Molecular Investigation of miRNA Biomarkers as Chemoresistance Regulators in Melanoma: A Protocol for Systematic Review and Meta-Analysis
Abstract
1. Introduction
1.1. Epidemiology
1.2. Rationale
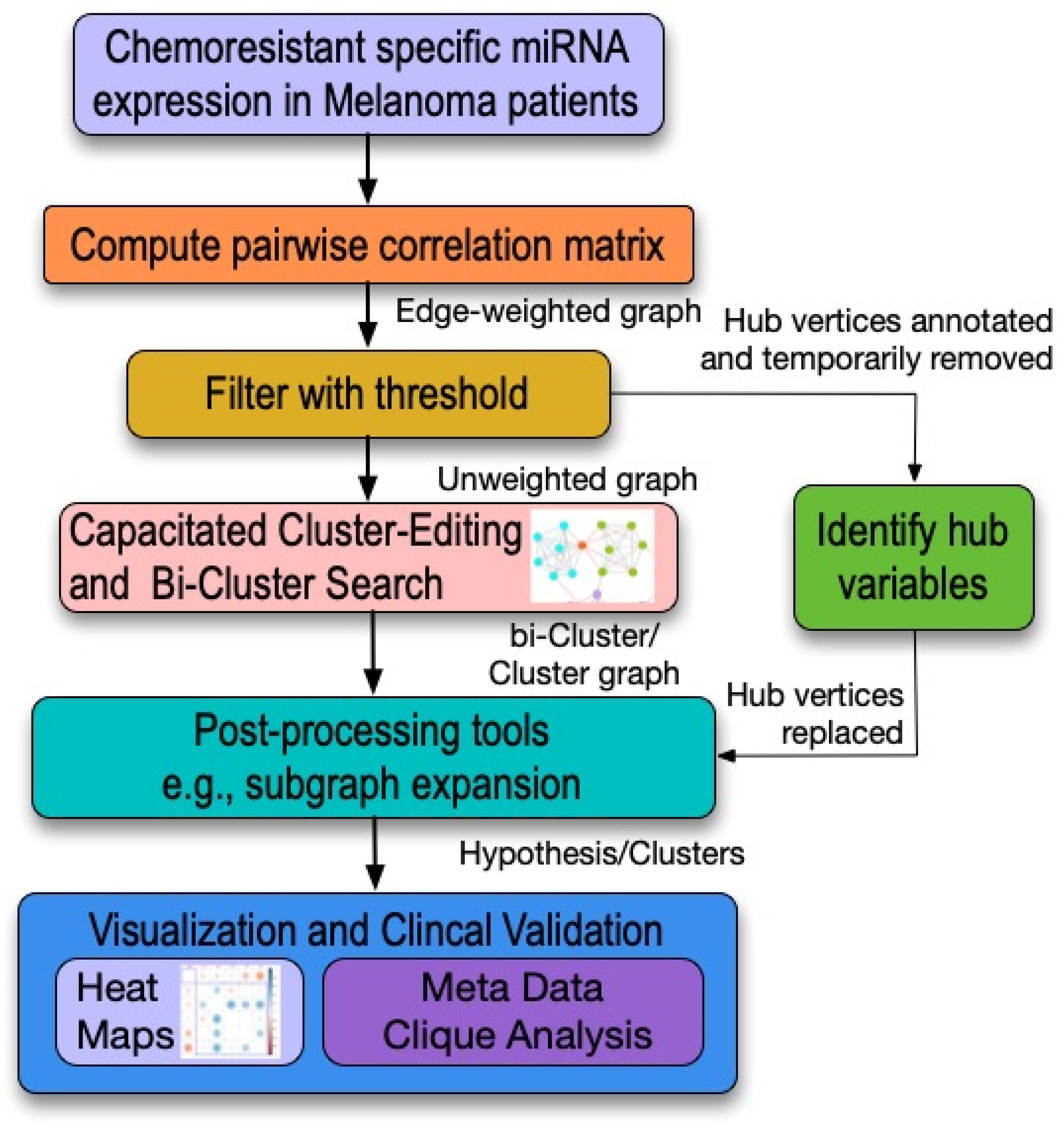
1.2.1. The Importance of This Study
1.2.2. What Will the Study’s Approach Be to This Problem?
1.2.3. How Will It Help?
2. Methods
2.1. Study Design
2.1.1. Eligibility Criteria
Inclusion Criteria
- Studies that deal with resistance in melanoma.
