Complete Chloroplast Genomes of Fagus sylvatica L. Reveal Sequence Conservation in the Inverted Repeat and the Presence of Allelic Variation in NUPTs
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Isolation and Sequencing
2.2. Chloroplast Genome Assemblies and Annotation
2.3. Assessment of Genome Variation
2.4. Detection of Heteroplasmy
2.5. Phylogenetic Analysis
3. Results
3.1. Assembly Size Variance and Genome Annotation
3.2. Repeat elements and SNPs
3.3. Within Individual Polymorphisms
3.4. Phylogenetic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Douglas, S.E. Plastid evolution: Origins, diversity, trends. Curr. Opin. Genet. Dev. 1998, 8, 655–661. [Google Scholar] [CrossRef]
- Wu, Z.Q.; Ge, S. The phylogeny of the BEP clade in grasses revisited: Evidence from the whole-genome sequences of chloroplasts. Mol. Phylogenet. Evol. 2012, 62, 573–578. [Google Scholar] [CrossRef]
- Birky, C.W., Jr. The inheritance of genes in mitochondria and chloroplasts: Laws, mechanisms, and models. Ann. Rev. Genet. 2001, 35, 125–148. [Google Scholar] [CrossRef] [PubMed]
- Wicke, S.; Schneeweiss, G.M.; de Pamphilis, C.W.; Muller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef] [PubMed]
- Straub, S.C.; Parks, M.; Weitemier, K.; Fishbein, M.; Cronn, R.C.; Liston, A. Navigating the tip of the genomic iceberg: Next-generation sequencing for plant systematics. Am. J. Bot. 2012, 99, 349–364. [Google Scholar] [CrossRef]
- Bock, D.G.; Andrew, R.L.; Rieseberg, L.H. On the adaptive value of cytoplasmic genomes in plants. Mol. Ecol. 2014, 23, 4899–4911. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Yu, X.; Fu, Q.; Zheng, Y.; Zhang, C. Complete chloroplast genome sequence of the three-flowered maple, Acer triflorum (Sapindaceae). Mitochondrial DNA Part B 2020, 5, 1859–1860. [Google Scholar] [CrossRef]
- Xue, S.; Shi, T.; Luo, W.; Ni, X.; Iqbal, S.; Ni, Z.; Huang, X.; Yao, D.; Shen, Z.; Gao, Z. Comparative analysis of the complete chloroplast genome among Prunus mume, P. armeniaca, and P. salicina. Hortic. Res. 2019, 6, 89. [Google Scholar] [CrossRef]
- Zong, D.; Gan, P.; Zhou, A.; Li, J.; Xie, Z.; Duan, A.; He, C. Comparative analysis of the complete chloroplast genomes of seven Populus species: Insights into alternative female parents of Populus tomentosa. PLoS ONE 2019, 14, e0218455. [Google Scholar] [CrossRef]
