Genome-Wide SNPs Detect Hybridisation of Marsupial Gliders (Petaurus breviceps breviceps × Petaurus norfolcensis) in the Wild
Abstract
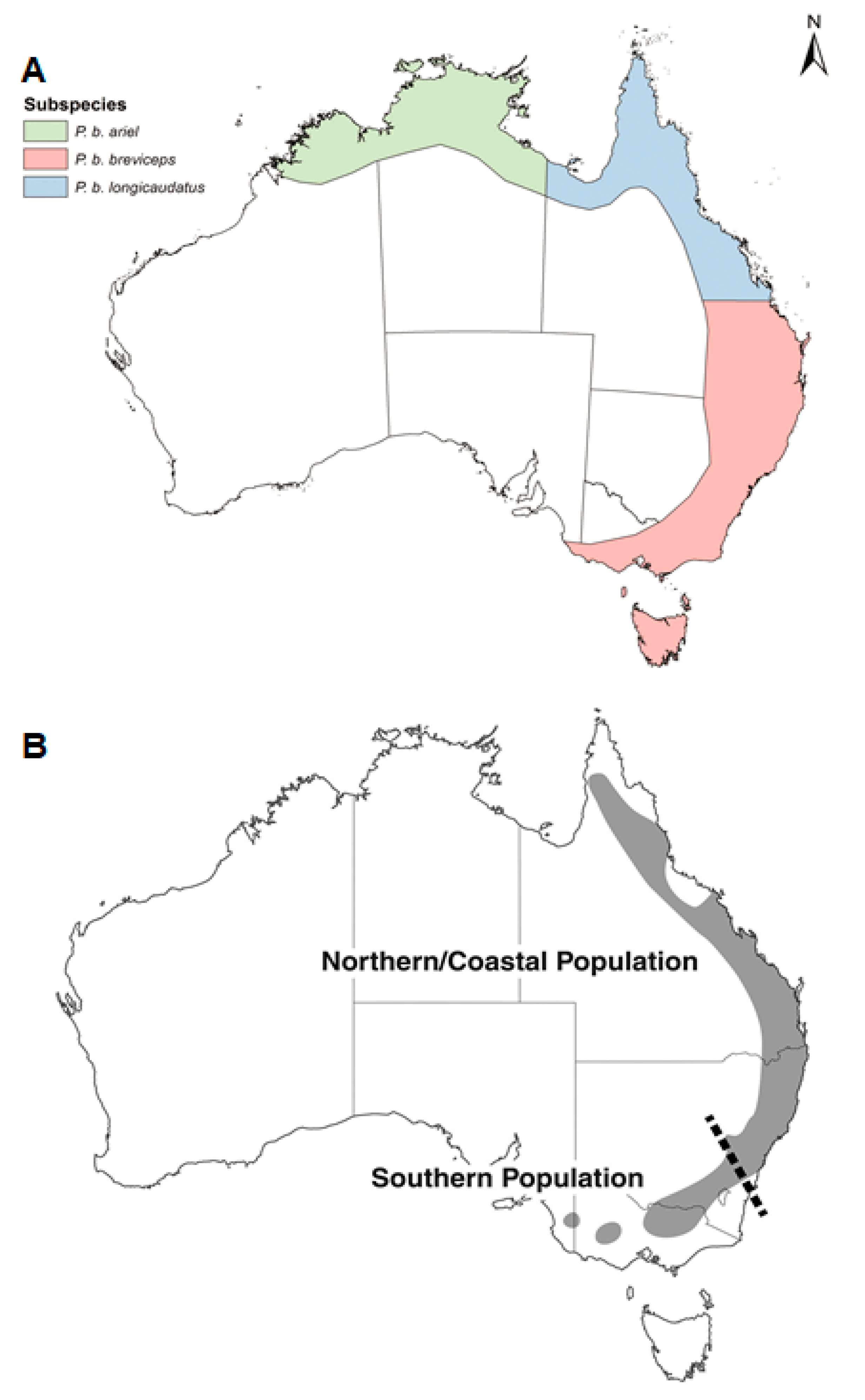
:1. Introduction
2. Materials and Methods
2.1. Genetic Sampling and Morphological Measurements
2.2. Sequencing Mitochondrial DNA
2.3. Next-Generation Sequencing
2.4. Statistical Analyses
3. Results
3.1. Morphological Features
3.2. Mitochondrial DNA Sequence
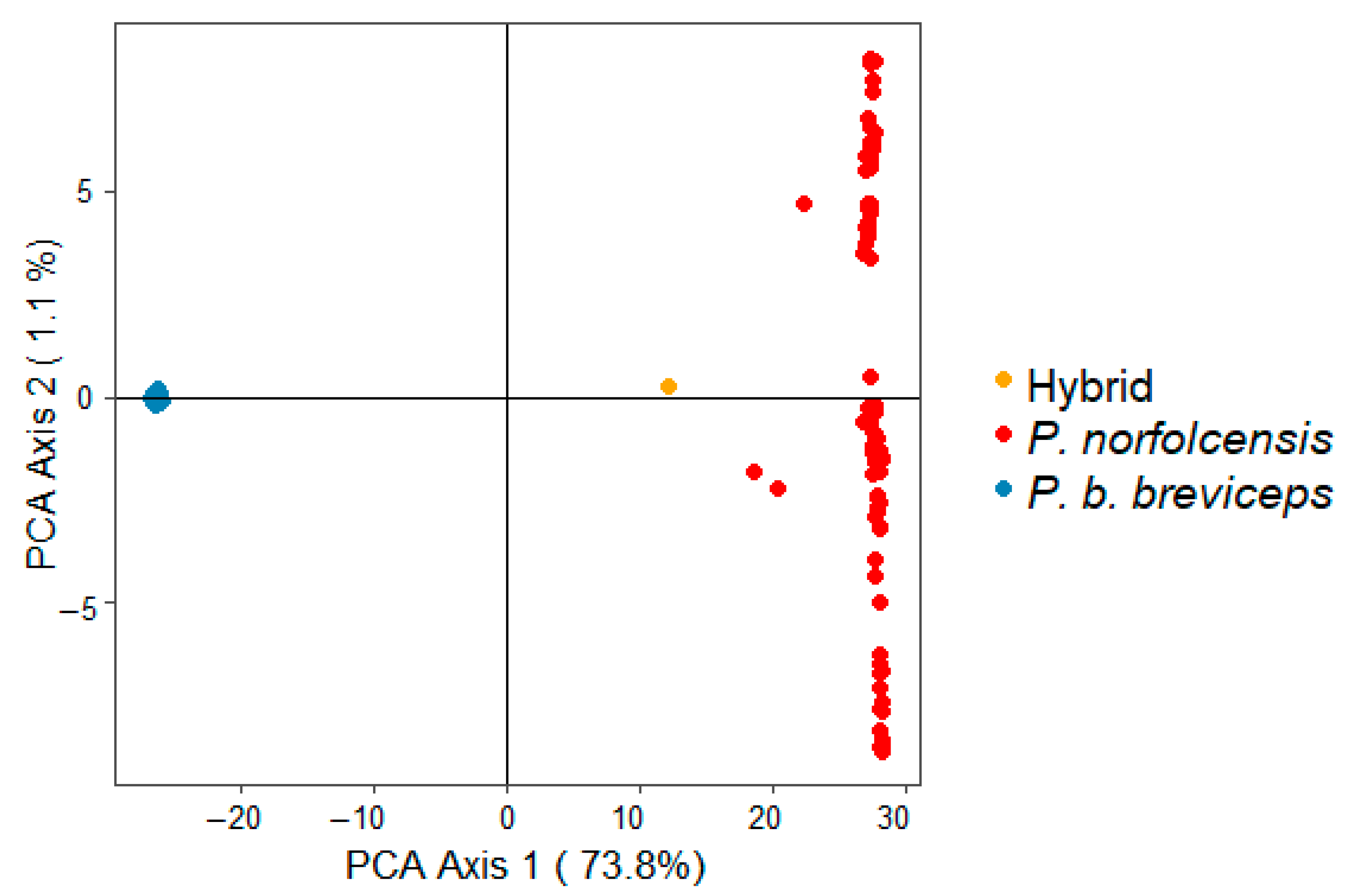
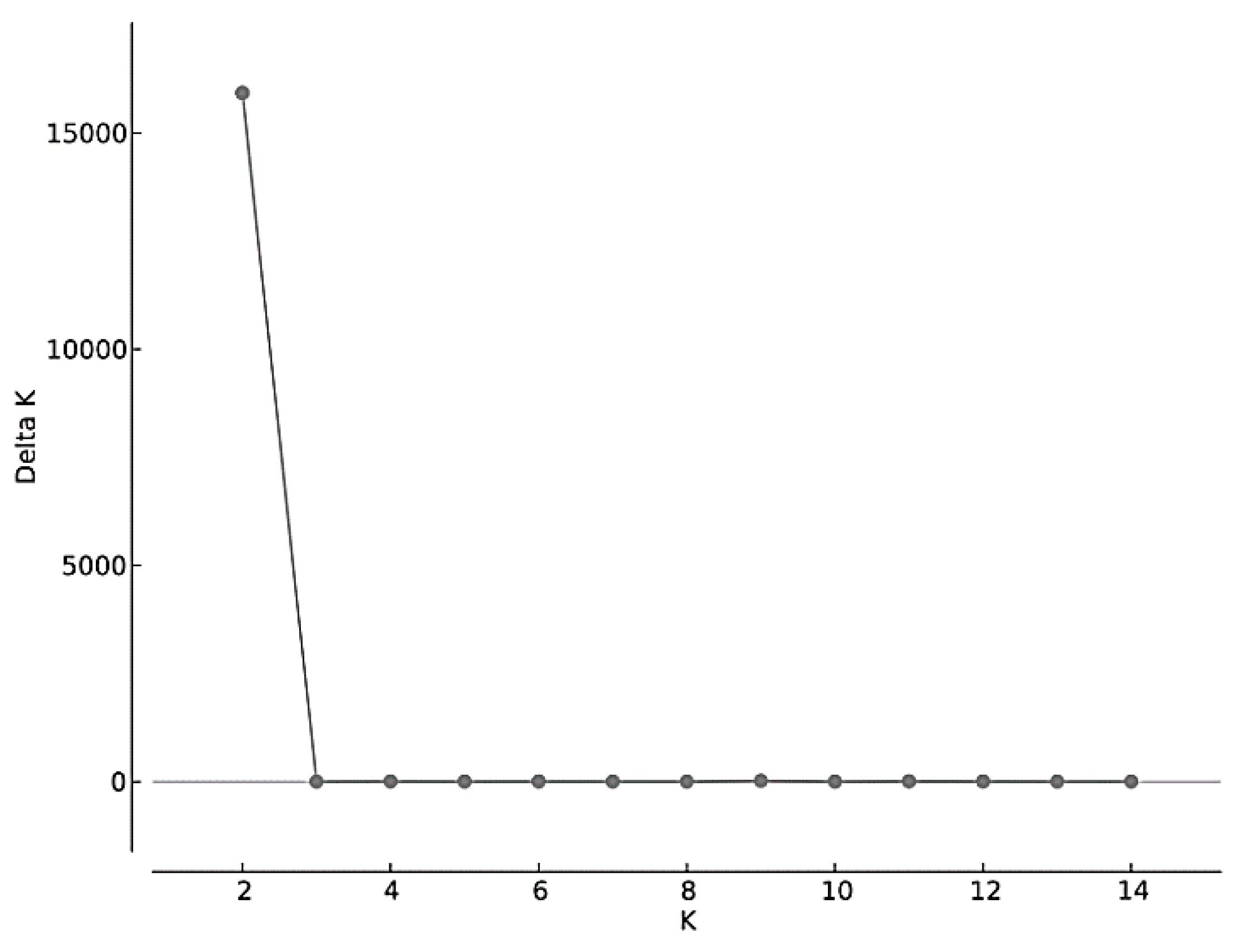
3.3. Genome Wide SNPs: Testing for Hybrid
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fox, B.; Murray, J. Laboratory hybridization of Australian Rattus fuscipes and Rattus lutreolus and its karyotypic confirmation. Aust. J. Zool. 1979, 27, 691–698. [Google Scholar] [CrossRef]
- Fleay, D. Gliders of the Gum Trees: The Most Beautiful and Enchanting Australian Marsupials; The Hawthorn Press: Hawthorn, VIC, Australia, 1947. [Google Scholar]
- Zuckerman, S. The breeding seasons of mammals in captivity. Proc. Zool. Soc. Lond. 1952, 122, 827–950. [Google Scholar] [CrossRef]