- Studies published until December 2021.
- Reporting of miRNA profiling platforms.
- Studies with appropriate patient data with therapeutic measures.
- Studies reporting the genes and/or pathways involved in chemoresistance or chemosensitivity.
- miRNA expression analysis using in vitro assays.
- Studies reporting the patient’s survival with 95% CI (confidence interval) values in hazard ratio (HR) or Kaplan–Meier (KM) curves for quantitative synthesis or meta-analysis.
Exclusion Criteria
- Letters to the editor, fact sheets, conference proceedings, unpublished materials, review articles, case studies, and studies conducted solely in patients or in vitro.
- Studies examining patient data from bioinformatic datasets.
- Duplicate publications from the same study will be treated as one study.
- Studies using non-human data.
Search Strategy and Study Selection
Data Extraction and Management
2.1.2. Data Collection Process
- From the studies, five major categories of data will be extracted: The study characteristics, including the author, geographic region, year of publication, study period, sample size, study design, sampling procedures, validity of confirmative diagnosis, method of data collection, and number of melanoma cancer cases/patients, as well as the International Classification of Disease (ICD) Code for the anatomical site of cancer under study.
- Clinical, pathological, and biological attributes, including comorbidity, risk factors, tumor histology (squamous, adenocarcinoma, clear cell, and undifferentiated), pathological grades (1, 2, and 3), tumor size, negative and positive lymph node metastasis, positive and negative vascular involvement, the lymphocyte infiltration (if any), histology grade (well, moderate, poor, and undetermined), P16 (positive and negative), deep stromal invasion (%), and specific body sites, such as the face (the temporal, frontal, periorbital, infraorbital, buccal, zygomatic, mental, or perioral region), nose, lip, ear, scalp, trunk, neck, and extremities [39].
- miRNA expression in melanoma patients and their responses towards their treatment.
- Hazard ratio (HR) and 95% confidence interval (CI) estimates of overall survival (OS), disease-free survival (DFS), and other endpoint measures.
- In vitro and in vivo studies.
Outcomes and Prioritization
Quality Assessment of Included Studies
- i.
- Information about the patient’s tissue collection.
- ii.
- Location of the study.
- iii.
- Gender.
- iv.
- Age.
- v.
- Exposure to sunlight.
- vi.
- Ulceration status.
- vii.
- miRNA analysis in melanoma patients.
- viii.
- List of melanoma cell lines used.
- ix.
- Tumor stage.
- x.
- Lymph node status.
- xi.
- miRNA profiling platform.
- xii.
- The form of therapy used.
- xiii.
- Genes and/or pathways involved in resistance.
Assessment of Risk of Bias in Individual Studies
Publication Bias
2.1.3. Statistical Analysis
Meta-Analysis
Subgroup Analyses
Meta-Regression
Network-Centric Model Analysis
Random Forest Analysis
2.2. Presenting and Reporting the Review Results
2.3. Ethics and Dissemination
2.4. Strengths and Limitations of This Study
- PRISMA-P (Preferred Reporting Items for Systematic Reviews and Meta-Analyses Protocol) recommendations are followed in the protocol.
- It will help researchers make informed decisions, due to specific evidence obtained from organized data.
- This study will help us obtain a clear picture of the role of miRNAs on chemoresistance for melanoma patients.
- Certain forms of data obtained from various literature may be challenging to incorporate due to statistical error and, hence, may hamper the outcome.
3. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
Abbreviations
References
- Ferlay, J. GLOBOCAN 2000. Cancer Incidence, Mortality and Prevalence Worldwide; Version 1.0. IARC Cancerbase; IARC: Lyon, France, 2001. [Google Scholar]
- Franceschi, S.; Levi, F.; Randimbison, L.; La Vecchia, C. Site distribution of different types of skin cancer: New aetiological clues. Int. J. Cancer 1996, 67, 24–28. [Google Scholar] [CrossRef]
- Godbole, V.; Toprani, H.; Shah, H. Skin cancer in Saurashtra. Indian J. Pathol. Bacteriol. 1968, 11, 183. [Google Scholar]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Chang, D.T.; Amdur, R.J.; Morris, C.G.; Mendenhall, W.M. Adjuvant radiotherapy for cutaneous melanoma: Comparing hypofractionation to conventional fractionation. Int. J. Radiat. Oncol. Biol. Phys. 2006, 66, 1051–1055. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, B.K.; Kricker, A. The epidemiology of UV induced skin cancer. J. Photochem. Photobiol. B Biol. 2001, 63, 8–18. [Google Scholar] [CrossRef]
- Haluska, F.G.; Hodi, F.S. Molecular genetics of familial cutaneous melanoma. J. Clin. Oncol. 1998, 16, 670–682. [Google Scholar] [CrossRef] [PubMed]
- Koh, H.K.; Kligler, B.E.; Lew, R.A. Sunlight and cutaneous malignant melanoma: Evidence for and against causation. Photochem. Photobiol. 1990, 51, 765–779. [Google Scholar] [CrossRef]
- Kerbel, R.S. A cancer therapy resistant to resistance. Nature 1997, 390, 335. [Google Scholar] [CrossRef]
- Wagle, N.; Emery, C.; Berger, M.F.; Davis, M.J.; Sawyer, A.; Pochanard, P.; Kehoe, S.M.; Johannessen, C.M.; MacConaill, L.E.; Hahn, W.C.; et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J. Clin. Oncol. 2011, 29, 3085. [Google Scholar] [CrossRef]
- Sharma, K.; Mohanti, B.K.; Rath, G.K. Malignant melanoma: A retrospective series from a regional cancer center in India. J. Cancer Res. Ther. 2009, 5, 173. [Google Scholar] [CrossRef]
- Müller, D.; Bosserhoff, A. Integrin β 3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene 2008, 27, 6698. [Google Scholar] [CrossRef]
- Dar, A.A.; Majid, S.; de Semir, D.; Nosrati, M.; Bezrookove, V.; Kashani-Sabet, M. miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J. Biol. Chem. 2011, 286, 16606–16614. [Google Scholar] [CrossRef]
- Pencheva, N.; Tran, H.; Buss, C.; Huh, D.; Drobnjak, M.; Busam, K.; Tavazoie, S.F. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell 2012, 151, 1068–1082. [Google Scholar] [CrossRef] [PubMed]
- Madhav, M.R.; Nayagam, S.G.; Biyani, K.; Pandey, V.; Kamal, D.G.; Sabarimurugan, S.; Ramesh, N.; Gothandam, K.M.; Jayaraj, R. Epidemiologic analysis of breast cancer incidence, prevalence, and mortality in India: Protocol for a systematic review and meta-analyses. Medicine (Baltimore) 2018, 97, e13680. [Google Scholar] [CrossRef] [PubMed]
- Poddar, A.; Aranha, R.R.; Muthukaliannan, G.K.; Nachimuthu, R.; Jayaraj, R. Head and neck cancer risk factors in India: Protocol for systematic review and meta-analysis. BMJ Open 2018, 8, e020014. [Google Scholar] [CrossRef]
- Jayaraj, R.; Kumarasamy, C.; Piedrafita, D. Systematic review and meta-analysis protocol for Fasciola DNA vaccines. Online J. Vet. Res. 2018, 22, 517. [Google Scholar]
- Kim, W.T.; Kim, W.-J. MicroRNAs in prostate cancer. Prostate Int. 2013, 1, 3–9. [Google Scholar] [CrossRef]
- Lin Teoh, S.; Das, S. The role of MicroRNAs in diagnosis, prognosis, metastasis and resistant cases in breast cancer. Curr. Pharm. Des. 2017, 23, 1845–1859. [Google Scholar] [CrossRef]