- Du, S. The complete chloroplast genome sequence of Populus wilsonii based on landscape design, and a comparative analysis with other Populus species. Mitochondrial DNA B Resour. 2020, 5, 2716–2718. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, T.; Duan, D.; Yang, J.; Feng, L.; Zhao, G. Comparative Analysis of the Complete Chloroplast Genomes of Five Quercus Species. Front. Plant. Sci. 2016, 7, 959. [Google Scholar] [CrossRef]
- Liu, X.; Chang, E.; Liu, J.; Jiang, Z. Comparative analysis of the complete chloroplast genomes of six white oaks with high ecological amplitude in China. J. For. Res. 2021. [Google Scholar] [CrossRef]
- Asaf, S.; Khan, A.L.; Khan, M.A.; Shahzad, R.; Lubna; Kang, S.M.; Al-Harrasi, A.; Al-Rawahi, A.; Lee, I.J. Complete chloroplast genome sequence and comparative analysis of loblolly pine (Pinus taeda L.) with related species. PLoS ONE 2018, 13, e0192966. [Google Scholar] [CrossRef] [PubMed]
- Sabir, J.S.; Arasappan, D.; Bahieldin, A.; Abo-Aba, S.; Bafeel, S.; Zari, T.A.; Edris, S.; Shokry, A.M.; Gadalla, N.O.; Ramadan, A.M.; et al. Whole mitochondrial and plastid genome SNP analysis of nine date palm cultivars reveals plastid heteroplasmy and close phylogenetic relationships among cultivars. PLoS ONE 2014, 9, e94158. [Google Scholar] [CrossRef] [PubMed]
- Li, F.W.; Harkess, A. A guide to sequence your favorite plant genomes. Appl. Plant Sci. 2018, 6, e1030. [Google Scholar] [CrossRef] [PubMed]
- Bondar, E.I.; Putintseva, Y.A.; Oreshkova, N.V.; Krutovsky, K.V. Siberian larch (Larix sibirica L.) chloroplast genome and development of polymorphic chloroplast markers. BMC Bioinform. 2019, 20, 38. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Ishizuka, W.; Hara, T.; Goto, S. Complete Chloroplast Genome of Japanese Larch (Larix kaempferi): Insights into Intraspecific Variation with an Isolated Northern Limit Population. Forests 2020, 11, 884. [Google Scholar] [CrossRef]
- Frey, J.E. Genetic flexibility of plant chloroplasts. Nature 1999, 398, 115–116. [Google Scholar] [CrossRef]
- Packham, J.R.; Thomas, P.A.; Atkinson, M.D.; Degen, T. Biological Flora of the British Isles: Fagus sylvatica. J. Ecol. 2012, 100, 1557–1608. [Google Scholar] [CrossRef]
