblies of the common bean are available, one for genotype G19833 of Andean origin [30] and one for genotype BAT93 of Mesoamerican origin [31]. AGO and DCL genes have been analyzed in the Andean G19833 genotype leading to the identification of 15 PvAGO genes and 6 PvDCL genes [32]. Consequently, except for the report of de Sousa Cardoso et al. [32], our knowledge of the RNA silencing mechanism in common bean remains quite poor.
The aims of this study were to identify and characterize, by in silico analysis, the genes involved in RNA silencing, including AGO, DCL, RDR, DRB, NRPD1, NRPE1, and NRPD2/NRPE2 in common bean. Taking advantage of the availability of two genome assemblies of contrasting origins (Andean and Mesoamerican), we wanted to address the evolution of these genes on a short time scale. Their expression patterns were investigated in different organs using online data and after infection with the fungus C. lindemuthianum by quantitative RT-PCR. The identification of these core components to trigger RNA silencing in this non-model plant species of worldwide economic relevance pave the way for further investigation.
2. Materials and Methods
2.1. Common Bean Genome Sequence Databases and Annotation Data
G19833 (v1.0) and BAT93 (v10) P. vulgaris genome assemblies and annotation data were downloaded from Phytozome (v10.0) (http://www.phytozome.net/, accessed on 1 December 2020) and from BAT93 genome data repository [31] (http://denovo.cnag.cat/genomes/bean/, accessed on 1 December 2020), respectively.
2.2. Identification of Argonaute, Dicer-like, RDR, DRB, NRPD1, NRPE1, and NRPD2 Genes in Common Bean Genomes
In order to identify DCL, AGO, RDR, DRB, NRPD1, NRPE1, NRPD2 genes, tBLASTn [33] search was performed on the G19833 and BAT93 genome sequences with the published Arabidopsis DCL [34], AGO [17], RDR [15], DRB [15], NRPD1, NRPE1, and NRPD2 [35] gene sequences as queries, using a cut-off E-value of 1e-10. Gene structure was determined by integrating evidence in the Artemis annotation platform [36], including (1) Genemark.hmm ab initio gene prediction [37], (2) Glycine max and P. vulgaris ESTs available from Genbank, aligned on the genomes using Sim4 [38], (3) similarities to protein sequences identified using BLASTx [33] on G. max (Wm82.a2v1) from Phytozome (v10.0) and Arabidopsis (TAIRv10) (https://www.arabidopsis.org, accessed on 1 December 2020), (4) contigs resulting from G19833 RNA-seq velvet assembly [30,39] aligned on both G19833 and BAT93 genomes using Sim4 [38]. Finally, the Pfam database (http://pfam.xfam.org/, accessed on 1 December 2020) was used to confirm each candidate gene by checking the presence of the typical domain of each family. DCL proteins should have an N-terminal helicase domain (DExD/H-box and helicase-C subdomains) followed by DUF283 (domain of unknown function, known also as Dicer dimerization domain), PAZ (Piwi-Argonaute-Zwille), tandem RNase III domains, and one or two C-terminal double-stranded RNA binding domains (dsRBDs) [14]. AGOs should have PAZ, MID (middle), and PIWI domains. RDRs should have a conserved RDRP domain. DRB proteins should have two double-stranded RNA binding motif domains.
Candidate proteins were named based on their phylogenetic proximity to known members in A. thaliana, soybean, and/or M. truncatula. The prefix PvA or PvM was added for sequences originating from G19833 (Andean) or BAT93 (Meso-American), respectively.
2.3. Protein Sequence Alignment and Phylogenetic Tree Building
The complete sequence of each putative AGO, DCL, RDR, and DRB proteins were aligned using Muscle [40], and the resulting alignments were manually optimized using SeaView [41]. For a given gene, when more than one isoform was identified, the longest was selected for the alignment. Aligned sequences were then analyzed using ProtTest3 [42] to estimate the best phylogenetic model. Maximum-likelihood trees were generated with PhyML [43]. Bootstrap values were computed with the consensus of 1000 trees generated with PhyML. The resulting phylogenetic trees were displayed using MEGA version 7 [44]. For phylogenetic analysis, the common bean sequences were completed with AGO sequences from soybean [17], DCL sequences from soybean, and M. truncatula [14], and RDR and DRB1 [also known as HYPONASTIC LEAVES 1 (HYL1)] sequences from soybean [15].
2.4. Characterization of the P. vulgaris DCL, AGO, RDR, DRB, NRPD, and NRPE Genes
The location of each PvA_AGO, PvA_DCL, PvA_RDR, PvA_DRB, PvA_NRPD, PvA_NRPE gene on G19833 chromosomes was determined by tBLASTn searching against the G19833 genome. Molecular weights (Mol. Wt.) and isoelectric points (pI) were determined using the Pepstats program from EMBOSS [45] analysis package. The number of isoforms in G19833 (v1.0) and BAT93 (v10) were obtained from corresponding official annotations in the Phytozome (V9.0) and BAT93 genome data repositories, respectively. Protein similarity and identity percentage were calculated with needleglobal pairwise alignment [45]. The number of introns in the CDS was obtained from manual reannotation performed in the Artemis platform [35].
2.5. RNA-Seq Data Analysis
RNA-seq data from G19833 genotype, were downloaded at https://www.ncbi.nlm.nih.gov/sra?linkname=bioproject_sra_all&from_uid=41439 (accessed on 1 December 2020), for 11 different organs including: roots_10DAP (days after planting), trifoliates_19DAP, young_pods, Leaves_10DAP, stem_10DAP, stem_19DAP, nodules_19DAP, roots_19DAP, mature_pods, flower_buds, flowers [30]. RNA-seq count data were transformed as moderated log-counts-per-million using the package EdgeR (version 3.16.4, [46]) in the statistical software ‘R’ (version 3.3.2, [47]). Then for each subset of genes, we used the MixOmics R package (version 6.1.1, [48]) to run a hierarchical clustering on both genes and organs using the Euclidean distance and Ward method.
2.6. Plant Materials, Infection with C. lindemuthianum, RNA Extraction, and RT-qPCR Analysis
Infections of the common bean Andean landrace JaloEEP558 with the incompatible strain 100 of C. lindemuthianum were carried out as previously described in Richard et al. [49]. A time-course gene expression analysis was conducted at 6, 24, 48, 72, and 96 hpi in JaloEEP558 seedlings infected with strain 100. For each time, one of the two cotyledonary leaves from three different inoculated plants and control plants were sampled and flash-frozen in liquid nitrogen for RNA isolation and RT-qPCR analysis.