- Zhang, W.; Zhou, J.; Zhu, X.; Yuan, H. MiR-126 reverses drug resistance to TRAIL through inhibiting the expression of c-FLIP in cervical cancer. Gene 2017, 627, 420–427. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, J. MicroRNAs are important regulators of drug resistance in colorectal cancer. Biol. Chem. 2017, 398, 929–938. [Google Scholar] [CrossRef]
- Yang, W.; Ma, J.; Zhou, W.; Cao, B.; Zhou, X.; Yang, Z.; Zhang, H.; Zhao, Q.; Fan, D.; Hong, L. Molecular mechanisms and theranostic potential of miRNAs in drug resistance of gastric cancer. Expert Opin. Ther. Targets 2017, 21, 1063–1075. [Google Scholar] [CrossRef]
- Haefliger, S.; Hudson, A.; Hayes, S.; Pavlakis, N.; Howell, V. P2. 01-012 Acquired Chemotherapy Resistance in vitro: miRNA Profiles of Chemotherapy Resistant Squamous Lung Cancer Cell Lines: Topic: Analysis of RNA. J. Thorac. Oncol. 2017, 12, S790–S791. [Google Scholar] [CrossRef][Green Version]
- Zhuang, Z.; Hu, F.; Hu, J.; Wang, C.; Hou, J.; Yu, Z.; Wang, T.T.; Liu, X.; Huang, H. MicroRNA-218 promotes cisplatin resistance in oral cancer via the PPP2R5A/Wnt signaling pathway. Oncol. Rep. 2017, 38, 2051–2061. [Google Scholar] [CrossRef]
- Tung, S.L.; Huang, W.C.; Hsu, F.C.; Yang, Z.P.; Jang, T.H.; Chang, J.W.; Chuang, C.M.; Lai, C.R.; Wang, L.H. miRNA-34c-5p inhibits amphiregulin-induced ovarian cancer stemness and drug resistance via downregulation of the AREG-EGFR-ERK pathway. Oncogenesis 2017, 6, e326. [Google Scholar] [CrossRef]
- Amponsah, P.S.; Fan, P.; Bauer, N.; Zhao, Z.; Gladkich, J.; Fellenberg, J.; Herr, I. microRNA-210 overexpression inhibits tumor growth and potentially reverses gemcitabine resistance in pancreatic cancer. Cancer Lett. 2017, 388, 107–117. [Google Scholar] [CrossRef]
- Armstrong, C.M.; Liu, C.; Lou, W.; Lombard, A.P.; Evans, C.P.; Gao, A.C. MicroRNA-181a promotes docetaxel resistance in prostate cancer cells. Prostate 2017, 77, 1020–1028. [Google Scholar] [CrossRef]
- Fattore, L.; Sacconi, A.; Mancini, R.; Ciliberto, G. MicroRNA-driven deregulation of cytokine expression helps development of drug resistance in metastatic melanoma. Cytokine Growth Factor Rev. 2017, 36, 39–48. [Google Scholar] [CrossRef]
- Joyce, K.M. Surgical management of melanoma. Exon Publ. 2017, 91–100. [Google Scholar]
- Grossman, D.; Altieri, D.C. Drug resistance in melanoma: Mechanisms, apoptosis, and new potential therapeutic targets. Cancer Metastasis Rev. 2001, 20, 3–11. [Google Scholar] [CrossRef]
- Royam, M.M.; Kumarasamy, C.; Baxi, S.; Gupta, A.; Ramesh, N.; Muthukaliannan, G.K.; Jayaraj, R. Current Evidence on miRNAs as Potential Theranostic Markers for Detecting Chemoresistance in Colorectal Cancer: A Systematic Review and Meta-Analysis of Preclinical and Clinical Studies. Mol. Diagn. Ther. 2019, 23, 65–82. [Google Scholar] [CrossRef]
- Wang, W.; Li, J.; Zhu, W.; Gao, C.; Jiang, R.; Li, W.; Hu, Q.; Zhang, B. MicroRNA-21 and the clinical outcomes of various carcinomas: A systematic review and meta-analysis. BMC Cancer 2014, 14, 819. [Google Scholar] [CrossRef]
- Nair, V.S.; Maeda, L.S.; Ioannidis, J.P. Clinical outcome prediction by microRNAs in human cancer: A systematic review. J. Natl. Cancer Inst. 2012, 104, 528–540. [Google Scholar] [CrossRef]
- Jayawardana, K.; Schramm, S.J.; Tembe, V.; Mueller, S.; Thompson, J.F.; Scolyer, R.A.; Mann, G.J.; Yang, J. Identification, review, and systematic cross-validation of microRNA prognostic signatures in metastatic melanoma. J. Investig. Dermatol. 2016, 136, 245–254. [Google Scholar] [CrossRef]