- Demesure, B.; Comps, B.; Petit, R.J. Chloroplast DNA phylogeography of the common beech (Fagus sylvatica L.) in Europe. Evolution 1996, 50, 2515–2520. [Google Scholar] [CrossRef]
- Sebastiani, F.; Carnevale, S.; Vendramin, G.G. A new set of mono- and dinucleotide chloroplast microsatellites in Fagaceae. Mol. Ecol. Notes 2004, 4, 259–261. [Google Scholar] [CrossRef]
- Vettori, C.; Vendramin, G.G.; Anzidei, M.; Pastorelli, R.; Paffetti, D.; Giannini, R. Geographic distribution of chloroplast variation in Italian populations of beech (Fagus sylvatica L.). Theor. Appl. Genet. 2004, 109, 1–9. [Google Scholar] [CrossRef]
- Magri, D.; Vendramin, G.G.; Comps, B.; Dupanloup, I.; Geburek, T.; Gomory, D.; Latalowa, M.; Litt, T.; Paule, L.; Roure, J.M.; et al. A new scenario for the quaternary history of European beech populations: Palaeobotanical evidence and genetic consequences. New Phytol. 2006, 171, 199–221. [Google Scholar] [CrossRef] [PubMed]
- Hatziskakis, S.; Papageorgiou, A.C.; Gailing, O.; Finkeldey, R. High chloroplast haplotype diversity in Greek populations of beech (Fagus sylvatica L.). Plant Biol. 2009, 11, 425–433. [Google Scholar] [CrossRef]
- Papageorgiou, A.C.; Tsiripidis, I.; Mouratidis, T.; Hatziskakis, S.; Gailing, O.; Eliades, N.G.H.; Vidalis, A.; Drouzas, A.D.; Finkeldey, R. Complex fine-scale phylogeographical patterns in a putative refugial region for Fagus sylvatica (Fagaceae). Bot. J. Linn. Soc. 2014, 174, 516–528. [Google Scholar] [CrossRef][Green Version]
- Mishra, B.; Ulaszewski, B.; Ploch, S.; Burczyk, J.; Thines, M. A Circular Chloroplast Genome of Fagus sylvatica Reveals High Conservation between Two Individuals from Germany and One Individual from Poland and an Alternate Direction of the Small Single-Copy Region. Forests 2021, 12, 180. [Google Scholar] [CrossRef]
- Meger, J.; Ulaszewski, B.; Vendramin, G.G.; Burczyk, J. Using reduced representation libraries sequencing methods to identify cpDNA polymorphisms in European beech (Fagus sylvatica L). Tree Genet. Genomes 2019, 15, 7. [Google Scholar] [CrossRef]
- Barzdajn, W.; Rzeznik, Z. Wstepne wyniki miedzynarodowego doswiadczenia proweniencyjnego z bukiem (Fagus sylvatica L.) serii 1993/1995 w Lesnym Zakladzie Doswiadczalnym Siemnianice. Sylwan 2002, 146, 149–164. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. Unraveling heteroplasmy patterns with NOVOPlasty. NAR Genom. Bioinform. 2020, 2, lqz011. [Google Scholar] [CrossRef]