Total RNA extraction and quantitative RT-PCR (RT-qPCR) experiments were performed as described in Richard et al. [49]. The expression analyses of the genes PvAGO1, PvAGO2a, PvDCL2a, PvDCL2b, PvAGO4a, PvAGO4b, and PvAGO4c were performed using the gene-specific primers listed in Supplementary Table S1. Gene expression was normalized with four reference genes (PvUkn1, PvUkn2, PvIDE, and PvAct11) [50] (Supplementary Table S1). For each gene, gene expression in mock condition was used to calibrate gene expression in infected plants at each time point. Relative gene expression in inoculated leaves compared to mock leaves was calculated using the method 2−ΔΔCt on three technical replicates and two biological replicates [51]. Statistical comparisons were carried out using unpaired t-tests between each mean value (at t = 6, 24, 48, 72, 96 hpi) and the corresponding mean value at t = 0 hpi.
3. Results
3.1. Six Putative DCL Genes Are Present in P. vulgaris Genome
The search for homologous DCL sequences in the P. vulgaris genome generated six full-length DCL genes recovered from both G19833 and BAT93 genomes (Table 1). These genes were named using the prefix PvA_ or PvM_ to indicate genotype G19833 (Andean) or BAT93 (Meso-American), respectively, or PvA/M to indicate a gene present in both genotypes. PvA/M prefix was then followed by an identifier for their Arabidopsis homologs determined by phylogenetic analysis (e.g., PvA_DCL1 corresponds to the AtDCL1 gene). For paralogs, a letter (a, b, c…) was used as the suffix. The same nomenclature was used for all genes involved in RNA silencing described in this study. Dicer-like 1–4 occurred as monophyletic groups containing DCLs from P. vulgaris, G. max, M. truncatula, and A. thaliana. Our manual annotation allowed us to identify PvM_DCL2c that was not present in the automatic annotation of BAT93 assembly. In P. vulgaris, for both BAT93 and G19833, DCL1, DCL3, and DCL4 occurred as single-copy genes, while DCL2 had three paralogs (PvA/M_DCL2a, PvA/M_DCL2b, PvA/M_DCL2c) (Figure 1, Table 1). The six DCL genes in the common bean presented high levels of protein identity between BAT93 and G19833 (> 97% protein identity). PvA_DCL2a and PvA_DCL2b were separated by 2.5 kb on chromosome 6, while PvA_DCL2c was located on chromosome 8 (Figure 2). Despite their tight physical linkage, DCL2a and DCL2b were phylogenetically separated (Figure 1), such that PvA/M_DCL2b and PvA/M_DCL2c grouped with GmDCL2b, while PvA/M_DCL2a grouped with GmDCL2a (Figure 1). Manual inspection of flanking genes in the P. vulgaris and G. max genomes showed that both copies of DCL2 (a and b) are located in a syntenic region (Supplementary Figure S1). Indeed, in both species, the duplicated DCL2 genes were flanked by genes encoding a histidinol dehydrogenase and a protein male sterile 5 on one side and by genes encoding a stress up-regulated Nod 19 and 3-hydroxyisobutyrate dehydrogenase on the other side (Supplementary Figure S1). Amplification of DCL2 genes has also been observed in M. truncatula, which has three copies [14]; however, these DCL2s formed a separate clade (Figure 1). The PvA/M_DCL proteins ranged in length from 1388 to 1975 amino acids (aa) (Table 1). As observed for other legume species, the smaller DCL proteins occur within the DCL2 clade [52].

Table 1.
Identification of AGO, DCL, RDR, and DRB genes in common bean.
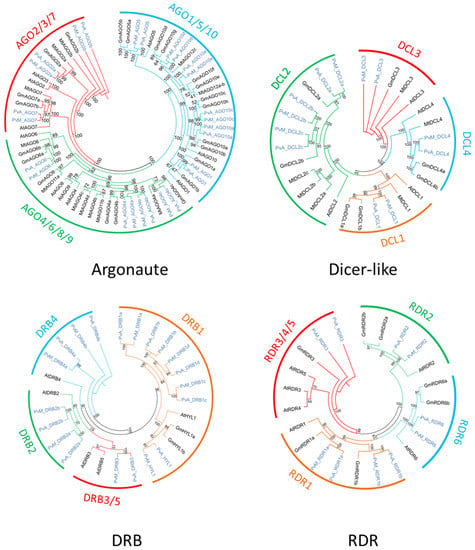
Figure 1.
Phylogenetic analysis of Argonaute, Dicer-like, DRB, and RDR family. Pv sequences are presented in light blue, while sequences from A. thaliana, soybean, and M. truncatula are presented in black.
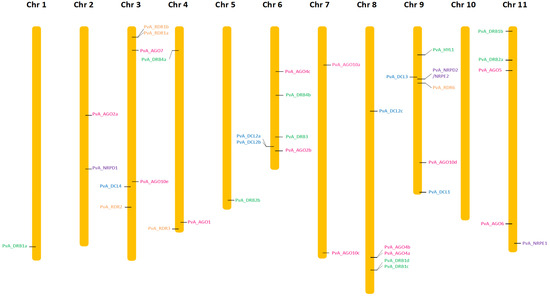
Figure 2.
Chromosomal localization of AGO (pink), DCL (light blue), DRB (green), RDR (orange), NRDP1, NRPE1, and NRPD2/NRPE2 (purple) genes in the common bean genome (G19833).
3.2. Thirteen AGO Genes in Common Bean Genome
The search for homologous AGO sequences in the P. vulgaris genome generated 13 full-length AGO genes recovered from both G19833 and BAT93 genomes (Table 1). Our manual annotation allowed us to correct PvM_AGO2a by fusing two distinct genes from BAT93 automatic annotation leading to a 971 aa long PvM_AGO2a protein, sharing 99.3% of protein identity with the G19833 homolog (Table 1). The length of the identified AGOs varied from 886 to 1063 aa. The Pv AGO genes were spread on 8 out of 11 common bean chromosomes, with two genes (PvA_AGO4a and PvA_AGO4b) organized in a tandem array on chromosome 8 and separated by ∼20 kb (Figure 2). The phylogenetic tree classified the AGOs proteins into three clades: AGO 1/5/10, AGO 4/6/8/9, and AGO 2/3/7 (Figure 1). For each 13 Pv AGO genes, a clear orthology relationship was identified between G19833 (PvA_AGO) and BAT93 (PvM_AGO), testifying to the absence of recent gene duplication or deletion for this AGO gene family (Figure 1). In particular, the gene duplication leading to PvA/M_AGO4a and PvA/M_AGO4b occurred prior to the Andean/Mesoamerican gene pool divergence.