- Mocellin, S.; Verdi, D.; Nitti, D. DNA repair gene polymorphisms and risk of cutaneous melanoma: A systematic review and meta-analysis. Carcinogenesis 2009, 30, 1735–1743. [Google Scholar] [CrossRef]
- Kalal, B.S.; Upadhya, D.; Pai, V.R. Chemotherapy resistance mechanisms in advanced skin cancer. Oncol. Rev. 2017, 11, 326. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834. [Google Scholar] [CrossRef]
- Iorio, M.V.; Visone, R.; Di Leva, G.; Donati, V.; Petrocca, F.; Casalini, P.; Taccioli, C.; Volinia, S.; Liu, C.G.; Alder, H.; et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007, 67, 8699–8707. [Google Scholar] [CrossRef]
- Ceylan, C.; Oztürk, G.; Alper, S. Non-Melanoma Skin Cancers between the Years of 1990 and 1999 in Izmir, Turkey: Demographic and Clinicopathological Characteristics. J. Dermatol. 2003, 30, 123–131. [Google Scholar] [CrossRef]
- Stroup, D.F.; Berlin, J.A.; Morton, S.C.; Olkin, I.; Williamson, G.D.; Rennie, D.; Moher, D.; Becker, B.J.; Sipe, T.A.; Thacker, S.B. Meta-analysis of observational studies in epidemiology: A proposal for reporting. JAMA 2000, 283, 2008–2012. [Google Scholar] [CrossRef]
- Wells, G.; Shea, B.; O’connell, D.; Peterson, J.; Welch, V.; Losos, M.; Tugwell, P. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-Analyses; Ottawa Hospital Research Institute: Ottawa, ON, Canada, 2009. [Google Scholar]
- Kumarasamy, C.; Devi, A.; Jayaraj, R. Prognostic value of microRNAs in head and neck cancers: A systematic review and meta-analysis protocol. Syst Rev. 2018, 7, 150. [Google Scholar] [CrossRef]
- Jayaraj, R.; Kumarasamy, C. Systematic review and meta-analysis of cancer studies evaluating diagnostic test accuracy and prognostic values: Approaches to improve clinical interpretation of results. Cancer Manag. Res. 2018, 10, 4669–4670. [Google Scholar] [CrossRef]
- Jayaraj, R.; Kumarasamy, C.; Ramalingam, S.; Devi, A. Systematic review and meta-analysis of risk-reductive dental strategies for medication related osteonecrosis of the jaw among cancer patients: Approaches and strategies. Oral Oncol. 2018, 85, 15–23. [Google Scholar] [CrossRef]
- Sabarimurugan, S.; Royam, M.M.; Das, A.; Das, S.; Gothandam, K.M.; Jayaraj, R. Systematic Review and Meta-analysis of the Prognostic Significance of miRNAs in Melanoma Patients. Mol. Diagn Ther. 2018, 22, 653–669. [Google Scholar] [CrossRef] [PubMed]
- Jayaraj, R.; Kumarasamy, C.; Madhav, M.R.; Pandey, V.; Sabarimurugan, S.; Ramesh, N.; Gothandam, K.M.; Baxi, S. Systematic Review and Meta-Analysis of Diagnostic Accuracy of miRNAs in Patients with Pancreatic Cancer. Dis. Markers 2018, 2018, 6904569. [Google Scholar] [CrossRef]
- Jayaraj, R.; Kumarasamy, C. Prognostic biomarkers for oral tongue squamous cell carcinoma: A systematic review and meta-analysis. Br. J. Cancer 2018, 118, e11. [Google Scholar] [CrossRef]
- Jayaraj, R.; Kumarasamy, C. Survival for HPV-positive oropharyngeal squamous cell carcinoma with surgical versus non-surgical treatment approach: A systematic review and meta-analysis. J. Oral. Oncol. 2018, 90, 137–138. [Google Scholar] [CrossRef]
- Jayaraj, R.; Kumarasamy, C.; Sabarimurugan, S.; Baxi, S. Commentary: Blood-Derived microRNAs for Pancreatic Cancer Diagnosis: A Narrative Review and Meta-Analysis. Front. Physiol. 2018, 9, 1896. [Google Scholar] [CrossRef]