- Twyford, A.D.; Ennos, R.A. Next-generation hybridization and introgression. Heredity 2012, 108, 179–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrison, R.G.; Larson, E.L. Hybridization, Introgression, and the Nature of Species Boundaries. J. Hered. 2014, 105, 795–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattucci, F.; Galaverni, M.; Lyons, L.A.; Alves, P.C.; Randi, E.; Velli, E.; Pagani, L.; Caniglia, R. Genomic approaches to identify hybrids and estimate admixture times in European wildcat populations. Sci. Rep. 2019, 9, 11612. [Google Scholar] [CrossRef] [Green Version]
- Cairns, K.M.; Shannon, L.M.; Koler-Matznick, J.; Ballard, J.W.O.; Boyko, A.R. Elucidating biogeographical patterns in Australian native canids using genome wide SNPs. PLoS ONE 2018, 13, e0198754. [Google Scholar] [CrossRef]
- Melville, J.; Haines, M.L.; Boysen, K.; Hodkinson, L.; Kilian, A.; Smith Date, K.L.; Potvin, D.A.; Parris, K.M. Identifying hybridization and admixture using SNPs: Application of the DArTseq platform in phylogeographic research on vertebrates. R. Soc. Open Sci. 2017, 4, 161061. [Google Scholar] [CrossRef] [Green Version]
