Metformin Triggers Apoptosis and Induction of the G0/G1 Switch 2 Gene in Macrophages
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Metformin Treatment
2.2. Plasmids and Constructs
2.3. DNA Transfection
2.4. Cell Proliferation Assay
2.5. Apoptotic DNA Ladder Detection
2.6. Flow Cytometry
2.7. Library Construction for RNA-Seq
2.8. Data Analysis for Gene Expression
2.9. Gene Set Enrichment Analysis (GSEA)
2.10. Reverse Transcription and Quantitative PCR
2.11. Statistical Analyses
3. Results
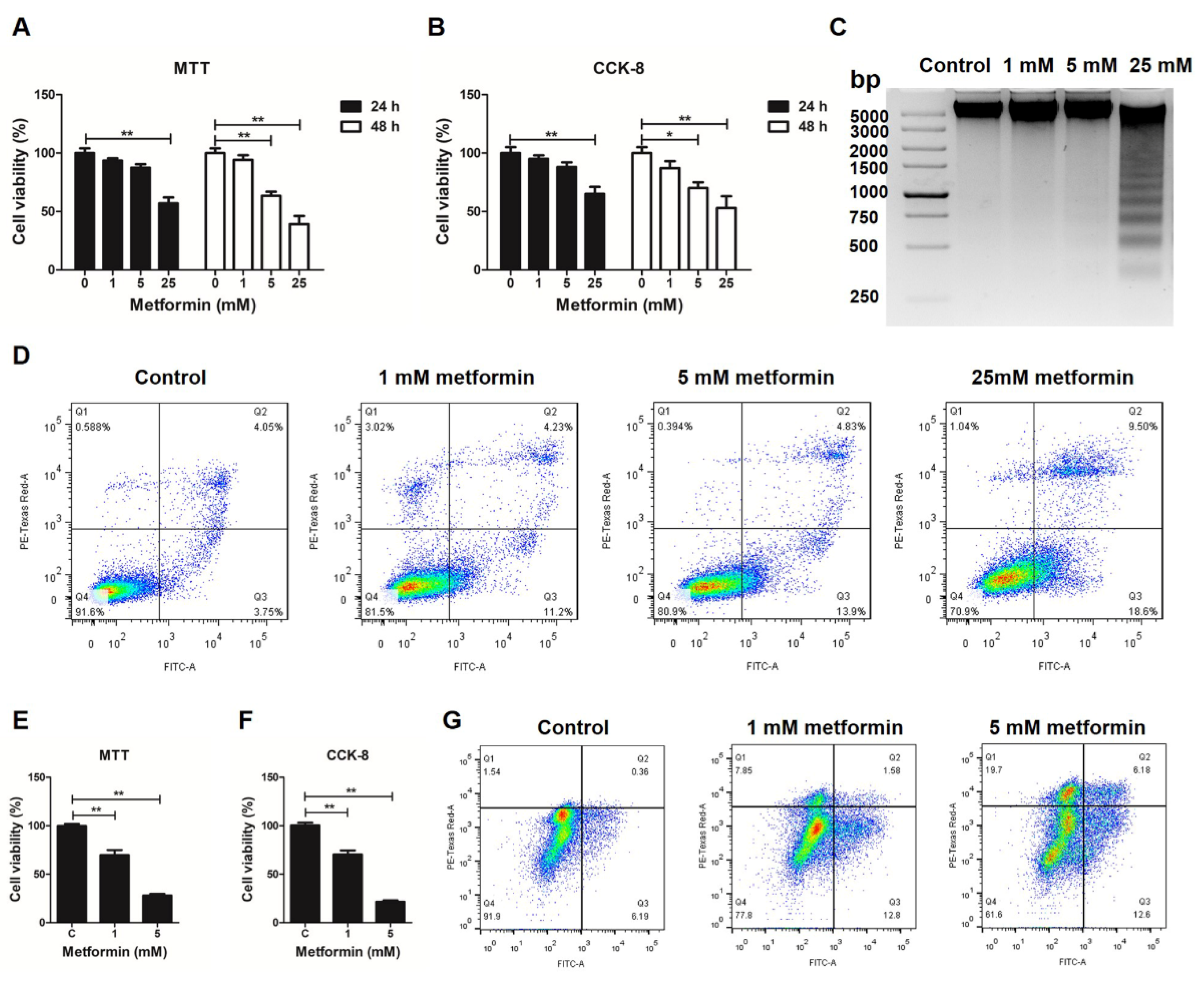
3.1. Metformin Induces Apoptosis in Macrophages
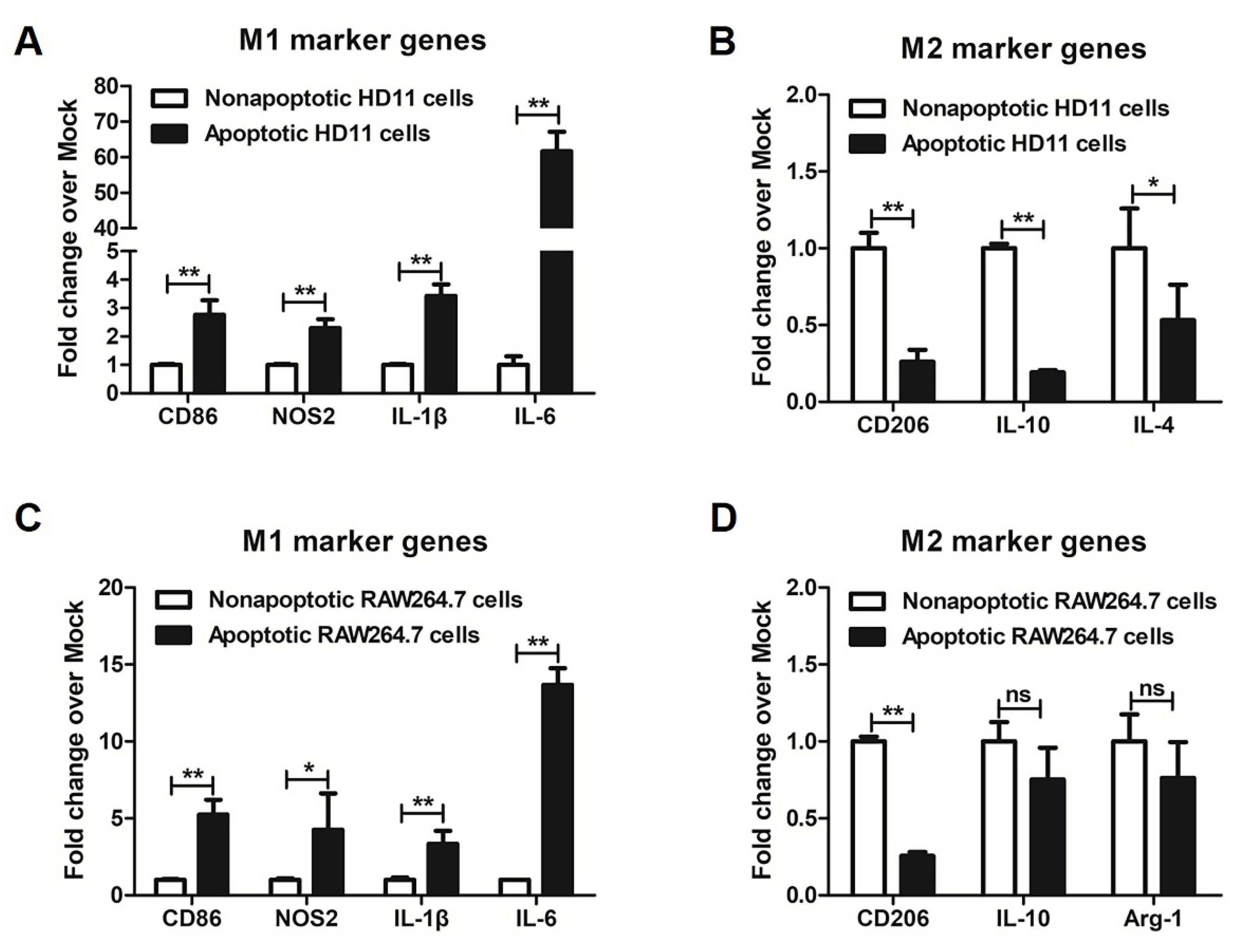
3.2. More M1 Marker Genes Were Detected in Metformin-Induced Apoptotic Macrophages
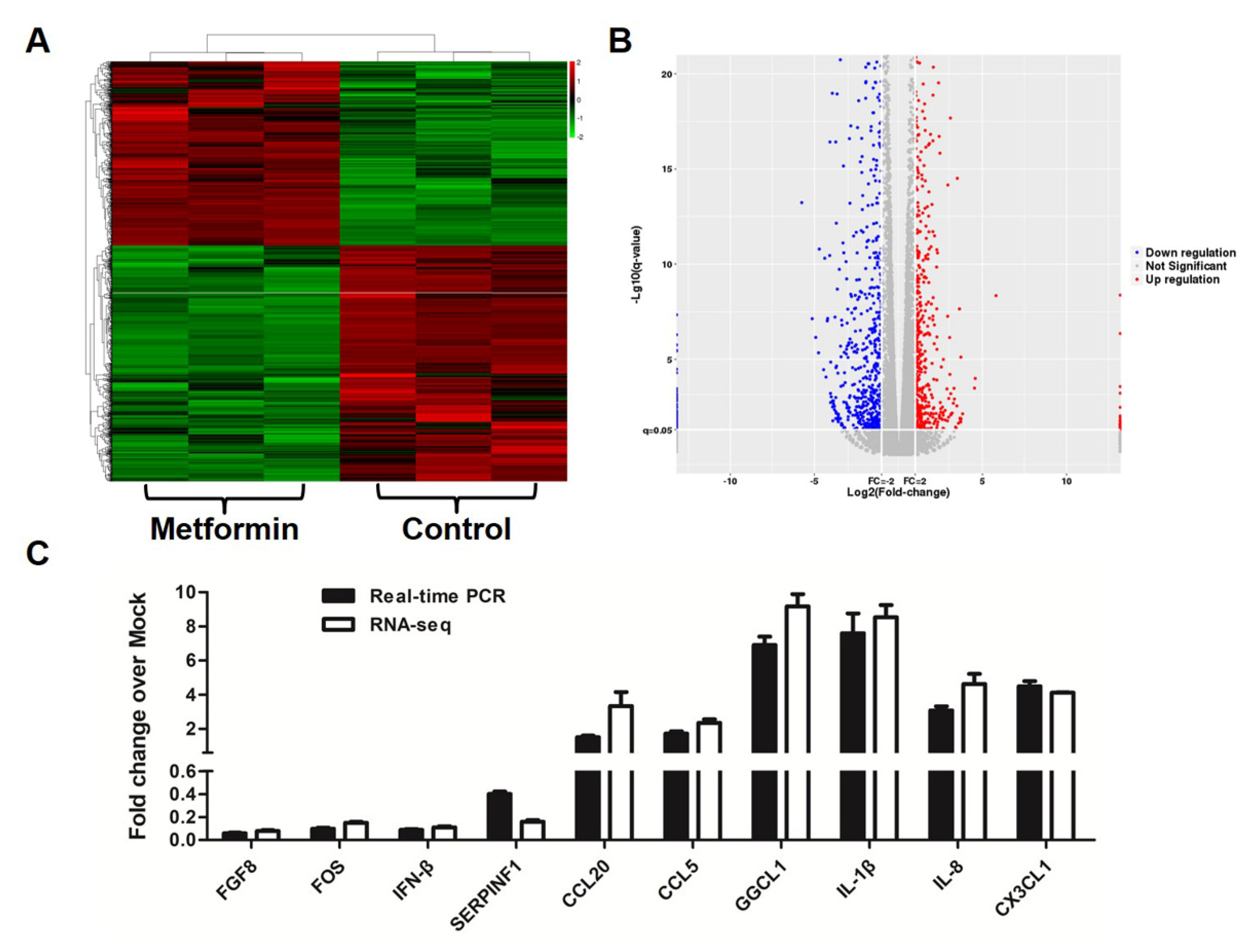
3.3. Metformin Affects Gene Expression Profiles in Macrophages
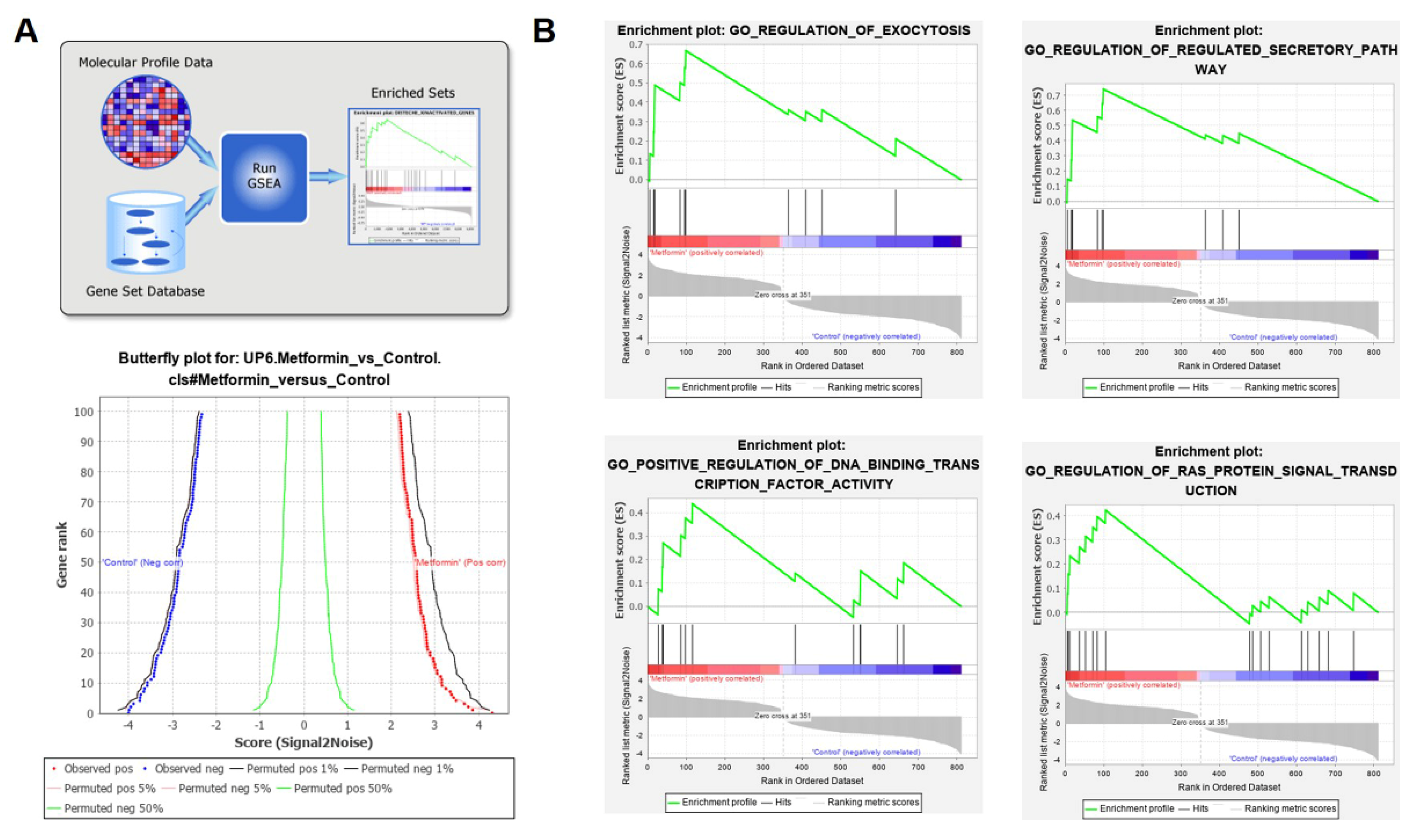
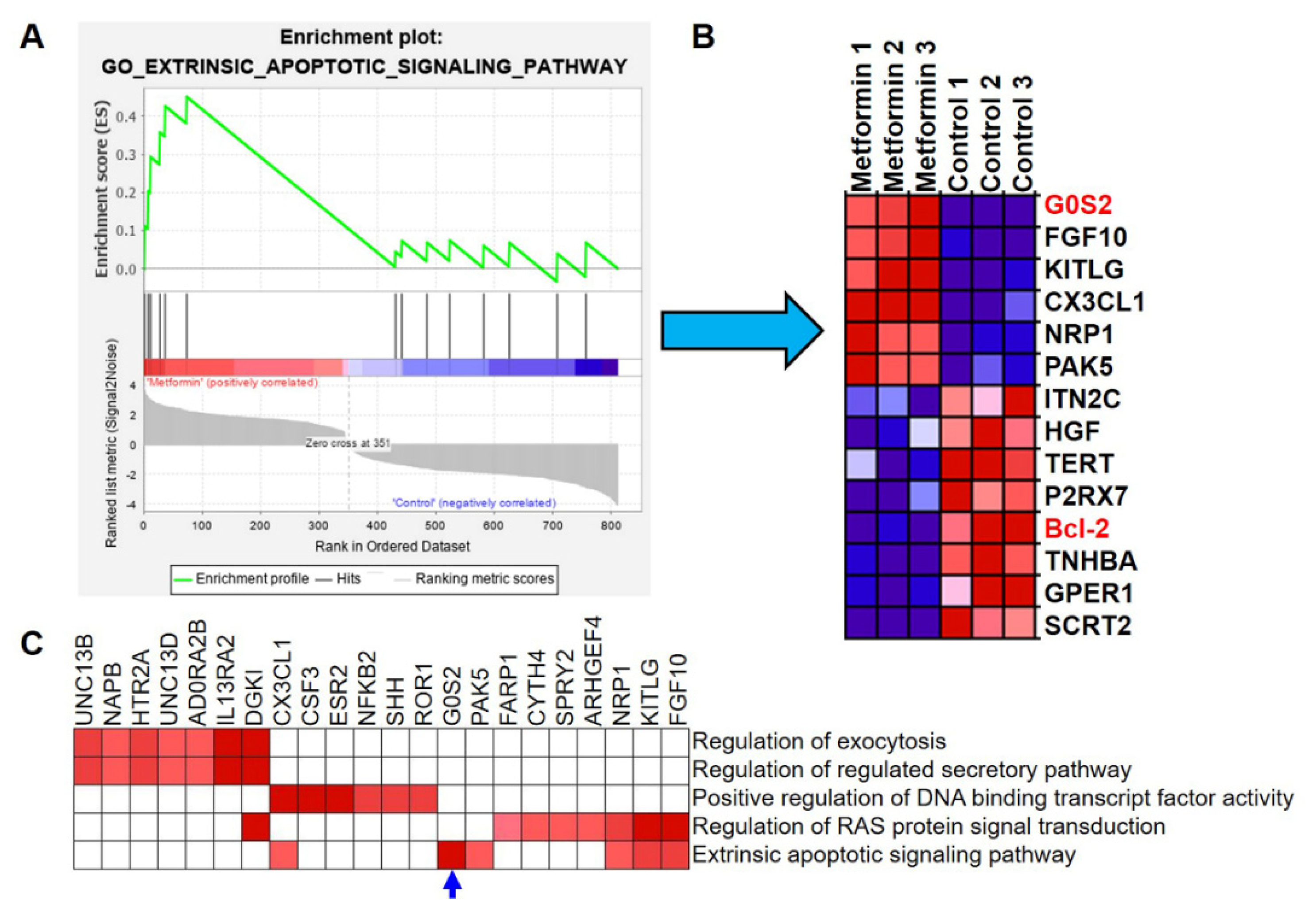
3.4. G0S2 Has the Most Significant and Positive Correlation with the Extrinsic Apoptotic Signaling Pathway in Metformin-Treated Macrophages
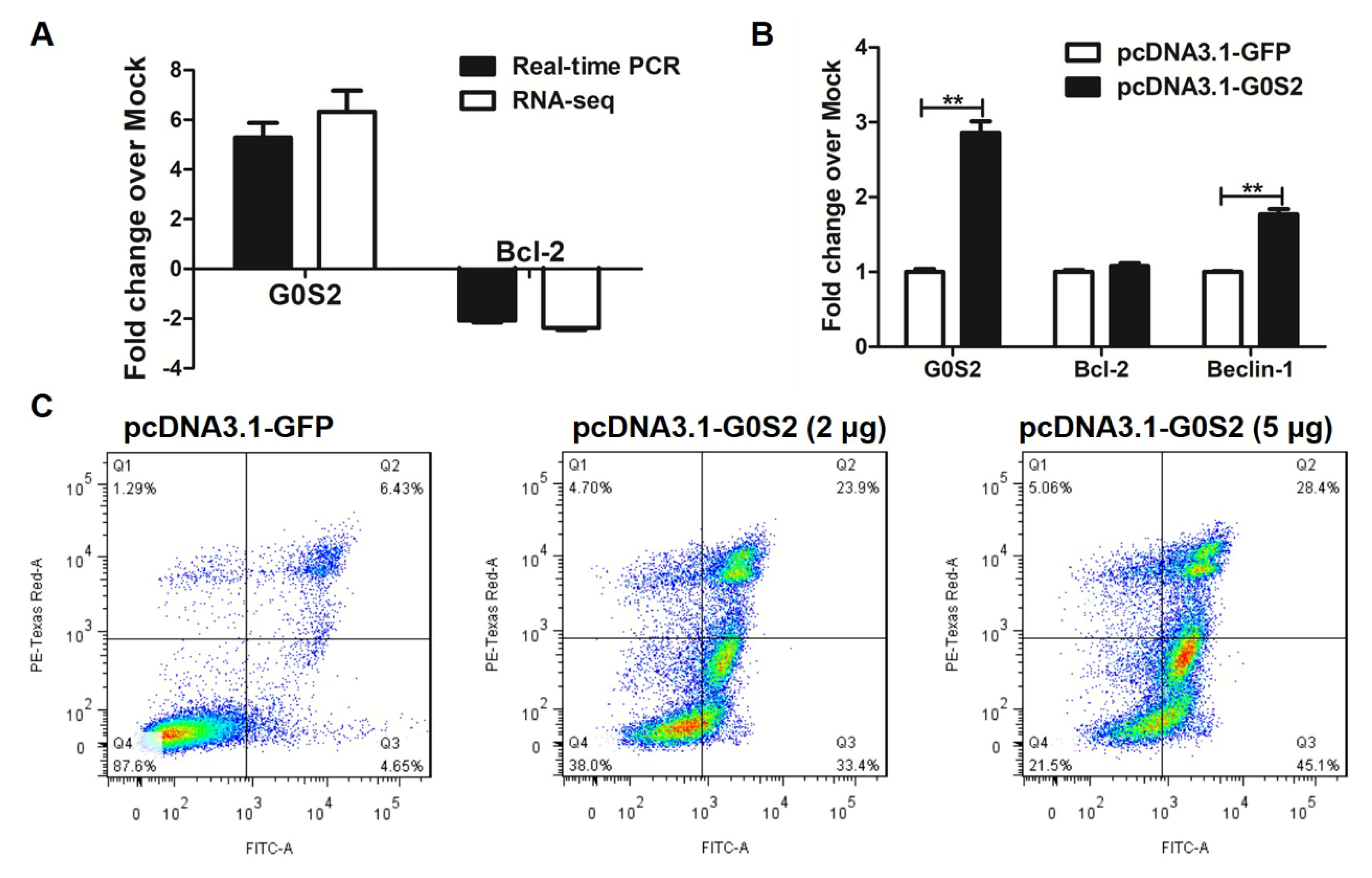
3.5. Metformin Promotes Apoptosis via G0S2 Induction
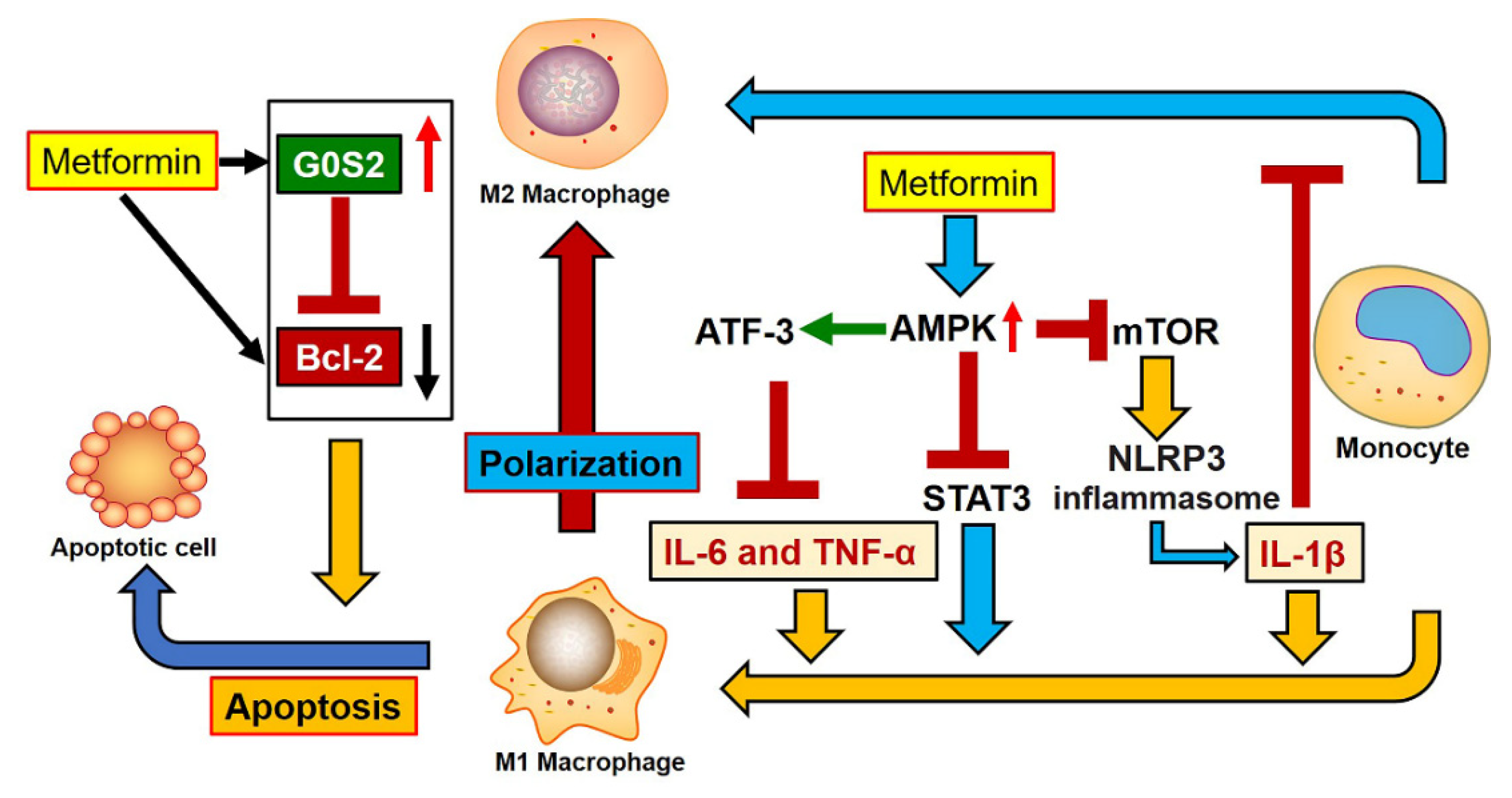
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Koelwyn, G.J.; Corr, E.M.; Erbay, E.; Moore, K.J. Regulation of macrophage immunometabolism in atherosclerosis. Nat. Immunol. 2018, 19, 526–537. [Google Scholar] [CrossRef] [Green Version]
- Wynn, T.A.; Chawla, A.; Pollard, J.W. Macrophage biology in development, homeostasis and disease. Nature 2013, 496, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef] [Green Version]