3.3. Five RDR Genes in the Common Bean Genome
Common bean G19833 and BAT93 genomes contain five RDR genes each (Table 1), located on chromosomes 3, 4, and 9 (Figure 2). Our manual annotation allowed us to correct PvM_RDR1a by fusing two distinct genes from BAT93 automatic annotation leading to an 1139 aa long PvM_RDR1a protein, sharing 99.3% of protein identity with its G19833 homolog (Table 1). The length of RDRs ranged from 980 aa to 1222 aa (Table 1). As previously observed [12,53], phylogenetic analysis grouped RDR into four clades (RDR1, RDR2, RDR3, RDR6) with clade RDR3 containing three Arabidopsis members (AtRDR3, AtRDR4, AtRDR5) out of the 6 AtRDR (Figure 1). Concerning P. vulgaris, each clade contained a single Pv RDR gene, except for clade 1, which contained two RDR1 paralogs (PvA/M_RDR1a and PvA/M_RDR1b) closely linked on chromosome 3 and separated by 10 kb (Figure 1 and Figure 2). Similarly, two RDR1 paralogs were also identified in chickpea and pigeonpea genomes [50], suggesting that they could correspond to an ancient gene duplication.
3.4. Ten DRB Genes in Common Bean Genome
Ten DRB genes were identified in both G19833 and BAT93 genomes (Table 1) with a clear orthology relationship, suggesting no recent duplication/deletion for this gene family in common bean (Figure 1). Our manual annotation led us to correct PvM_DRB4b by fusing two distinct genes from BAT93 automatic annotation leading to a 248 aa long PvM_DRB4b protein, sharing 99.6% of protein identity with its G19833 homolog (Table 1). Compared to Arabidopsis, the common bean genome experienced an amplification of the DRB1 gene family (five members) as well as the DRB4 gene family (two members). A clear ortholog of AtHYL1, a key interactor of DCL1 in miRNA biogenesis [54], referred to as PvA/M_HYL1, was identified on common bean chromosome 9 (Figure 1 and Figure 2). The 10 common bean DRB genes were spread on seven chromosomes, with PvA/M_DRB1d and PvA/M_RDB1c genes tightly linked on chromosome 8.
3.5. Common Bean Pol IV and Pol V
In order to gain insight into the Pol IV and Pol V complex in the common bean genome, the largest and second-largest subunits of Pol IV and Pol V were searched by seeking AtNRPD1, AtNRPE1, and AtNRPD2/NRPE2 against common bean BAT93 and G19833 genomes with tBLASTn. Common bean encodes one NRPD1, one NRPE1, and one NRPD2/NRPE2, and hence they are named PvA/M_NRPD1, PvA/M_NRPE1, and PvA/M_NRPD2/NRPE2 (Table 1). These three proteins present a high level of identity (> 99%) between BAT93 and G19833. They are located on chromosome 2 (PvA_NRPD1), 11 (PvA_NRPE1), and 9 (PvA_NRPD2/NRPE2) (Figure 2).
3.6. In Silico Expression Pattern of AGO, DCL, RDR, DRB, NRPD1, NRPE1, and NRPD2 Candidate Genes
In order to analyze the transcript abundance of these 37 genes in different organs of common bean, we performed a comprehensive gene expression in silico analysis using online RNAseq data for genotype G19833. The results are shown in Figure 3. After moderated log-counts-per-million transformation, we applied hierarchical clustering (with Euclidean distance and Ward method) on the 37 genes. The genes can be organized into three clusters. Cluster 1 corresponds to genes presenting a low expression level. This cluster comprises several DRB genes (PvA_DRB4b, 1d, 1b, 1a), two AGO genes (PvA_AGO2a, 2b), one DCL gene (PvA_DCL2c), and one RDR gene (PvA_RDR1a). Cluster 3 corresponds to genes that are highly expressed and comprises four AGO genes (PvA_AGO1, 4c, 5, 4b), two DRB genes (PvA_DRB2a, 2b), one DCL gene (PvA_DCL1), as well as PvA_NRPE1 and PvA_NRPD2. In particular, PvA_AGO1 seems to be highly expressed in all tested organs. Finally, the remaining 20 genes correspond to genes presenting an intermediary expression level (cluster 2; Figure 3). For most genes of this cluster, the expression level seems relatively homogenous in the 11 analyzed organs, except PvA_RDR3, which seems up-regulated in the nodules.
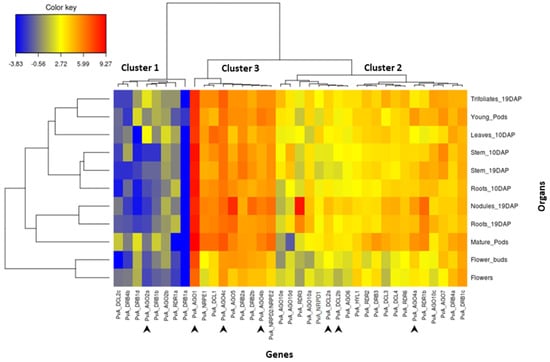
Figure 3.
Heat map showing the expression pattern of PvA_AGO, PvA_DCL, PvA_RDR, PvA_DRB, PvA_NRDP1, PvA_NRPE1, and PvA_NRPD2/NRPE2 genes in 11 common bean organs from genotype G19833. The color scale fold-change values are shown on the left of the heat map. DAP = days after planting. Arrows indicate genes analyzed in RT-qPCR experiments after C. lindemuthianum infection.
3.7. Expression Pattern Analysis after Fungus Infection
In order to investigate the role of RNA silencing in pathogen defense in common bean, we studied the expression profile of seven genes, including AGO1, AGO2a, DCL2a, DCL2b, AGO4a, AGO4b, and AGO4c (indicated by the arrows in Figure 3). The expression of these genes in response to infection with the hemibiotrophic fungus C. lindemuthianum was studied using RT-qPCR at 6, 24, 48, 72, 96 hpi in a resistant genotype (incompatible interaction). Significantly, temporal gene expression analysis revealed that DCL2a and DCL2b are both ca. nine-fold up-regulated after infection compared to the mock control at 72 hpi. Similarly, a clear upregulation is observed at 72 hpi for both AGO4a and AGO2a, and also for AGO4b but with a lower extend (Figure 4). Conversely, the expression of AGO1 and AGO4c was not modified upon C. lindemuthianum infection (Figure 4).