- Jayaraj, R.; Kumarasamy, C. Conceptual interpretation of analysing and reporting of results on systematic review and meta-analysis of optimal extent of lateral neck dissection for well-differentiated thyroid carcinoma with metastatic lateral neck lymph nodes. Oral Oncol. 2019, 89, 153–154. [Google Scholar] [CrossRef] [PubMed]
- Begg, C.B.; Mazumdar, M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994, 50, 1088–1101. [Google Scholar] [CrossRef]
- Duval, S.; Tweedie, R. Trim and fill: A simple funnel-plot–based method of testing and adjusting for publication bias in meta-analysis. Biometrics 2000, 56, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Orwin, R.G. A fail-safe N for effect size in meta-analysis. J. Educ. Stat. 1983, 8, 157–159. [Google Scholar] [CrossRef]
- Jayaraj, R.; Kumarasamy, C.; Gothandam, K.M. Letter to the editor "Prognostic value of microRNAs in colorectal cancer: A meta-analysis". Cancer Manag Res. 2018, 10, 3501–3503. [Google Scholar] [CrossRef]
- Jayaraj, R.; Kumarasamy, C. Letter to the Editor about the Article:" Performance of different imaging techniques in the diagnosis of head and neck cancer mandibular invasion: A systematic review and meta-analysis". J. Oncol. 2018, 89, 159–160. [Google Scholar] [CrossRef]
- Jayaraj, R.; Kumarasamy, C.; Sabarimurugan, S.; Baxi, S. Letter to the Editor in response to the article," The epidemiology of oral human papillomavirus infection in healthy populations: A systematic review and meta-analysis". Oral Oncol. 2018, 84, 121–122. [Google Scholar] [CrossRef]
- Jayaraj, R.; Kumarasamy, C.; Samiappan, S.; Swaminathan, P. Letter to the Editor regarding, The prognostic role of PD-L1 expression for survival in head and neck squamous cell carcinoma: A systematic review and meta-analysis. Oral Oncol. 2018, 77, 92–97. [Google Scholar] [CrossRef]
- Jayaraj, R.; Kumarasamy, C.; Madurantakam Royam, M.; Devi, A.; Baxi, S. Letter to the editor: Is HIF-1alpha a viable prognostic indicator in OSCC? A critical review of a meta-analysis study. World J. Surg. Oncol. 2018, 16, 111. [Google Scholar] [CrossRef]
- Higgins, J.P.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. BMJ Br. Med. J. 2003, 327, 557. [Google Scholar] [CrossRef]
- Cochran, W.G. The combination of estimates from different experiments. Biometrics 1954, 10, 101–129. [Google Scholar] [CrossRef]
- Borenstein, M.; Hedges, L.V.; Higgins, J.P.; Rothstein, H.R. Introduction to Meta-Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- Renehan, A.G.; Zwahlen, M.; Minder, C.; T O’Dwyer, S.; Shalet, S.M.; Egger, M.J.T.L. Insulin-like growth factor (IGF)-I, IGF binding protein-3, and cancer risk: Systematic review and meta-regression analysis. Lancet 2004, 363, 1346–1353. [Google Scholar] [CrossRef]
- Sterne, J.A.; Jüni, P.; Schulz, K.F.; Altman, D.G.; Bartlett, C.; Egger, M. Statistical methods for assessing the influence of study characteristics on treatment effects in ‘meta-epidemiological’research. Stat. Med. 2002, 21, 1513–1524. [Google Scholar] [CrossRef] [PubMed]
- Thompson, S.G.; Sharp, S.J. Explaining heterogeneity in meta-analysis: A comparison of methods. Stat. Med. 1999, 18, 2693–2708. [Google Scholar] [CrossRef]
- Ben-Dor, A.; Shamir, R.; Yakhini, Z. Clustering gene expression patterns. J. Comput. Biol. 1999, 6, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Jay, J.J.; Eblen, J.D.; Zhang, Y.; Benson, M.; Perkins, A.D.; Saxton, A.M.; Voy, B.H.; Chesler, E.J.; Langston, M.A. A Systematic Comparison of Genome-Scale Clustering Algorithms; Springer: Berlin/Heidelberg, Germany, 2012; Volume 13. [Google Scholar]