- Blandino, G.; Valerio, M.; Cioce, M.; Mori, F.; Casadei, L.; Pulito, C.; Sacconi, A.; Biagioni, F.; Cortese, G.; Galanti, S.; et al. Metformin elicits anticancer effects through the sequential modulation of DICER and c-MYC. Nat. Commun. 2012, 3, 865. [Google Scholar] [CrossRef] [PubMed]
- Vancura, A.; Bu, P.; Bhagwat, M.; Zeng, J.; Vancurova, I. Metformin as an Anticancer Agent. Trends Pharmacol. Sci. 2018, 39, 867–878. [Google Scholar] [CrossRef] [PubMed]
- Martin-Montalvo, A.; Mercken, E.M.; Mitchell, S.J.; Palacios, H.H.; Mote, P.L.; Scheibye-Knudsen, M.; Gomes, A.P.; Ward, T.M.; Minor, R.K.; Blouin, M.J.; et al. Metformin improves healthspan and lifespan in mice. Nat. Commun. 2013, 4, 2192. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Otin, C.; Galluzzi, L.; Freije, J.M.P.; Madeo, F.; Kroemer, G. Metabolic Control of Longevity. Cell 2016, 166, 802–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pryor, R.; Norvaisas, P.; Marinos, G.; Best, L.; Thingholm, L.B.; Quintaneiro, L.M.; De Haes, W.; Esser, D.; Waschina, S.; Lujan, C.; et al. Host-Microbe-Drug-Nutrient Screen Identifies Bacterial Effectors of Metformin Therapy. Cell 2019, 178, 1299–1312.e29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontaine, E. Metformin-Induced Mitochondrial Complex I Inhibition: Facts, Uncertainties, and Consequences. Front. Endocrinol. 2018, 9, 753. [Google Scholar] [CrossRef] [PubMed]
- Foretz, M.; Guigas, B.; Viollet, B. Understanding the glucoregulatory mechanisms of metformin in type 2 diabetes mellitus. Nat. Rev. Endocrinol. 2019, 15, 569–589. [Google Scholar] [CrossRef] [Green Version]