- Manos, P.S.; Stanford, A.M. The historical biogeography of Fagaceae: Tracking the tertiary history of temperate and subtropical forests of the Northern Hemisphere. Int. J. Plant Sci. 2001, 162, S77–S93. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997v1. [Google Scholar]
- Milne, I.; Bayer, M.; Cardle, L.; Shaw, P.; Stephen, G.; Wright, F.; Marshall, D. Tablet—Next generation sequence assembly visualization. Bioinformatics 2010, 26, 401–402. [Google Scholar] [CrossRef] [PubMed]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Worth, J.R.P.; Liu, L.; Wei, F.J.; Tomaru, N. The complete chloroplast genome of Fagus crenata (subgenus Fagus) and comparison with F. engleriana (subgenus Engleriana). PeerJ 2019, 7, e7026. [Google Scholar] [CrossRef]
- Yang, Y.; Zhu, J.; Feng, L.; Zhou, T.; Bai, G.; Yang, J.; Zhao, G. Plastid Genome Comparative and Phylogenetic Analyses of the Key Genera in Fagaceae: Highlighting the Effect of Codon Composition Bias in Phylogenetic Inference. Front. Plant Sci. 2018, 9, 82. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Takayama, K.; Youn, J.S.; Pak, J.H.; Kim, S.C. Plastome Characterization and Phylogenomics of East Asian Beeches with a Special Emphasis on Fagus multinervis on Ulleung Island, Korea. Genes 2020, 11, 1338. [Google Scholar] [CrossRef]
- Mader, M.; Schroeder, H.; Schott, T.; Schoning-Stierand, K.; Leite Montalvao, A.P.; Liesebach, H.; Liesebach, M.; Fussi, B.; Kersten, B. Mitochondrial Genome of Fagus sylvatica L. as a Source for Taxonomic Marker Development in the Fagales. Plants 2020, 9, 1274. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Thiel, T.; Munch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; Team, U. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Schirmer, M.; D’Amore, R.; Ijaz, U.Z.; Hall, N.; Quince, C. Illumina error profiles: Resolving fine-scale variation in metagenomic sequencing data. BMC Bioinform. 2016, 17, 125. [Google Scholar] [CrossRef]
- Mishra, B.; Ulaszewski, B.; Meger, J.; Pfenninger, M.; Gupta, D.K.; Wötzel, S.; Ploch, S.; Burczyk, J.; Thines, M. A chromosome-level genome assembly of the European Bee (Fagus sylvatica L) reveals anomalies for organelle DNA integration, repeat content and distribution of SNPs. bioRxiv 2021. [Google Scholar] [CrossRef]