- Levin, D. Hybridization and Extinction: In protecting rare species, conservationists should consider the dangers of interbreeding, which compound the more well-known threats to wildlife on JSTOR. Am. Sci. 2002, 90, 254–261. [Google Scholar] [CrossRef]
- Shurtliff, Q.R. Mammalian hybrid zones: A review. Mamm. Rev. 2013, 43, 1–21. [Google Scholar] [CrossRef]
- Turner, L.M.; White, M.A.; Tautz, D.; Payseur, B.A. Genomic Networks of Hybrid Sterility. PLoS Genet. 2014, 10, e1004162. [Google Scholar] [CrossRef] [Green Version]
- Simberloff, D. Hybridization between native and introduced wildlife species: Importance for conservation. Wildl. Biol. 1996, 2, 143–150. [Google Scholar] [CrossRef]
- Wintle, B.; Bekessy, S. Let’s Get this Straight, Habitat Loss is the Number-One Threat to Australia’s Species. Available online: http://theconversation.com/lets-get-this-straight-habitat-loss-is-the-number-one-threat-to-australias-species-85674 (accessed on 26 March 2018).
- Woinarski, J.C.Z.; Burbidge, A.; Harrison, P.L. The Action Plan for Australian Mammals 2012; CSIRO Publishing: Collingwood, VIC, Australia, 2014; ISBN 9780643108738. [Google Scholar]