- Abu-Khzam, F.N. On the complexity of multi-parameterized cluster editing. J. Discret. Algorithms 2017, 45, 26–34. [Google Scholar] [CrossRef]
- Dehne, F.; Langston, M.A.; Luo, X.; Pitre, S.; Shaw, P.; Zhang, Y. The Cluster Editing Problem: Implementations and Experiments; Springer: Berlin/Heidelberg, Germany, 2006; pp. 13–24. [Google Scholar]
- Abu-Khzam, F.N.; Egan, J.; Gaspers, S.; Shaw, A.; Shaw, P. Cluster Editing with Vertex Splitting; Springer: Berlin/Heidelberg, Germany, 2018; pp. 1–13. [Google Scholar]
- Barr, J.; Shaw, P. AI Application to Data Analysis, Automatic File Processing; IEEE: Piscataway, NJ, USA, 2018; pp. 100–105. [Google Scholar]
- Landry, M.; Angela, B. Machine Learning with R and H2O; H2O.ai, Inc.: Mountain View, CA, USA, 2018. [Google Scholar]
- Shamseer, L.; Moher, D.; Clarke, M.; Ghersi, D.; Liberati, A.; Petticrew, M.; Shekelle, P.; Stewart, L.A. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: Elaboration and explanation. BMJ 2015, 349, g7647. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Prisma Group. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed]

| Search Number | Parameter |
|---|---|
| 1 | Melanoma “[Topic]” OR miRNA “[Topic]” |
| 2 | Melanoma “[Topic]” OR miRNA “[Topic]” OR patient “[Topic]” OR clinical study “[Topic]” |
| 3 | Melanoma “[Topic]” OR miRNA “[Topic]” OR microRNA “[Topic]” AND resistance “[Topic]” OR patient “[Topic]” OR clinical study “[Topic]” |
| 4 | Melanoma “[Topic]” OR miRNA “[Topic]” OR microRNA “[Topic]” AND chemoresistance (Chemoresist*) “[Topic]” OR patient “[Topic]” OR clinical study “[Topic]” |
| 5 | Melanoma “[Topic]” OR miRNA “[Topic]” OR microRNA “[Topic]” AND chemosensitivity (Chemosens*) “[Topic]” OR patient “[Topic]” OR clinical study “[Topic]” |
| 6 | 1 AND 2 AND 3 AND 4 AND 5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaw, P.; Raymond, G.; Tzou, K.S.; Baxi, S.; Mani, R.R.; Kumar Govind, S.; Chandramoorthy, H.C.; Sivanandy, P.; Rajagopal, M.; Samiappan, S.; et al. Molecular Investigation of miRNA Biomarkers as Chemoresistance Regulators in Melanoma: A Protocol for Systematic Review and Meta-Analysis. Genes 2022, 13, 115. https://doi.org/10.3390/genes13010115
Shaw P, Raymond G, Tzou KS, Baxi S, Mani RR, Kumar Govind S, Chandramoorthy HC, Sivanandy P, Rajagopal M, Samiappan S, et al. Molecular Investigation of miRNA Biomarkers as Chemoresistance Regulators in Melanoma: A Protocol for Systematic Review and Meta-Analysis. Genes. 2022; 13(1):115. https://doi.org/10.3390/genes13010115
Chicago/Turabian StyleShaw, Peter, Greg Raymond, Katherine S. Tzou, Siddhartha Baxi, Ravishankar Ram Mani, Suresh Kumar Govind, Harish C. Chandramoorthy, Palanisamy Sivanandy, Mogana Rajagopal, Suja Samiappan, and et al. 2022. "Molecular Investigation of miRNA Biomarkers as Chemoresistance Regulators in Melanoma: A Protocol for Systematic Review and Meta-Analysis" Genes 13, no. 1: 115. https://doi.org/10.3390/genes13010115
APA StyleShaw, P., Raymond, G., Tzou, K. S., Baxi, S., Mani, R. R., Kumar Govind, S., Chandramoorthy, H. C., Sivanandy, P., Rajagopal, M., Samiappan, S., Krishnan, S., & Jayaraj, R. (2022). Molecular Investigation of miRNA Biomarkers as Chemoresistance Regulators in Melanoma: A Protocol for Systematic Review and Meta-Analysis. Genes, 13(1), 115. https://doi.org/10.3390/genes13010115