- Tavaré, S. Some probabilistic and statistical problems in the analysis of DNA sequences. Lect. Math. Life Sci. 1986, 17, 57–86. [Google Scholar]
- Soubrier, J.; Steel, M.; Lee, M.S.; Der Sarkissian, C.; Guindon, S.; Ho, S.Y.; Cooper, A. The influence of rate heterogeneity among sites on the time dependence of molecular rates. Mol. Biol. Evol. 2012, 29, 3345–3358. [Google Scholar] [CrossRef]
- Yang, Z. A space-time process model for the evolution of DNA sequences. Genetics 1995, 139, 993–1005. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, M.S.; Anderson, C.D. PASSaGE: Pattern Analysis, Spatial Statistics and Geographic Exegesis; Version 2. Methods Ecol. Evol. 2011, 2, 229–232. [Google Scholar] [CrossRef]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
- Magri, D. Patterns of post-glacial spread and the extent of glacial refugia of European beech (Fagus sylvatica). J. Biogeogr. 2008, 35, 450–463. [Google Scholar] [CrossRef]
- Sjölund, M.J.; González-Díaz, P.; Moreno-Villena, J.J.; Jump, A.S. Understanding the legacy of widespread population translocations on the post-glacial genetic structure of the European beech, Fagus sylvatica L. J. Biogeogr. 2017, 44, 2475–2487. [Google Scholar] [CrossRef]
- Heuertz, M.; Fineschi, S.; Anzidei, M.; Pastorelli, R.; Salvini, D.; Paule, L.; Frascaria-Lacoste, N.; Hardy, O.J.; Vekemans, X.; Vendramin, G.G. Chloroplast DNA variation and postglacial recolonization of common ash (Fraxinus excelsior L.) in Europe. Mol. Ecol. 2004, 13, 3437–3452. [Google Scholar] [CrossRef]
- Weising, K.; Gardner, R.C. A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome 1999, 42, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, B.; Gilbert, L.E.; Ruhlman, T.A.; Jansen, R.K. Clade-Specific Plastid Inheritance Patterns Including Frequent Biparental Inheritance in Passiflora Interspecific Crosses. Int. J. Mol. Sci. 2021, 22, 2278. [Google Scholar] [CrossRef] [PubMed]
- Scarcelli, N.; Mariac, C.; Couvreur, T.L.; Faye, A.; Richard, D.; Sabot, F.; Berthouly-Salazar, C.; Vigouroux, Y. Intra-individual polymorphism in chloroplasts from NGS data: Where does it come from and how to handle it? Mol. Ecol. Resour. 2016, 16, 434–445. [Google Scholar] [CrossRef] [PubMed]