- Cremona, T.; Baker, A.M.; Cooper, S.J.B.; Montague-Drake, R.; Stobo-Wilson, A.M.; Carthew, S.M. Integrative taxonomic investigation of Petaurus breviceps (Marsupialia: Petauridae) reveals three distinct species. Zool. J. Linn. Soc. 2020, 191, 503–527. [Google Scholar] [CrossRef]
- Crane, M.; Lindenmayer, D.B.; Banks, S.C. Conserving and restoring endangered southern populations of the Squirrel Glider (Petaurus norfolcensis) in agricultural landscapes. Ecol. Manag. Restor. 2017, 18, 15–25. [Google Scholar] [CrossRef]
- Malekian, M.; Cooper, S.J.B.; Norman, J.A.; Christidis, L.; Carthew, S.M. Molecular systematics and evolutionary origins of the genus Petaurus (Marsupialia: Petauridae) in Australia and New Guinea. Mol. Phylogenet. Evol. 2010, 54, 122–135. [Google Scholar] [CrossRef]
- NSW Scientific Committee. NSW Scientific Committee established under the Threatened Species Conservation Act 1995 Squirrel Glider Petaurus Norfolcensis Review of Current Information in NSW. 2008. Available online: https://www.environment.nsw.gov.au/-/media/OEH/Corporate-Site/Documents/Animals-and-plants/Scientific-Committee/sc-squirrel-glider-petaurus-norfolcensis-review-report.pdf?la=en&hash=63FC3FA938DBA28BC42FA33AAFB9D5AF321E166E (accessed on 15 July 2021).