- Cameron, A.R.; Logie, L.; Patel, K.; Erhardt, S.; Bacon, S.; Middleton, P.; Harthill, J.; Forteath, C.; Coats, J.T.; Kerr, C.; et al. Metformin selectively targets redox control of complex I energy transduction. Redox Biol. 2018, 14, 187–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; An, H.; Liu, T.; Qin, C.; Sesaki, H.; Guo, S.; Radovick, S.; Hussain, M.; Maheshwari, A.; Wondisford, F.E.; et al. Metformin Improves Mitochondrial Respiratory Activity through Activation of AMPK. Cell Rep. 2019, 29, 1511–1523.e5. [Google Scholar] [CrossRef] [PubMed]
- Martin-Rodriguez, S.; de Pablos-Velasco, P.; Calbet, J.A.L. Mitochondrial Complex I Inhibition by Metformin: Drug-Exercise Interactions. Trends Endocrinol. Metab. 2020, 31, 269–271. [Google Scholar] [CrossRef]
- Chiang, C.F.; Chao, T.T.; Su, Y.F.; Hsu, C.C.; Chien, C.Y.; Chiu, K.C.; Shiah, S.G.; Lee, C.H.; Liu, S.Y.; Shieh, Y.S. Metformin-treated cancer cells modulate macrophage polarization through AMPK-NF-kappaB signaling. Oncotarget 2017, 8, 20706–20718. [Google Scholar] [CrossRef] [Green Version]
- Qing, L.; Fu, J.; Wu, P.; Zhou, Z.; Yu, F.; Tang, J. Metformin induces the M2 macrophage polarization to accelerate the wound healing via regulating AMPK/mTOR/NLRP3 inflammasome singling pathway. Am. J. Transl. Res. 2019, 11, 655–668. [Google Scholar]
- Jing, Y.; Wu, F.; Li, D.; Yang, L.; Li, Q.; Li, R. Metformin improves obesity-associated inflammation by altering macrophages polarization. Mol. Cell. Endocrinol. 2018, 461, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Vasamsetti, S.B.; Karnewar, S.; Kanugula, A.K.; Thatipalli, A.R.; Kumar, J.M.; Kotamraju, S. Metformin inhibits monocyte-to-macrophage differentiation via AMPK-mediated inhibition of STAT3 activation: Potential role in atherosclerosis. Diabetes 