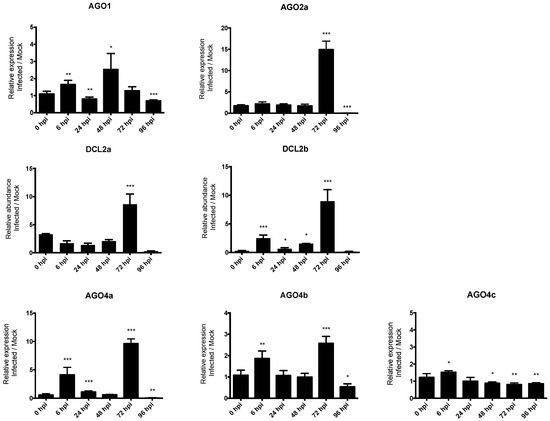
Figure 4.
The expression levels of P. vulgaris AGO and DCL genes in response to C. lindemuthianum infection. Bars show means +/− SD. * p < 0.05; ** p < 0.01; *** p < 0.001, unpaired t-test.
4. Discussion
Several studies have pointed out that genes involved in silencing evolve rapidly with a great variation of number even between closely related species. However, our comprehensive analysis of various gene families involved in RNA silencing in two common bean genomes of contrasting origins allowed us to identify the same number of AGO (13), DCL (6), DRB (10), RDR (5), NRPD1 (1), NRPE1 (1), and NRPD2/NRPE2 (1) genes in both G19833 (Andean) and BAT93 (Meso-American) genomes, suggesting that no recent gene duplication/deletion occurred after gene pool divergence. Indeed, for each of the 37 genes analyzed in the present study, orthologs presenting a high percentage of protein identity (> 94%) were unambiguously identified between BAT93 and G19833 (Table 1, Figure 1). This suggests that even if the genome assembly of BAT93 is of lower quality compared to G19833, and does not allow repeated sequence analysis [55], this quality is sufficient for gene analysis. However, we can not exclude that some genes were missing, and higher quality genome assembly based on long-read sequencing will be needed to unambiguously address this question. These 37 genes are distributed on all Pv chromosomes, except chromosome 10. Importantly, our manual annotation led us to correct misannotated genes, in particular in BAT93 (Table 1). Even if no recent gene dynamics were identified after gene pool divergence, an interesting pattern of evolution was identified for DCL and AGO gene families in the common bean.
DCL genes, and in particular DCL2 genes, present a complex pattern of evolution in legume species. Unlike the single-copy genes of DCL1, DCL3, and DCL4, in Pv and Mt, there were three copies of DCL2. Soybean contains seven DCL genes in its ancient polyploid (paleopolyploid) genome [56] (Figure 1). In soybean, the most recent genome doubling event occurred approximately 9–14 million years ago, and the soybean genome maintains at least one gene duplicate for ca. 75% of its genes, termed homoeologous gene pairs [57]. GmDCL4a/GmDCL4b and GmDCL1a/GmDCL1b correspond to such homoeologous gene pairs, while GmDCL3 is present as a single copy. By contrast, GmDCL2a and GmDCL2b are locally duplicated, separated by 5 kb, on chromosome 9. In soybean, the age of this GmDCL2a/GmDCL2b duplication was estimated to be 19.4 Mya [56], indicating that it predates the whole genome duplication of soybean at 9–14 Mya [56], and the split for common bean and soybean 19 Mya [58,59]. Consequently, this strongly suggests that in the putative Pv Gm common ancestor, a locally duplicated pair of DCL2 genes were present. In agreement with this, we found in common bean two DCL2 genes, Pv_DCL2a and Pv_DCL2b, organized in tandem array in the corresponding syntenic region with soybean (Figure 2 and Figure S1). In the Pv genome, an additional paralog, DCL2c, present on chromosome 8, was putatively derived from Pv_DCL2b by a yet unknown mechanism that could involve transposable elements identified in the vicinity of DCL2c. Three copies of DCL2 were also identified in Mt [14]. However, this amplification appears to be independent of that observed in the common bean (Figure 1). In contrast, only a single DCL2 has been identified in various other legume species, including chickpea (Cicer arietinum), pigeonpea (Cajanus cajan), and each of the two genomes composing the allotetraploid groundnut (Arachis duranensis and Arachis ipaensis) [52]. These independent amplifications of the DCL2 genes in specific legume species could lead to their functional diversification and probably reflect their functional importance. In Mt, a nodule-specific role for DCL2 has been proposed [14], while in soybean, DCL2 genes regulate traits such as seed color via the production of 22 nucleotide siRNA from long inverted repeats [60]. In another study, GmDCL2 paralogs exhibited a wide range of transcriptional changes in response to stress, suggesting DCL2s may play an important role in stress response [56]. Congruent with these findings, we found that Pv_DCL2a and Pv_DCL2b, mildly expressed in most organs (Figure 3), are up-regulated at 72 hpi in leaves infected by C. lindemuthianum (Figure 4).
We identified 13 AGO genes in Pv, while 15 AGO genes were reported in a previous analysis performed on the G19833 genome. Indeed, compared to de Sousa Cardoso et al. [32], our manual annotation led us to discard one AGO10 and one AGO2 gene. Similarly, 13 AGO genes were also identified in both chickpea and pigeonpea [52]. Within angiosperms, several AGO subgroups have expanded differently in monocots and eudicots, with lineage-specific gene duplications [61]. For example, the grasses exhibit an expanded AGO 1/5/10 clade [17]. More precisely, maize and rice harbor many AGO5 paralogs, and a grass-specific AGO18 family, a deep branch of the AGO1/5/10 clade, has been discovered and played important roles during plant reproduction and viral defense [17]. In common bean, expansion of the AGO1/5/10 clade was also observed, but it was the result of AGO10 gene amplification since four AGO10 genes were identified in the common bean genome (Table 1). Similar amplification of AGO10 was also observed in soybean, where eight paralogs were identified in its paleopolyploid genome [17]. Each Pv AGO10 gene clearly corresponds to two Gm orthologs (Figure 1), strongly suggesting that AGO10 amplification occurred prior to the soybean/common bean divergence. In soybean, the expansion of the AGO10 family presumably co-evolved with the expansion of the miR165/166 family since 21 copies of miR165/166 are annotated in the soybean genome [17]. Likewise, expansions of miR165/166 genes, with 10 copies, have also been identified in the Pv genome (Geffroy V. and Meyers B.C.; unpublished results). In addition to AGO10, expansion was also observed for AGO2 (two members) and AGO4 (three members) in the Pv genome. AtAGO4 primarily binds 24-nt, repeat and heterochromatin-associated siRNAs, and functions in RNA-directed DNA methylation [62], while AtAGO2 functions in antibacterial immunity [63]. In common bean, AGO2 and AGO4 genes have non-redundant expression profiles (Figure 3), suggesting that they may have acquired divergent functions. PvA_AGO1 seems to be highly expressed in all common bean-tested organs. In agreement with our results, AGO1 expression is detected in many organs, such as leaves, roots, and flowers, in Arabidopsis [64], rice [12], B. napus [65], and the emerging medicinal model plant Salvia miltiorrhiza [66].