- Suckling, G.C. Population ecology of the sugar glider, Petaurus breviceps, in a system of fragmented habitats. Wildl. Res. 1984, 11, 49–75. [Google Scholar] [CrossRef]
- Lindenmayer, D. Gliders of Australia: A Natural History; UNSW Press: Sydney, NSW, Australia, 2002; ISBN 086840523X. [Google Scholar]
- Smith, M.J. Observations on Growth of Petaurus Breviceps and P. Novfolcensis (Petauridae: Marsupialia) in Captivity. Wildl. Res. 1979, 6, 141–150. [Google Scholar] [CrossRef]
- Quin, D.G. Population ecology of the squirrel glider (Petaurus norfolcensis) and the sugar glider (P. breviceps) (Maruspialia: Petauridae) at Limeburners Creek, on the central north coast of New South Wales. Wildl. Res. 1995, 22, 471–505. [Google Scholar] [CrossRef]
- Smith, A.P. Diet and Feeding Strategies of the Marsupial Sugar Glider in Temperate Australia. J. Anim. Ecol. 1982, 51, 149. [Google Scholar] [CrossRef]
- Ball, T.; Adams, E.; Goldingay, R.L. Diet of the squirrel glider in a fragmented landscape near Mackay, central Queensland. Aust. J. Zool. 2009, 57, 295–304. [Google Scholar] [CrossRef]
- Knipler, M.; Dowton, M.; Clulow, J.; Meyer, N.; Mikac, K.M. Genome-wide SNPs Detect Fine-scale Genetic Structure in Threatened Populations of Squirrel Glider Petaurus norfolcensis. Res. Sq. 2021. under review. [Google Scholar] [CrossRef]
- Kocher, T.D.; Thomas, W.K.; Meyer, A.; Edwards, S.V.; Paabo, S.; Villablanca, F.X.; Wilson, A.C.; Pääbo, S.; Villablanca, F.X.; Wilson, A.C. Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proc. Natl. Acad. Sci. USA 1989, 86, 6196–6200. [Google Scholar] [CrossRef] [Green Version]
- Irwin, D.M.; Kocher, T.D.; Wilson, A.C. Evolution of the cytochrome b gene of mammals. J. Mol. Evol. 1991, 32, 128–144. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, A.; Walker, F.M.; Van der Ree, R.; Cesarini, S.; Taylor, A.C. Threatened populations of the Australian squirrel glider (Petaurus norfolcensis) show evidence of evolutionary distinctiveness on a Late Pleistocene timescale. Conserv. Genet. 2010, 11, 2393–2407. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kilian, A.; Wenzl, P.; Huttner, E.; Carling, J.; Xia, L.; Blois, H.; Caig, V.; Heller-Uszynska, K.; Jaccoud, D.; Hopper, C.; et al. Diversity arrays technology: A generic genome profiling technology on open platforms. Methods Mol. Biol. 2012, 888, 67–89. [Google Scholar] [CrossRef] [PubMed]
- Gruber, B.; Unmack, P.J.; Berry, O.F.; Georges, A. Dartr: An r package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Mol. Ecol. Resour. 2018, 18, 691–699. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.gbif.org/tool/81287/r-a-language-and-environment-for-statistical-computing (accessed on 1 April 2021).