2015, 64, 2028–2041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hattori, Y.; Hattori, K.; Hayashi, T. Pleiotropic benefits of metformin: Macrophage targeting its anti-inflammatory mechanisms. Diabetes 2015, 64, 1907–1909. [Google Scholar] [CrossRef] [Green Version]
- Jin, S.; Meng, C.; He, Y.; Wang, X.; Zhang, Q.; Wang, Z.; Huang, W.; Wang, H. Curcumin prevents osteocyte apoptosis by inhibiting M1-type macrophage polarization in mice model of glucocorticoid-associated osteonecrosis of the femoral head. J. Orthop. Res. 2020, 38, 2020–2030. [Google Scholar] [CrossRef]
- Jiang, Q.; Pan, D.; Yang, Y.; Hu, Y.; Fang, L.; Shang, P.; Xia, Y.; Li, D. Luteolin Regulates Macrophage Polarization via the PI3K/Akt Pathway to Inhibit the Apoptosis Stimulated by Angiotensin II. Curr. Pharm. Biotechnol. 2018, 19, 428–437. [Google Scholar] [CrossRef]
- Cheon, S.Y.; Kim, E.J.; Kim, J.M.; Kam, E.H.; Ko, B.W.; Koo, B.N. Regulation of Microglia and Macrophage Polarization via Apoptosis Signal-Regulating Kinase 1 Silencing after Ischemic/Hypoxic Injury. Front. Mol. Neurosci. 2017, 10, 261. [Google Scholar] [CrossRef] [Green Version]
- Gautier, E.L.; Ivanov, S.; Lesnik, P.; Randolph, G.J. Local apoptosis mediates clearance of macrophages from resolving inflammation in mice. Blood 2013, 122, 2714–2722. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstrale, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lu, L.; Zhong, X.; Yue, Y.; Hong, Y.; Li, Y.; Li, Y. Metformin reduced NLRP3 inflammasome activity in Ox-LDL stimulated macrophages through adenosine monophosphate activated protein kinase and protein phosphatase 2A. Eur. J. Pharmacol. 2019, 852, 99–106. [Google Scholar] [CrossRef]