Functional analysis of genes involved in RNA silencing revealed that most of them play multiple roles, not only in growth and development but also in immune defense against pathogens [1,67,68,69]. The importance of RNA silencing in plant viral defense has been well documented for a long time [63]. In addition to viral defense, more and more evidence is accumulating, showing that RNA silencing also plays a role in plant interactions with bacterial pathogens [70]. More recently, the potential role of RNA silencing in plant defense has also been reported for several fungal pathogens, including Verticillium dahliae [71], Verticillium longisporum [72], Magnaporthe oryzae [73], and Botrytis cinerea [74]. The importance of RNA silencing in plant defense is illustrated by the fact that it has stimulated a counter-defense system from the pathogens to overcome it. Indeed, it is now well-known that pathogens of a different nature (viruses, bacteria, fungi, oomycetes, and phytoplasma) have evolved effectors that are able to target and suppress the host plant RNA silencing pathway [67,75,76,77,78]. Suppressors of RNA silencing were first discovered in viruses (VSRs, for viral suppressors of RNA silencing) [4]. At present, there is no evidence of putative suppressors of silencing acting in C. lindemuthianum. However, there is growing evidence that this is a common mechanism exploited by fungal pathogens to promote their infection [79]. Consequently, such suppressors probably exist in C. lindemuthianum, although not yet identified.
To investigate the contribution of some of the genes involved in RNA silencing in the defense response in common bean, we performed quantitative RT-PCR-based expression analysis on leaves of resistant bean plants inoculated with the hemibiotrophic fungus C. lindemuthianum (in an incompatible context). Whereas expression levels of PvA_AGO2a, PvA_AGO4a, and PvA_DCL2 (a and b) is low to moderate without any biotic stress (Figure 3), a strong up-regulation of these genes was observed mainly at 72 hpi (Figure 4). On the contrary, expression of PvA_AGO1 and PvA_AGO4c, which are ubiquitously and highly expressed in uninfected plants, was not significantly modified after infection. This suggests that after fungal infection, PvA_AGO2, PvA_AGO4, and Pv_DCL2 may play a prominent role in the sRNA-based regulation of defense gene expression in the common bean. Interestingly, the Argonaute proteins, AGO4 and AGO2, have both been linked to antibacterial defense. AGO4, a component of the RdDM pathway that directs DNA methylation at specific loci, mediates resistance to P. syringae, independently of the other components of the RdDM pathway [80]. AGO2 functions in antibacterial immunity by binding a specific miRNA to modulate the exocytosis of antimicrobial PR proteins [81]. In the literature, different pathosystems involving either hemibiotrophic pathogens or incompatible plant-microbe interactions present similar results. Notably, in susceptible wild tobacco plants challenged by the hemibiotrophic fungus Fusarium brachygibbosum as well as in resistant cowpea plants in response to CPSMV (Cowpea severe mosaic virus) infection, an increased expression of AGO4 has been reported, whereas no change in expression was observed for AGO1 [82,83]. Moreover, an up-regulation of AGO2 expression after infection was reported in Arabidopsis after infection by the biotrophic bacteria P. syringae, in the oil crop B. napus infected by the fungal necrotrophic Sclerotinia sclerotiorum, and in the cowpea Vigna unguiculata infected by CPSMV [20,82,84]. Concerning the role of Dicer proteins in plant defense, little is known about DCL2, except that it is involved in the processing of viral dsRNA. However, it has also been observed that the quantity of DCL2 transcripts increases at the local site of infection by the CLRDV (Cotton leafroll dwarf virus) in a resistant genotype cotton Gossypium hirsutum [85]. This is in agreement with our results, where up-regulation of both PvA_DCL2a and Pv_DCL2b was observed in incompatible interaction with C. lindemuthianum. This suggests that in the common bean, an increased expression of specific genes involved in RNA silencing, acting in both the miRNA and the siRNA pathways, could counteract the infectious process of C. lindemuthianum. However, how these genes could regulate resistance in the common bean requires further investigation.
5. Conclusions
This work will further provide a solid foundation for future functional analysis of AGO, DCL, RDR, DRB, NRPD1, NRPE1, and NRPD2 genes in the common bean genome. For example, taking advantage of the work presented here, we would like to utilize virus-induced gene silencing (VIGS) to silence NRPD1 and NRPE1, in order to gain insight into the mechanisms involved in the unusual methylation pattern observed for NLR genes in the common bean [86,87,88]. Furthermore, our work will also help to design specific primers for RT-qPCR experiments. Finally, given the genomic location of the 37 genes studied (Table 1, Figure 2), and considering that RNA silencing is involved in a large number of traits, our work may also provide candidate genes for QTL analysis.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes13010064/s1, Figure S1: Syntenic analysis between common bean and soybean of the genomic region containing DCL2a and DCL2b genes. Table S1: List of primer sequences used in this study.