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the Analysis of Population Structure. Evolution 1984, 38, 1358. [Google Scholar] [CrossRef] [PubMed]
- Wright, S. THE Genetical structure of populations. Ann. Eugen. 1949, 15, 323–354. [Google Scholar] [CrossRef]
- Jombart, T. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [Green Version]
- Moehring, A.J. Heterozygosity and its unexpected correlations with hybrid sterility. Evolution 2011, 65, 2621. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, R.; Randi, E.; Mattucci, F.; Kurushima, J.D.; Lyons, L.A.; Alves, P.C. Toward a genome-wide approach for detecting hybrids: Informative SNPs to detect introgression between domestic cats and European wildcats (Felis silvestris). Heredity 2015, 115, 195–205. [Google Scholar] [CrossRef] [Green Version]
- Steyer, K.; Tiesmeyer, A.; Muñoz-Fuentes, V.; Nowak, C. Low rates of hybridization between European wildcats and domestic cats in a human-dominated landscape. Ecol. Evol. 2018, 8, 2290–2304. [Google Scholar] [CrossRef] [Green Version]
- Van Wyk, A.M.; Dalton, D.L.; Hoban, S.; Bruford, M.W.; Russo, I.-R.M.; Birss, C.; Grobler, P.; van Vuuren, B.J.; Kotzé, A. Quantitative evaluation of hybridization and the impact on biodiversity conservation. Ecol. Evol. 2017, 7, 320–330. [Google Scholar] [CrossRef] [Green Version]
- Haines, M.L.; Luikart, G.; Amish, S.J.; Smith, S.; Latch, E.K. Evidence for adaptive introgression of exons across a hybrid swarm in deer. BMC Evol. Biol. 2019, 19, 199. [Google Scholar] [CrossRef]
- Porras-Hurtado, L.; Ruiz, Y.; Santos, C.; Phillips, C.; Carracedo, Á.; Lareu, M.V. An overview of STRUCTURE: Applications, parameter settings, and supporting software. Front. Genet. 2013, 4, 98. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Anderson, E.C.; Thompson, E.A. A model-based method for identifying species hybrids using multilocus genetic data. Genetics 2002, 160, 1217–1229. [Google Scholar] [CrossRef]
- Dufresnes, C.; Remollino, N.; Stoffel, C.; Manz, R.; Weber, J.-M.; Fumagalli, L. Two decades of non-invasive genetic monitoring of the grey wolves recolonizing the Alps support very limited dog introgression. Sci. Rep. 2019, 9, 148. [Google Scholar] [CrossRef] [Green Version]
- Neaves, L.E.; Zenger, K.R.; Cooper, D.W.; Eldridge, M.D.B. Molecular detection of hybridization between sympatric kangaroo species in south-eastern Australia. Heredity 2010, 104, 502–512. [Google Scholar] [CrossRef]
- Goodman, S.J.; Barton, N.H.; Swanson, G.; Abernethy, K.; Pemberton, J.M. Introgression through rare hybridization: A genetic study of a hybrid zone between red and sika deer (genus Cervus) in Argyll, Scotland. Genetics 1999, 152, 355–371. [Google Scholar] [CrossRef]
- Transport for NSW Traffic Volume Viewer. Available online: https://roads-waterways.transport.nsw.gov.au/about/corporate-publications/statistics/traffic-volumes/aadt-map/index.html#/?z=16&lat=-32.92009726354131&lon=151.70382561311817&pco=1&pcl=1&sco=1&scl=1&nd=1&v=0&st=1&id=05204&yr=2018 (accessed on 16 July 2021).