- Kim, J.; Kwak, H.J.; Cha, J.Y.; Jeong, Y.S.; Rhee, S.D.; Kim, K.R.; Cheon, H.G. Metformin suppresses lipopolysaccharide (LPS)-induced inflammatory response in murine macrophages via activating transcription factor-3 (ATF-3) induction. J. Biol. Chem. 2014, 289, 23246–23255. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Wei, J.; Wu, Y.; Wang, Z.; Guo, Y.; Lee, P.; Li, X. Metformin induces ER stress-dependent apoptosis through miR-708-5p/NNAT pathway in prostate cancer. Oncogenesis 2015, 4, e158. [Google Scholar] [CrossRef] [Green Version]
- Loubiere, C.; Clavel, S.; Gilleron, J.; Harisseh, R.; Fauconnier, J.; Ben-Sahra, I.; Kaminski, L.; Laurent, K.; Herkenne, S.; Lacas-Gervais, S.; et al. The energy disruptor metformin targets mitochondrial integrity via modification of calcium flux in cancer cells. Sci. Rep. 2017, 7, 5040. [Google Scholar] [CrossRef] [Green Version]
- Solanki, S.; Dube, P.R.; Tano, J.Y.; Birnbaumer, L.; Vazquez, G. Reduced endoplasmic reticulum stress-induced apoptosis and impaired unfolded protein response in TRPC3-deficient M1 macrophages. Am. J. Physiol. Cell Physiol. 2014, 307, C521–C531. [Google Scholar] [CrossRef] [Green Version]
- Solanki, S.; Dube, P.R.; Birnbaumer, L.; Vazquez, G. Reduced Necrosis and Content of Apoptotic M1 Macrophages in Advanced Atherosclerotic Plaques of Mice With Macrophage-Specific Loss of Trpc3. Sci. Rep. 2017, 7, 42526. [Google Scholar] [CrossRef] [PubMed]