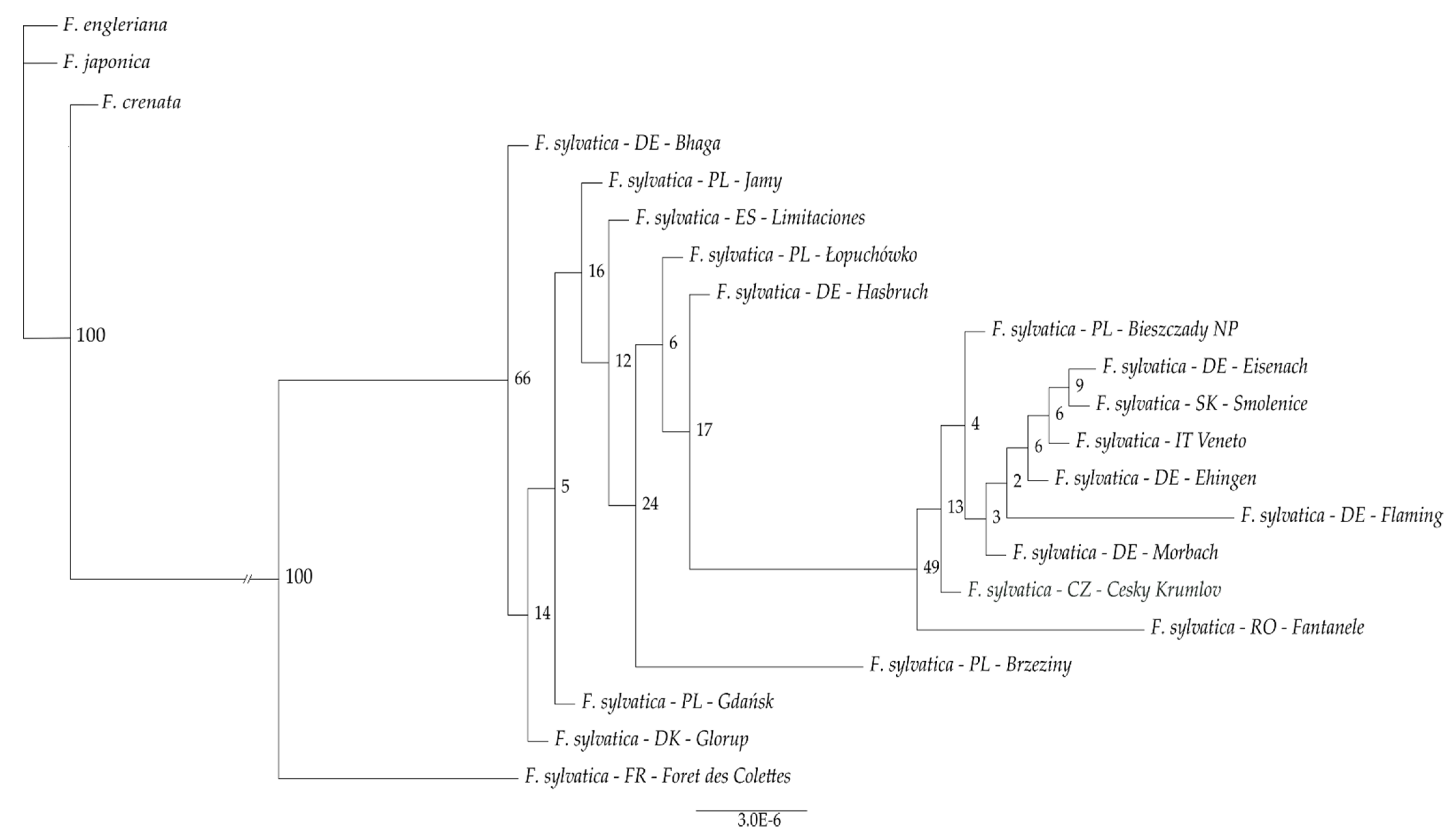
| No. | Origin or Individual Name | Country | Longitude | Latitude | Number of Read Pairs | NCBI Accession Number | SRA Accession Number |
|---|---|---|---|---|---|---|---|
| 1 | Bhaga | Germany | 51.169167 N | 8.963056 E | [26] | MW531753 | N/A |
| 2 | Jamy | Poland | 53.586019 N | 18.935019 E | [26] | MW537046 | SAMN08948264 |
| 3 | Gdańsk | Poland | 54.383262 N | 18.516724 E | 3,777,769 | MW566769 | SAMN18917950 |
| 4 | Foret des Colettes | France | 46.183328 N | 2.949992 E | 4,899,373 | MW566771 | SAMN18917951 |
| 5 | Limitaciones | Spain | 42.818059 N | 2.249663 W | 6,210,877 | MW566772 | SAMN18917952 |
| 6 | Glorup | Denmark | 55.184748 N | 10.681238 E | 20,891,953 | MW566770 | SAMN18917953 |
| 7 | Łopuchówko | Poland | 52.583300 N | 17.083339 E | 5,114,816 | MW566774 | SAMN18917954 |
| 8 | Hasbruch | Germany | 53.120708 N | 8.4302740 E | 4,650,347 | MW566776 | SAMN18917955 |
| 9 | Bieszczady NP | Poland | 49.117093 N | 22.579103 E | 3,046,013 | MW566773 | SAMN18917956 |
| 10 | Eisenach | Germany | 50.087605 N | 10.106152 E | 4,461,792 | MW566778 | SAMN18917957 |
| 11 | Morbach | Germany | 50.740891 N | 6.980116 E | 5,833,195 | MW566784 | SAMN18917958 |
| 12 | Ehingen | Germany | 48.399106 N | 9.500861 E | 5,632,928 | MW566775 | SAMN18917959 |
| 13 | Veneto | Italy | 46.133489 N | 12.216683 E | 7,741,036 | MW566783 | SAMN18917960 |
| 14 | Cesky Krumlov | Czechia | 48.850035 N | 14.250406 E | 7,853,097 | MW566777 | SAMN18917961 |
| 15 | Brzeziny | Poland | 51.836489 N | 19.601247 E | 7,349,714 | MW566779 | SAMN18917962 |
| 16 | Smolenice | Slovakia | 48.485171 N | 17.372687 E | 5,072,400 | MW566782 | SAMN18917963 |
| 17 | Fantanele | Romania | 46.416750 N | 26.466475 E | 6,584,825 | MW566780 | SAMN18917964 |
| 18 | Fläming | Germany | 52.133389 N | 12.583406 E | 7,423,489 | MW566781 | SAMN18917965 |
| Main Genome Elements | ||||||
|---|---|---|---|---|---|---|
| Origin or Individual Name | Read Coverage | NCBI Accession Number | Total Size (bp) | LSC (bp) | SSC (bp) | IR-A/IR-B) (bp) |
| Bhaga | - | MW531753 | 158,458 | 87,702 | 19,010 | 25,873 |
| Jamy | - | MW537046 | 158,462 | 87,705 | 19,011 | 25,873 |
| Gdańsk | 253x | MW566769 | 158,456 | 87,699 | 19,011 | 25,873 |
| Colettes | 498x | MW566771 | 158,391 | 87,634 | 19,011 | 25,873 |