- Yadav, A.D.; Sahu, J.D.; Dubey, A.D.; Gadpayle, R.D.; Kiran Barwa, D.; Kashyap, K.; Yadav, A.; Jain, A.; Sahu, J.; Dubey, A.; et al. An overview on species hybridization in animals. Int. J. Fauna Biol. Stud. 2019, 6, 36–42. [Google Scholar]
- Grabenstein, K.C.; Taylor, S.A. Breaking Barriers: Causes, Consequences, and Experimental Utility of Human-Mediated Hybridization. Trends Ecol. Evol. 2018, 33, 198–212. [Google Scholar] [CrossRef]
- Haldane, J.B.S. Sex ratio and unisexual sterility in hybrid animals. J. Genet. 1922, 12, 101–109. [Google Scholar] [CrossRef] [Green Version]
- Schilthuizen, M.; Giesbers, M.C.W.G.; Beukeboom, L.W. Haldane’s rule in the 21st century. Heredity 2011, 107, 95–102. [Google Scholar] [CrossRef] [Green Version]
- Borodin, P.; Barreiros-Gomez, S.; Zhelezova, A.; Bonvicino, C.; D’Andrea, P. Reproductive isolation due to the genetic incompatibilities between Thrichomys pachyurus and two subspecies of Thrichomys apereoides (Rodentia, Echimyidae). Genome 2006, 49, 159–167. [Google Scholar] [CrossRef]
- Schulte, U.; Veith, M.; Hochkirch, A. Rapid genetic assimilation of native wall lizard populations (Podarcis muralis) through extensive hybridization with introduced lineages. Mol. Ecol. 2012, 21, 4313–4326. [Google Scholar] [CrossRef]
- Lucchini, V.; Galov, A. Randi Evidence of genetic distinction and long-term population decline in wolves (Canis lupus) in the Italian Apennines. Mol. Ecol. 2004, 13, 523–536. [Google Scholar] [CrossRef]
- Corbin, K.W. Genic Heterozygosity in the White-Crowned Sparrow: A Potential Index to Boundaries between Subspecies. Auk 1981, 98, 669–680. [Google Scholar] [CrossRef]



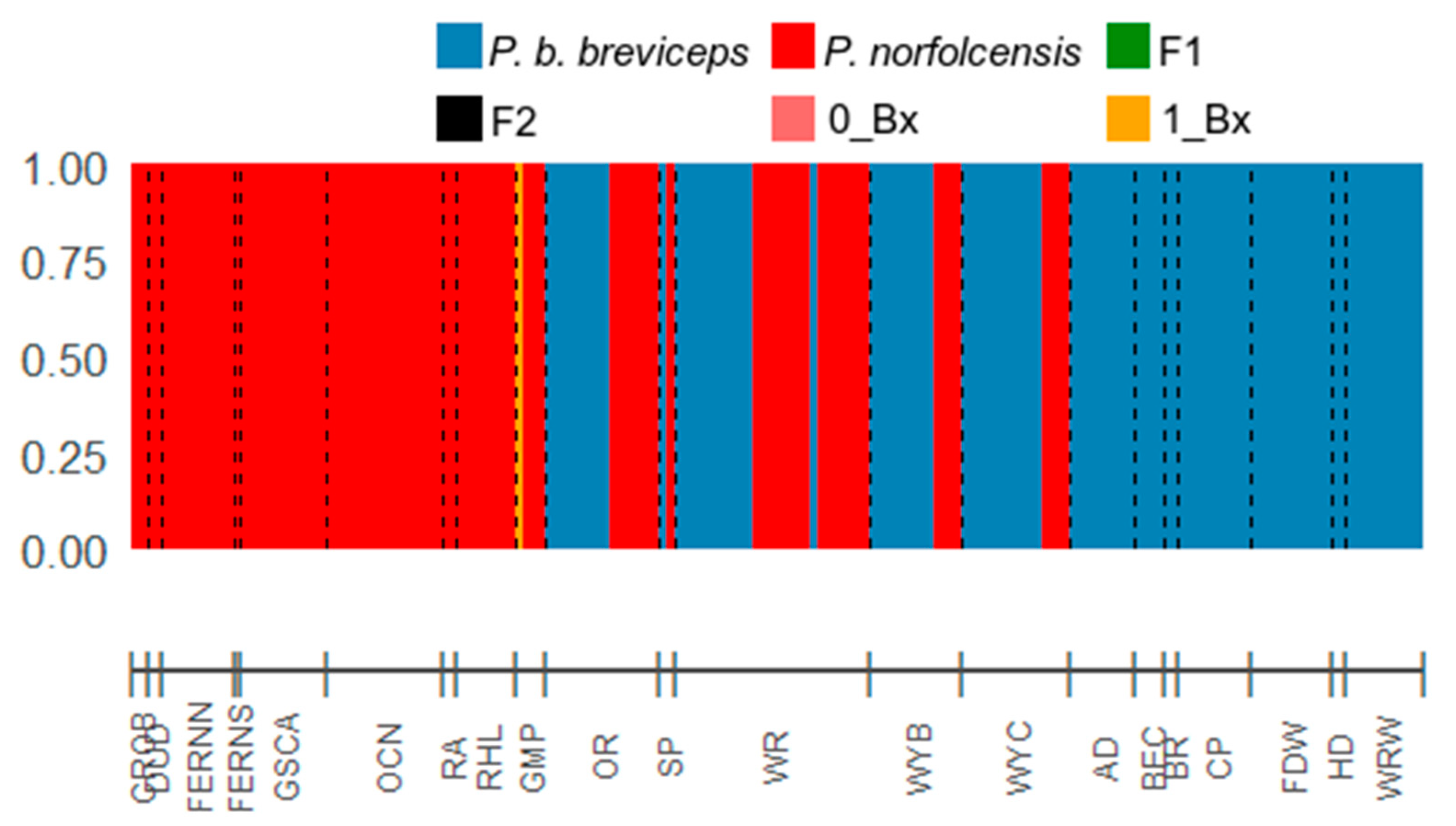
| Genotype Frequency Classes | Product of | Expected Genotype Frequencies | |||
|---|---|---|---|---|---|
| p2 | pq | pq | q2 | ||