- Hristodorov, D.; Mladenov, R.; Fischer, R.; Barth, S.; Thepen, T. Fully human MAP-fusion protein selectively targets and eliminates proliferating CD64(+) M1 macrophages. Immunol. Cell Biol. 2016, 94, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Letai, A.; Sarosiek, K. Regulation of apoptosis in health and disease: The balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol. 2019, 20, 175–193. [Google Scholar] [CrossRef] [PubMed]
- Ashkenazi, A.; Fairbrother, W.J.; Leverson, J.D.; Souers, A.J. From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors. Nat. Rev. Drug Discov. 2017, 16, 273–284. [Google Scholar] [CrossRef]
- Cory, S.; Adams, J.M. The Bcl2 family: Regulators of the cellular life-or-death switch. Nat. Rev. Cancer 2002, 2, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, D.; Bartok, E.; Gaidt, M.M.; Bock, F.J.; Herrmann, J.; Seeger, J.M.; Broz, P.; Beckmann, R.; Kashkar, H.; Tait, S.W.G.; et al. BAX/BAK-Induced Apoptosis Results in Caspase-8-Dependent IL-1beta Maturation in Macrophages. Cell Rep. 2018, 25, 2354–2368.e5. [Google Scholar] [CrossRef] [Green Version]
- Welch, C.; Santra, M.K.; El-Assaad, W.; Zhu, X.; Huber, W.E.; Keys, R.A.; Teodoro, J.G.; Green, M.R. Identification of a protein, G0S2, that lacks Bcl-2 homology domains and interacts with and antagonizes Bcl-2. Cancer Res. 2009, 69, 6782–6789. [Google Scholar] [CrossRef] [Green Version]
- Rothlin, C.V.; Hille, T.D.; Ghosh, S. Determining the effector response to cell death. Nat. Rev. Immunol. 2021, 21, 292–304. [Google Scholar] [CrossRef] [PubMed]
| Primer Name | Nucleotide Sequence 5′-3′ |
|---|---|
| Chicken CD86 fwd | GGATGTCTTACAGGATGCT |
| Chicken CD86 rev | CTGCTCTCCAAGGTGAAG |
| Chicken NOS2 fwd | CCACTCATTCTCCAAGCAA |
| Chicken NOS2 rev | AGGCAGAGCATACCACTT |
| Chicken IL-6 fwd | GTCGAGTCTCTGTGCTAC |
| Chicken IL-6 rev | CTTCAGATTGGCGAGGAG |