Author Contributions
V.G. designed the project; J.C.A.-D., M.M.S.R., V.T., C.P.-L.-R., G.R., A.G. and V.G. performed the bioinformatics analysis; J.C.A.-D. performed the experiments; J.C.A.-D., M.M.S.R., V.T., G.T., C.P.-L.-R., G.R., S.P., A.G. and V.G. analyzed and interpreted the data; V.G. funding acquisition; J.C.A.-D., A.G. and V.G. wrote the manuscript with significant input from all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by grants from IDEEV and INRAE. IPS2 benefits from the support of Saclay Plant Sciences-SPS (ANR-17-EUR-0007).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We are grateful to Randall Wisser for critical reading of the manuscript. We thank Blake C. Meyers for helpful discussions on small RNA.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Seo, J.K.; Wu, J.; Lii, Y.; Li, Y.; Jin, H. Contribution of small RNA pathway components in plant immunity. Mol. Plant -Microbe Interact. 2013, 26, 617–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Ferrer, V.; Voinnet, O. Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 2009, 60, 485–510. [Google Scholar] [CrossRef] [Green Version]
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baulcombe, D. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Carmell, M.A.; Hannon, G.J. RNase III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol. 2004, 11, 214–218. [Google Scholar] [CrossRef]
- Bologna, N.G.; Voinnet, O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 2014, 65, 473–503. [Google Scholar] [CrossRef]
- Hiraguri, A.; Itoh, R.; Kondo, N.; Nomura, Y.; Aizawa, D.; Murai, Y.; Koiwa, H.; Seki, M.; Shinozaki, K.; Fukuhara, T. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol. Biol. 2005, 57, 173–188. [Google Scholar] [CrossRef]
- Clavel, M.; Pélissier, T.; Montavon, T.; Tschopp, M.A.; Pouch-Pélissier, M.N.; Descombin, J.; Jean, V.; Dunoyer, P.; Bousquet-Antonelli, C.; Deragon, J.M. Evolutionary history of double-stranded RNA binding proteins in plants: Identification of new cofactors involved in easiRNA biogenesis. Plant Mol. Biol. 2016, 91, 131–147. [Google Scholar] [CrossRef]
- Matzke, M.A.; Mosher, R.A. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014, 15, 394–408. [Google Scholar] [CrossRef]
- Haag, J.R.; Pikaard, C.S. Multisubunit RNA polymerases IV and V: Purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 2011, 12, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Henderson, I.R.; Zhang, X.; Lu, C.; Johnson, L.; Meyers, B.C.; Green, P.J.; Jacobsen, S.E. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 2006, 38, 721–725. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, M.; Arora, R.; Lama, T.; Nijhawan, A.; Khurana, J.P.; Tyagi, A.K.; Kapoor, S. Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA Polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genom. 2008, 9, 451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, M.; Yang, G.S.; Chen, W.T.; Mao, Z.C.; Kang, H.X.; Chen, G.H.; Yang, Y.H.; Xie, B.Y. Genome-wide identification of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families and their expression analyses in response to viral infection and abiotic stresses in Solanum lycopersicum. Gene 2012, 501, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Tworak, A.; Urbanowicz, A.; Podkowinski, J.; Kurzynska-Kokorniak, A.; Koralewska, N.; Figlerowicz, M. Six Medicago truncatula Dicer-like protein genes are expressed in plant cells and upregulated in nodules. Plant Cell Rep. 2016, 35, 1043–1052. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lu, T.; Dou, Y.; Yu, B.; Zhang, C. Identification of RNA silencing components in soybean and sorghum. BMC Bioinform. 2014, 15, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, T.; Li, G.; Mi, S.; Li, S.; Hannon, G.J.; Wang, X.J.; Qi, Y. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 2007, 21, 1190–1203. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Xia, R.; Meyers, B.C.; Walbot, V. Evolution, functions, and mysteries of plant ARGONAUTE proteins. Curr. Opin. Plant Biol. 2015, 27, 84–90. [Google Scholar] [CrossRef] [Green Version]
- Qian, Y.; Cheng, Y.; Cheng, X.; Jiang, H.; Zhu, S.; Cheng, B. Identification and characterization of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families in maize. Plant Cell Rep. 2011, 30, 1347–1363. [Google Scholar] [CrossRef]
- Fang, X.; Qi, Y. RNAi in plants: An argonaute-centered view. Plant Cell 2015, 28, 272–285. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Zheng, W.; Zhong, Z.; Chen, X.; Wang, A.; Wang, Z. Genome-wide analysis of RNA-interference pathway in Brassica napus, and the expression profile of BnAGOs in response to Sclerotinia sclerotiorum infection. Eur. J. Plant Pathol. 2016, 146, 565–579. [Google Scholar] [CrossRef]
- Mirzaei, K.; Bahramnejad, B.; Shamsifard, M.H.; Zamani, W. In silico identification, phylogenetic and bioinformatic analysis of argonaute genes in plants. Int. J. Genom. 2014. [Google Scholar] [CrossRef]
- Fernandes-Brum, C.N.; Rezende, P.M.; Ribeiro, T.H.C.; De Oliveira, R.R.; De Sousa Cardoso, T.C.; Do Amaral, L.R.; De Souza Gomes, M.; Chalfun, A. A genome-wide analysis of the RNA-guided silencing pathway in coffee reveals insights into its regulatory mechanisms. PLoS ONE 2017, 12, e0176333. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, W.; Guo, M.; Liu, S.; Liu, L.; Yu, Y.; Mo, B.; Chen, X.; Gao, L. Origin, evolution and diversification of plant ARGONAUTE proteins. Plant J. 2021, 1–12. [Google Scholar] [CrossRef]
- Cao, S.; Loladze, A.; Yuan, Y.; Wu, Y.; Zhang, A.; Chen, J.; Huestis, G.; Cao, J.; Chaikam, V.; Olsen, M.; et al. Genome-Wide Analysis of Tar Spot Complex Resistance in Maize Using Genotyping-by-Sequencing SNPs and Whole-Genome Prediction. Plant Genome 2017, 10. [Google Scholar] [CrossRef] [Green Version]
- Miklas, P.N.; Kelly, J.D.; Beebe, S.E.; Blair, M.W. Common bean breeding for resistance against biotic and abiotic stresses: From classical to MAS breeding. Euphytica 2006, 147, 105–131. [Google Scholar] [CrossRef]
- Meziadi, C.; Richard, M.M.S.; Derquennes, A.; Thareau, V.; Blanchet, S.; Gratias, A.; Pflieger, S.; Geffroy, V. Development of molecular markers linked to disease resistance genes in common bean based on whole genome sequence. Plant Sci. 2016, 242, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Arumuganathan, K.; Earle, E.D. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 1991, 9, 208–218. [Google Scholar] [CrossRef]
- Bitocchi, E.; Rau, D.; Bellucci, E.; Rodriguez, M.; Murgia, M.L.; Gioia, T.; Santo, D.; Nanni, L.; Attene, G.; Papa, R. Beans (Phaseolus ssp.) as a model for understanding crop evolution. Front. Plant Sci. 2017, 8, 722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mamidi, S.; Rossi, M.; Moghaddam, S.M.; Annam, D.; Lee, R.; Papa, R.; McClean, P.E. Demographic factors shaped diversity in the two gene pools of wild common bean Phaseolus vulgaris L. Heredity 2013, 110, 267–276. [Google Scholar] [CrossRef] [Green Version]
- Schmutz, J.; McClean, P.E.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C.; et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef] [Green Version]
- Vlasova, A.; Capella-Gutiérrez, S.; Rendón-Anaya, M.; Hernández-Oñate, M.; Minoche, A.E.; Erb, I.; Câmara, F.; Prieto-Barja, P.; Corvelo, A.; Sanseverino, W.; et al. Genome and transcriptome analysis of the Mesoamerican common bean and the role of gene duplications in establishing tissue and temporal specialization of genes. Genome Biol. 2016, 17, 32. [Google Scholar] [CrossRef] [Green Version]
- de Sousa Cardoso, T.C.; Portilho, L.G.; de Oliveira, C.L.; Mckeown, P.C.; Maluf, W.R.; Gomes, L.A.A.; Teixeira, T.A.; do Amaral, L.R.; Spillane, C.; de Souza Gomes, M. Genome-wide identification and in silico characterisation of microRNAs, their targets and processing pathway genes in Phaseolus vulgaris L. Plant Biol. 2016, 18, 206–219. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustos-Sanmamed, P.; Bazin, J.; Hartmann, C.; Crespi, M.; Lelandais-Brière, C. Small RNA pathways and diversity in model legumes: Lessons from genomics. Front. Plant Sci. 2013, 10, 236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matzke, M.A.; Kanno, T.; Matzke, A.J.M. RNA-Directed DNA Methylation: The Evolution of a Complex Epigenetic Pathway in Flowering Plants. Annu. Rev. Plant Biol. 2015, 66, 243–267. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, K.; Parkhill, J.; Crook, J.; Horsnell, T.; Rice, P.; Rajandream, M.A.; Barrell, B. Artemis: Sequence visualization and annotation. Bioinformatics 2000, 16, 944–945. [Google Scholar] [CrossRef] [Green Version]
- Borodovsky, M.; Lomsadze, A. Eukaryotic gene prediction using GeneMark.hmm-E and GeneMark-ES. Curr Protoc Bioinform. 2011, 35, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Florea, L.; Hartzell, G.; Zhang, Z.; Rubin, G.M.; Miller, W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998, 8, 967–974. [Google Scholar] [CrossRef] [Green Version]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [Green Version]
- Gouy, M.; Guindon, S.; Gascuel, O. Sea view version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Tamura, K.; Nei, M. MEGA: Molecular evolutionary genetics analysis software for microcomputers. Bioinformatics 1994, 10, 189–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice, P.; Longden, L.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 267–277. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- R Development Core Team. R: A Language and Environment for Statistical Computing; Version 2.2; R Foundation for Statistical Computing: Vienna, Austria, 2005; ISBN 3-900051-07-0. Available online: http://www.R-project.org (accessed on 1 December 2020).