| Pure P. b. breviceps | 2 × P. b. breviceps | 1.00 | 0.00 | 0.00 | 0.00 |
| Pure P. norfolcensis | 2 × P. norfolcensis | 0.00 | 0.00 | 0.00 | 1.00 |
| First generation hybrid (F1) | P. b. breviceps × P. norfolcensis | 0.00 | 0.50 | 0.50 | 0.00 |
| Second generation hybrid (F2) | F1 × F1 | 0.25 | 0.25 | 0.25 | 0.25 |
| P. b. breviceps backcross (0_Bx) | F1 × P. b. breviceps | 0.50 | 0.25 | 0.25 | 0.00 |
| P. norfolcensis backcross (1_Bx) | F1 × P. norfolcensis | 0.00 | 0.25 | 0.25 | 0.50 |
| Species | P. b. breviceps | P. norfolcensis | Hybrid |
|---|---|---|---|
| P. b. breviceps | - | * | * |
| P. norfolcensis | 0.767 | - | * |
| Hybrid | 0.671 | 0.346 | - |
| Species | Hs | Ho |
|---|---|---|
| P. b. breviceps | 0.084 | 0.082 |
| P. norfolcensis | 0.092 | 0.096 |
| Hybrid | 0.114 | 0.220 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knipler, M.L.; Dowton, M.; Mikac, K.M. Genome-Wide SNPs Detect Hybridisation of Marsupial Gliders (Petaurus breviceps breviceps × Petaurus norfolcensis) in the Wild. Genes 2021, 12, 1327. https://doi.org/10.3390/genes12091327
Knipler ML, Dowton M, Mikac KM. Genome-Wide SNPs Detect Hybridisation of Marsupial Gliders (Petaurus breviceps breviceps × Petaurus norfolcensis) in the Wild. Genes. 2021; 12(9):1327. https://doi.org/10.3390/genes12091327
Chicago/Turabian StyleKnipler, Monica L., Mark Dowton, and Katarina Maryann Mikac. 2021. "Genome-Wide SNPs Detect Hybridisation of Marsupial Gliders (Petaurus breviceps breviceps × Petaurus norfolcensis) in the Wild" Genes 12, no. 9: 1327. https://doi.org/10.3390/genes12091327
APA StyleKnipler, M. L., Dowton, M., & Mikac, K. M. (2021). Genome-Wide SNPs Detect Hybridisation of Marsupial Gliders (Petaurus breviceps breviceps × Petaurus norfolcensis) in the Wild. Genes, 12(9), 1327. https://doi.org/10.3390/genes12091327