| Limitaciones | 491x | MW566772 | 158,461 | 87,704 | 19,011 | 25,873 |
| Glorup | 356x | MW566770 | 158,461 | 87,704 | 19,011 | 25,873 |
| Łopuchówko | 212x | MW566774 | 158,461 | 87,704 | 19,011 | 25,873 |
| Hasbruch | 267x | MW566776 | 158,462 | 87,705 | 19,011 | 25,873 |
| Bieszczady NP | 211x | MW566773 | 158,426 | 87,669 | 19,011 | 25,873 |
| Eisenach | 105x | MW566778 | 158,456 | 87,699 | 19,011 | 25,873 |
| Morbach | 350x | MW566784 | 158,463 | 87,706 | 19,011 | 25,873 |
| Ehingen | 91x | MW566775 | 158,446 | 87,689 | 19,011 | 25,873 |
| Veneto | 625x | MW566783 | 158,463 | 87,706 | 19,011 | 25,873 |
| Cesky Krumlov | 300x | MW566777 | 158,462 | 87,705 | 19,011 | 25,873 |
| Brzeziny | 521x | MW566779 | 158,462 | 87,705 | 19,011 | 25,873 |
| Smolenice | 86x | MW566782 | 158,430 | 87,674 | 19,010 | 25,873 |
| Fantanele | 157x | MW566780 | 158,462 | 87,705 | 19,011 | 25,873 |
| Fläming | 306x | MW566781 | 158,464 | 87,705 | 19,013 | 25,873 |
| Mononucleotide | Dinucleotide | Pentanucleotide | Complex | Total | |
|---|---|---|---|---|---|
| Monomorphic | 93 | 2 | 4 | 27 | 126 |
| Polymorphic | 4 | - | - | 8 | 12 |
| Total | 97 | 2 | 4 | 35 | 138 |
| No. | Starting Position (bp) * | Type | Region | Marker Ratio | Flanking Annotation |
|---|---|---|---|---|---|
| 1 | 4363 | Complex | SSC | 17/1 | ndhA (exon II) ↔ ndhA (exon I) |
| 2 | 8012 | Complex | SSC | 16/1/1 | psaC ↔ ndhD |
| 3 | 11,476 | Mononucleotide (A) | SSC | 17/1 | trnL ↔ rpl32 |
| 4 | 12,583 | Mononucleotide (T) | SSC | 17/0 ** | rpl32 ↔ ndhF |
| 5 | 46,142 | Complex | LSC | 16/1/1 | matK ↔ trnQ |
| 6 | 46,952 | Complex | LSC | 11/2/2/1/1/1 | matK ↔ trnQ |
| 7 | 50,589 | Mononucleotide (A) | LSC | 17/1 | trnG (exon I) ↔ trnG (exon II) |
| 8 | 55,923 | Complex | LSC | 16/2 | atpH ↔ atpI |
| 9 | 70,097 | Complex | LSC | 16/2 | rpoB ↔ trnC |
| 10 | 92,043 | Mononucleotide (A) | LSC | 16/2 | trnG (exon II) ↔ trnG (exon I) |
| 11 | 105,126 | Complex | LSC | 12/5/1 | ycf4 ↔ cemA |
| 12 | 107,580 | Complex | LSC | 17/1 | petA ↔ psbJ |
| No. | Position (bp) * | Marker Type | Region | Consensus | Alternative | Area | Marker Ratio | Flanking Annotation |
|---|---|---|---|---|---|---|---|---|
| 1 | 12,587 | SNP | SSC | T | C | noncoding | 17/1 | rpl32 ↔ ndhF |