- Le Cao, K.-A.; Rohart, F.; Gonzalez, I.; Dejean, S.; Abadi, A.J.; Gautier, B.; Bartolo, F.; Monget, P.; Coquery, J.; Yao, F.Z.; et al. MixOmics: Omics Data Integration Project. R Packag. Version 6.1.1. 2016. Available online: https://cran.r-project.org/package=mixOmics (accessed on 1 December 2020).
- Richard, M.M.S.; Gratias, A.; Alvarez Diaz, J.C.; Thareau, V.; Pflieger, S.; Meziadi, C.; Blanchet, S.; William, M.; Bitocchi, E.; Papa, R.; et al. A common bean truncated CRINKLY4 kinase controls gene-for-gene resistance to the fungus Colletotrichum lindemuthianum. J. Exp. Bot. 2021, 72, 3569–3581. [Google Scholar] [CrossRef]
- Borges, A.; Tsai, S.M.; Caldas, D.G.G. Validation of reference genes for RT-qPCR normalization in common bean during biotic and abiotic stresses. Plant Cell Rep. 2012, 31, 827–838. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Garg, V.; Agarwal, G.; Pazhamala, L.T.; Nayak, S.N.; Kudapa, H.; Khan, A.W.; Doddamani, D.; Sharma, M.; Kavi Kishor, P.B.; Varshney, R.K. Genome-wide identification, characterization, and expression analysis of small RNA biogenesis purveyors reveal their role in regulation of biotic stress responses in three legume crops. Front. Plant Sci. 2017, 8, 488. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, X.; Muhammad, I.; Ge, B.; Hong, B. Multiplex reverse transcription loop-mediated isothermal amplification for the simultaneous detection of CVB and CSVd in chrysanthemum. J. Virol. Methods 2014, 210, 26–31. [Google Scholar] [CrossRef]
- Vazquez, F.; Gasciolli, V.; Crété, P.; Vaucheret, H. The Nuclear dsRNA Binding Protein HYL1 Is Required for MicroRNA Accumulation and Plant Development, but Not Posttranscriptional Transgene Silencing. Curr. Biol. 2004, 14, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Richard, M.M.S.; Chen, N.W.G.; Thareau, V.; Pflieger, S.; Blanchet, S.; Pedrosa-Harand, A.; Iwata, A.; Chavarro, C.; Jackson, S.A.; Geffroy, V. The subtelomeric khipu satellite repeat from Phaseolus vulgaris: Lessons learned from the genome analysis of the andean genotype G19833. Front. Plant Sci. 2013, 4, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curtin, S.J.; Kantar, M.B.; Yoon, H.W.; Whaley, A.M.; Schlueter, J.A.; Stupar, R.M. Co-expression of soybean Dicer-like genes in response to stress and development. Funct. Integr. Genom. 2012, 12, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavin, M.; Herendeen, P.S.; Wojciechowski, M.F. Evolutionary rates analysis of leguminosae implicates a rapid diversification of lineages during the tertiary. Syst. Biol. 2005, 54, 575–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefanović, S.; Pfeil, B.E.; Palmer, J.D.; Doyle, J.J. Relationships among phaseoloid legumes based on sequences from eight chloroplast regions. Syst. Bot. 2009, 34, 115–128. [Google Scholar] [CrossRef]
- Jia, J.; Ji, R.; Li, Z.; Yu, Y.; Nakano, M.; Long, Y.; Feng, L.; Qin, C.; Lu, D.; Zhan, J.; et al. Soybean DICER-LIKE2 regulates seed coat color via production of primary 22-nucleotide small interfering RNAs from long inverted repeats. Plant Cell 2020, 32, 3662–3673. [Google Scholar] [CrossRef]
- Rodríguez-Leal, D.; Castillo-Cobián, A.; Rodríguez-Arévalo, I.; Vielle-Calzada, J.P. A primary sequence analysis of the ARGONAUTE protein family in plants. Front. Plant Sci. 2016, 7, 1347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Havecker, E.R.; Wallbridge, L.M.; Hardcastle, T.J.; Bush, M.S.; Kelly, K.A.; Dunn, R.M.; Schwach, F.; Doonan, J.H.; Baulcombe, D.C. The arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 2010, 22, 321–334. [Google Scholar] [CrossRef] [Green Version]
- Pumplin, N.; Voinnet, O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013, 11, 745–760. [Google Scholar] [CrossRef] [PubMed]
- Bohmert, K.; Camus, I.; Bellini, C.; Bouchez, D.; Caboche, M.; Banning, C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998, 17, 170–180. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, Y.L.; Zhao, J.H.; Wang, S.; Jin, Y.; Chen, Z.Q.; Fang, Y.Y.; Hua, C.L.; Ding, S.W.; Guo, H.S. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2016, 2, 16153. [Google Scholar] [CrossRef]
- Shao, F.; Lu, S. Genome-wide identification, molecular cloning, expression profiling and posttranscriptional regulation analysis of the Argonaute gene family in Salvia miltiorrhiza, an emerging model medicinal plant. BMC Genom. 2013, 14, 512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Wang, A. RNA-Targeted Antiviral Immunity: More Than Just RNA Silencing. Trends Microbiol. 2019, 27, 792–805. [Google Scholar] [CrossRef]
- Muhammad, T.; Zhang, F.; Zhang, Y.; Liang, Y. RNA Interference: A Natural Immune System of Plants to Counteract Biotic Stressors. Cells 2019, 8, 792–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deleris, A.; Halter, T.; Navarro, L. DNA Methylation and Demethylation in Plant Immunity. Annu. Rev. Phytopathol. 2016, 54, 579–603. [Google Scholar] [CrossRef]