| Chicken CD206 fwd | GAGGACTGCGTTGTTATGA |
| Chicken CD206 rev | TCTTCTGTCGGTGCTTCT |
| Chicken IL-10 fwd | GCTCTGAACTGCTGGATG |
| Chicken IL-10 rev | ATGCTCTGCTGATGACTG |
| Chicken IL-4 fwd | AATGACATCCAGGGAGAGG |
| Chicken IL-4 rev | CAGGTTCTTGTGGCAGTG |
| Chicken CCL5 fwd | GCTCTGTCCCTCTCCATCCT |
| Chicken CCL5 rev | GTTGAAGCAGCACACGGTTG |
| Chicken IL-1β fwd | TAGATGTCGTGTGTGATGAG |
| Chicken IL-1β rev | GTAGAAGATGAAGCGGGTC |
| Chicken IL-8 fwd | ACGCTGGTAAAGATGGGGAA |
| Chicken IL-8 rev | GCACACCTCTCTTCCATCC |
| Chicken CX3CL1 fwd | GGTGGAGAAGATCGTGAAG |
| Chicken CX3CL1 rev | CTGGAGGTGAAGGTGGTA |
| Chicken CCL20 fwd | TCAAGAGGATGTCAATGTGA |
| Chicken CCL20 rev | AGAGATAGTGGTGAGTAAGC |
| Chicken GGCL1 fwd | CGAGCAAGGTGATGATGTA |
| Chicken GGCL1 rev | TTGGCACAGCACTTCTTC |
| Chicken IFN-β fwd | GCCCACACACTCCAAAACACTG |
| Chicken IFN-β rev | TTGATGCTGAGGTGAGCGTTG |
| Chicken FGF8 fwd | ACTGATCGGCAAGAGTAAC |
| Chicken FGF8 rev | CTCGTACTTGGCGTTCTG |
| Chicken FOS fwd | CTACTGTGTTCCTGGCAAT |
| Chicken FOS rev | ACATTCAGACCACCTCAAC |
| Chicken SERPINF1 fwd | CCACAGCCAACTAGAGAAG |
| Chicken SERPINF1 rev | AGCAAGGAGAATACTGACATC |
| Chicken Bcl-2 fwd | AGACCTACCTGCTTACACT |
| Chicken Bcl-2 rev | GCTTACTCTGACTGCTCTC |
| Chicken G0S2 fwd | TCAGCCAGAAGCCCAACAGG |
| Chicken G0S2 rev | GATGACCACGCCGAAGAACG |
| ALVJ env fwd | TGCGTGCGTGGTATTATTTC |
| ALVJ env rev | AATGGTGAGGTCGCTGACTGT |
| Chicken β-Actin fwd | GAGAAATTGTGCGTGACATCA |
| Chicken β-Actin rev | CCTGAACCTCTCATTGCCA |
| Chicken GAPDH fwd | GAGAAACCAGCCAAGTATGA |
| Chicken GAPDH rev | CTGGTCCTCTGTGTATCCTA |
| Mouse CD86 fwd | GACTCTACGACTTCACAATG |
| Mouse CD86 rev | TGTTAATGTCTGTTGGAGGA |