| 2 | 46,985 | SNP | LSC | G | A | noncoding | 17/1 | tRNA-K ↔tRNA-Q |
| 3 | 71,204 | SNP | LSC | G | T | noncoding | 9/9 | tRNA-C ↔ petN |
| 4 | 80,558 | Indel | LSC | T | - | noncoding | 17/1 | psbZ ↔ tRNA-G |
| 5 | 112,198 | SNP | LSC | A | C | noncoding | 17/1 | psaJ ↔ rpl3 |
| LSC | SSC | IR-A | IR-B | |
|---|---|---|---|---|
| Avg. variant depth | 349x | 360x | 477x | 477x |
| Avg. alternative var. depth | 18.7x | 16.1x | 18.4x | 18.5x |
| Number of uniqe positions | 5348 | 1161 | 1257 | 1262 |
| SNP | 76.8% | 80.9% | 83.7% | 84.1% |
| Indel | 10.2% | 8.8% | 9.6% | 9.4% |
| Complex | 8.2% | 6.2% | 3.1% | 3.1% |
| MNP | 0.2% | 0.3% | 0.8% | 0.7% |
| Mix | 4.6% | 3.9% | 2.7% | 2.7% |
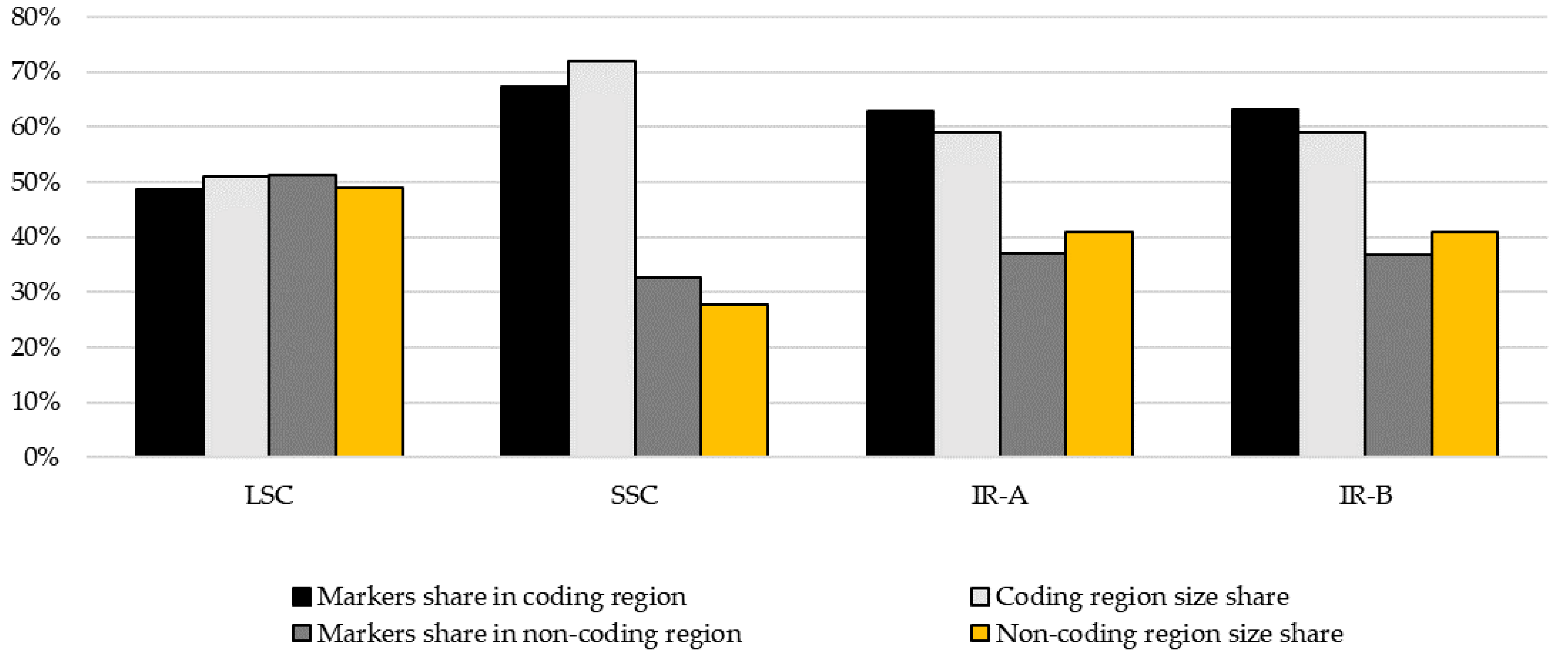
| Coding | 48.6% | 67.3% | 62.9% | 63.1% |
| Non-coding | 51.4% | 32.7% | 37.1% | 36.9% |
| Class | Boundry max (km) | Number of Pairs | Mantel r | p |
|---|---|---|---|---|
| 1 | 250 | 11 | 0.286 | 0.011 |
| 2 | 500 | 31 | 0.106 | 0.361 |
| 3 | 750 | 46 | 0.121 | 0.144 |
| 4 | 1000 | 27 | −0.016 | 0.760 |
| 5 | 1250 | 15 | −0.004 | 0.900 |
| 6 | 1500 | 11 | −0.023 | 0.374 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ulaszewski, B.; Meger, J.; Mishra, B.; Thines, M.; Burczyk, J. Complete Chloroplast Genomes of Fagus sylvatica L. Reveal Sequence Conservation in the Inverted Repeat and the Presence of Allelic Variation in NUPTs. Genes 2021, 12, 1357. https://doi.org/10.3390/genes12091357
Ulaszewski B, Meger J, Mishra B, Thines M, Burczyk J. Complete Chloroplast Genomes of Fagus sylvatica L. Reveal Sequence Conservation in the Inverted Repeat and the Presence of Allelic Variation in NUPTs. Genes. 2021; 12(9):1357. https://doi.org/10.3390/genes12091357
Chicago/Turabian StyleUlaszewski, Bartosz, Joanna Meger, Bagdevi Mishra, Marco Thines, and Jarosław Burczyk. 2021. "Complete Chloroplast Genomes of Fagus sylvatica L. Reveal Sequence Conservation in the Inverted Repeat and the Presence of Allelic Variation in NUPTs" Genes 12, no. 9: 1357. https://doi.org/10.3390/genes12091357
APA StyleUlaszewski, B., Meger, J., Mishra, B., Thines, M., & Burczyk, J. (2021). Complete Chloroplast Genomes of Fagus sylvatica L. Reveal Sequence Conservation in the Inverted Repeat and the Presence of Allelic Variation in NUPTs. Genes, 12(9), 1357. https://doi.org/10.3390/genes12091357