- Voinnet, O. Post-transcriptional RNA silencing in plant-microbe interactions: A touch of robustness and versatility. Curr. Opin. Plant Biol. 2008, 11, 464–470. [Google Scholar] [CrossRef]
- Ellendorff, U.; Fradin, E.F.; De Jonge, R.; Thomma, B.P.H.J. RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J. Exp. Bot. 2009, 60, 591–602. [Google Scholar] [CrossRef] [Green Version]
- Shen, D.; Suhrkamp, I.; Wang, Y.; Liu, S.; Menkhaus, J.; Verreet, J.A.; Fan, L.; Cai, D. Identification and characterization of microRNAs in oilseed rape (Brassica napus) responsive to infection with the pathogenic fungus Verticillium longisporum using Brassica AA (Brassica rapa) and CC (Brassica oleracea) as reference genomes. New Phytol. 2014, 204, 577–594. [Google Scholar] [CrossRef]
- Li, Y.; Lu, Y.G.; Shi, Y.; Wu, L.; Xu, Y.J.; Huang, F.; Guo, X.Y.; Zhang, Y.; Fan, J.; Zhao, J.Q.; et al. Multiple rice MicroRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol. 2014, 164, 1077–1092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, W.; Wu, F. Characterization of miRNAs associated with Botrytis cinerea infection of tomato leaves. BMC Plant Biol. 2015, 15, 1. [Google Scholar] [CrossRef] [PubMed]
- Mosher, R.A.; Baulcombe, D.C. Bacterial pathogens encode suppressors of RNA-mediated silencing. Genome Biol. 2008, 9, 237. [Google Scholar] [CrossRef]
- Qiao, Y.; Liu, L.; Xiong, Q.; Flores, C.; Wong, J.; Shi, J.; Wang, X.; Liu, X.; Xiang, Q.; Jiang, S.; et al. Oomycete pathogens encode RNA silencing suppressors. Nat. Genet. 2013, 45, 330–333. [Google Scholar] [CrossRef] [Green Version]
- Yin, C.; Ramachandran, S.R.; Zhai, Y.; Bu, C.; Pappu, H.R.; Hulbert, S.H. A novel fungal effector from Puccinia graminis suppressing RNA silencing and plant defense responses. New Phytol. 2019, 222, 1561–1572. [Google Scholar] [CrossRef] [PubMed]
- Licheng, W.; Wenbao, C.; Huan, M.; Jingyuan, L.; Xingan, H.; Yunfeng, W. Identification of RNA silencing suppressor encoded by wheat blue dwarf (WBD) phytoplasma. Plant Biol. J. 2021, 5, 843–849. [Google Scholar] [CrossRef]
- Hua, C.; Zhao, J.H.; Guo, H.S. Trans-Kingdom RNA Silencing in Plant–Fungal Pathogen Interactions. Mol. Plant 2018, 11, 235–244. [Google Scholar] [CrossRef] [Green Version]
- Agorio, A.; Vera, P. ARGONAUTE4 Is Required for Resistance to Pseudomonas syringae in Arabidopsis. Plant Cell 2007, 19, 3778–3790. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhao, H.; Gao, S.; Wang, W.-C.; Katiyar-Agarwal, S.; Huang, H.-D.; Raikhel, N.; Jin, H. Arabidopsis Argonaute 2 Regulates Innate Immunity. Mol. Cell 2011, 42, 356–366. [Google Scholar] [CrossRef] [Green Version]
- Martins, T.F.; Souza, P.F.N.; Alves, M.S.; Silva, F.D.A.; Arantes, M.R.; Vasconcelos, I.M.; Oliveira, J.T.A. Identification, characterization, and expression analysis of cowpea (Vigna unguiculata [L.] Walp.) miRNAs in response to cowpea severe mosaic virus (CPSMV) challenge. Plant Cell Rep. 2020, 39, 1061–1078. [Google Scholar] [CrossRef]
- Pradhan, M.; Pandey, P.; Baldwin, I.T.; Pandey, S.P. Argonaute4 modulates resistance to fusarium brachygibbosum infection by regulating jasmonic acid signaling. Plant Physiol. 2020, 184, 1128–1152. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, G.; Tang, F.; Shao, J.; Lu, Y.; Bao, Y.; Yao, H.; Lu, C. Pre-absorbed immunoproteomics: A novel method for the detection of Streptococcus suis surface proteins. PLoS ONE 2011, 6, e21234. [Google Scholar] [CrossRef] [PubMed]
- Moura, M.O.; Fausto, A.K.S.; Fanelli, A.; Guedes, F.A.D.F.; Silva, T.D.F.; Romanel, E.; Vaslin, M.F.S. Genome-wide identification of the Dicer-like family in cotton and analysis of the DCL expression modulation in response to biotic stress in two contrasting commercial cultivars. BMC Plant Biol. 2019, 19, 503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pflieger, S.; Richard, M.M.S.; Blanchet, S.; Meziadi, C.; Geffroy, V. VIGS technology: An attractive tool for functional genomics studies in legumes. Funct. Plant Biol. 2013, 40, 1234–1248. [Google Scholar] [CrossRef] [PubMed]
- Pflieger, S.; Blanchet, S.; Meziadi, C.; Richard, M.M.S.; Thareau, V.; Mary, F.; Mazoyer, C.; Geffroy, V. The “one-step” Bean pod mottle virus (BPMV)-derived vector is a functional genomics tool for efficient overexpression of heterologous protein, virus-induced gene silencing and genetic mapping of BPMV R-gene in common bean (Phaseolus vulgaris L.). BMC Plant Biol. 2014, 14, 232. [Google Scholar] [CrossRef] [PubMed]
- Richard, M.M.S.; Gratias, A.; Thareau, V.; Kim, K.D.; Balzergue, S.; Joets, J.; Jackson, S.A.; Geffroy, V. Genomic and epigenomic immunity in common bean: The unusual features of NB-LRR gene family. DNA Res. 2018, 25, 161–172. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).