| Mouse NOS2 fwd | TACTGCTGGTGGTGACAA |
| Mouse NOS2 rev | CTGAAGGTGTGGTTGAGTT |
| Mouse IL-6 fwd | GCCAGAGTCCTTCAGAGA |
| Mouse IL-6 rev | GATGGTCTTGGTCCTTAGC |
| Mouse IL-1β fwd | CTACAGGCTCCGAGATGA |
| Mouse IL-1β rev | CGTTGCTTGGTTCTCCTT |
| Mouse CD206 fwd | GGCAAGTATCCACAGCAT |
| Mouse CD206 rev | GGTTCCATCACTCCACTC |
| Mouse IL-10 fwd | GGTTGCCAAGCCTTATCG |
| Mouse IL-10 rev | TCCACTGCCTTGCTCTTA |
| Mouse Arg1 fwd | GCAGAGGTCCAGAAGAATG |
| Mouse Arg1 rev | GGAGTGTTGATGTCAGTGT |
| Mouse GAPDH fwd | GTGAAGGTCGGTGTGAAC |
| Mouse GAPDH rev | CTTGACTGTGCCGTTGAA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, X.; Luo, H.; Dou, C.; Chen, X.; Huang, Y.; Wang, L.; Xue, S.; Sun, Z.; Chen, S.; Xu, Q.; et al. Metformin Triggers Apoptosis and Induction of the G0/G1 Switch 2 Gene in Macrophages. Genes 2021, 12, 1437. https://doi.org/10.3390/genes12091437
Hu X, Luo H, Dou C, Chen X, Huang Y, Wang L, Xue S, Sun Z, Chen S, Xu Q, et al. Metformin Triggers Apoptosis and Induction of the G0/G1 Switch 2 Gene in Macrophages. Genes. 2021; 12(9):1437. https://doi.org/10.3390/genes12091437
Chicago/Turabian StyleHu, Xuming, Huan Luo, Chunfeng Dou, Xujing Chen, Yi Huang, Liping Wang, Songlei Xue, Zhen Sun, Shihao Chen, Qi Xu, and et al. 2021. "Metformin Triggers Apoptosis and Induction of the G0/G1 Switch 2 Gene in Macrophages" Genes 12, no. 9: 1437. https://doi.org/10.3390/genes12091437
APA StyleHu, X., Luo, H., Dou, C., Chen, X., Huang, Y., Wang, L., Xue, S., Sun, Z., Chen, S., Xu, Q., Geng, T., Zhao, X., & Cui, H. (2021). Metformin Triggers Apoptosis and Induction of the G0/G1 Switch 2 Gene in Macrophages. Genes, 12(9), 1437. https://doi.org/10.3390/genes12091437








