Abstract
The Amazonian symbol fish Arapaima gigas is the only living representative of the Arapamidae family. Environmental pressures and illegal fishing threaten the species’ survival. To protect wild populations, a national regulation must be developed for the management of A. gigas throughout the Amazon basin. Moreover, the reproductive genetic management and recruitment of additional founders by aquaculture farms are needed to mitigate the damage caused by domestication. To contribute to the sustainable development, we investigated the genetic diversity of wild and cultivated populations of A. gigas and developed a panel composed by 12 microsatellite markers for individual and population genetic tracing. We analyzed 368 samples from three wild and four farmed populations. The results revealed low rates of genetic diversity in all populations, loss of genetic diversity and high inbreeding rates in farmed populations, and genetic structuring among wild and farmed populations. Genetic tracing using the 12 microsatellite markers was effective, and presented a better performance in identifying samples at the population level. The 12-microsatellite panel is appliable to the legal aspects of the trade of the A. gigas, such as origin discrimination, reproductive genetic management by DNA profiling, and evaluation and monitoring of genetic diversity.
1. Introduction
The Amazon symbol fish Arapaima gigas (Schinz, 1822) is the only living representative of the Arapamidae family. It is commonly referred to as pirarucu, a name that comes from two indigenous native terms, pira (which means fish) and urucum (which is a red-colored seasoning called annatto), because of the species’ red tail color [1].
The species was given the title of “Amazonian’s codfish”, and features such as its large size (weighing up to 200 kg) and excellent flavored meat make the A. gigas’ marketing extremely profitable, compared with other species. Additionally, it has great cultural and economic value for riverside communities that survive from the A. gigas’ fishing [2].
The A. gigas’ marketing is mostly supplied by specimens coming from fishing, and the largest fishing landing harbor is located in the state of Amazonas, with production coming from federal and state environmental conservation units, indigenous lands and regions with current fishing agreements due to predatory fishing and risk of extinction of the species. Another smaller portion of A. gigas’ commercial trade originates from farming, in various aquaculture production systems.
According to the last farming census, 3.246 tons of A. gigas meat was produced, which represents 0.43% of all aquaculture production in Brazil [3]. Despite the low production, the species is indicated among the native fish with the greatest potential for aquaculture because of its high growth (reaching 10 kg in one year of cultivation), high fillet yield and its rusticity [4].
In recent years, commercial sales to consumers of A. gigas products and by-products have broadened in Brazil and abroad. The fillet of A. gigas’ meat is already in high demand by the international gourmet market [5]. Additionally, its leather has a high market value for the confection of accessories, such as bags and shoes, and it will likely exceed meat exports in the next years.
In a recent study [6], there is warning of the risk of new threats to wild stocks of A. gigas, due to the increase in commercial activity to mainly serve the rising leather market. Data from the United States Law Enforcement Management Information System (LEMIS) show that, among the incidents involving arapaima leather products, 89% were originated from wild-caught animals, and they all originated from Brazil [6].
It is important to emphasize that wild stocks of A. gigas in Brazil were on the verge of collapse in the 60s and 70s due to overfishing; consequently, part of the genetic diversity has been lost. Despite positive signs of recovery in terms of population size in managed areas, there are still data deficits regarding the diversity and genetic structure of wild populations of A. gigas—until now there is also an absence of these data for the farmed ones. The genetic diversity is the starting point of adaptation, evolution, and survival, without which the population is more susceptible to extinction [7]. In farming conditions, it is the source of genetic variation for future breeding programs [8], thus the knowledge of the genetic diversity is essential for the sustainability of species.
Nowadays, one of the main threats to the wild populations of A. gigas is the illegal trade, by which animals are caught in prohibited areas and traded illegally or, even, with an indication that they come from a farming environment [9]. The Convention on International Trade in Endangered Species of Wild Fauna and Flora—CITES—recommends the use of forensic genetics in surveillance practices as a gold standard against this issue, since genetic evidence has a crucial advantage over other types of documentaries, tangible, physical or biological evidence because it confers accurate confirmation for identifying or tracing [10].
This process is based on the identification of individuals and populations by genetic tracing. At the individual level, the process is based on parental screening for the identification of individuals. At the population level, it is based on identifying allelic frequencies using probabilistic or deterministic approaches [11]. For the A. gigas, the genetic tracing can be useful against the illegal trade through genetic proof of the origin of the marketed animals, and it could also help in the implantation of breeding genetic management in the aquaculture farms based on the identification of DNA profiles.
In studies of diversity and genetic tracing of aquatic species, microsatellite molecular markers are the most used genomic markers, mainly due to their high polymorphism rates, their abundance throughout the genome, and the practicality and affordable cost of analysis using these markers. In this context, aiming to contribute to the economic development and the sustainable management, the objective of this study is to investigate the genetic diversity of wild and cultivated populations of A. gigas and the potential of a panel composed by 12 microsatellite markers, combined in a multiplex-PCR system, for the individual and population genetic tracing of this species.
2. Materials and Methods
2.1. Ethics Statement
This research was approved by the Ethics Committee on Animal Use (CEUA) of the Federal Rural University of Amazon (UFRA), protocol number 055/2017 (CEUA)–23084.017501/2017-02 (UFRA).
2.2. Sample Collection and DNA Extraction
We collected 2 g of muscle tissue of 368 specimens of A. gigas belonging to seven populations, including farmed and wild ones. One-hundred and ninety of these samples were obtained from four farmed populations located at the state of Pará, in the Northern region of Brazil (Tucumã—TUC and Moju—MOJ), and from the states of Maranhão and Ceará, in the Brazilian Northeast (Imperatriz—IMP and Pentecostes—PEN). The other 178 samples come from three wild populations, located at the states of Pará and Amazonas, in the Northern region of Brazil (Mamiráua—MAM, Santarém—SAN and Mexiana—MEX) (Figure 1).
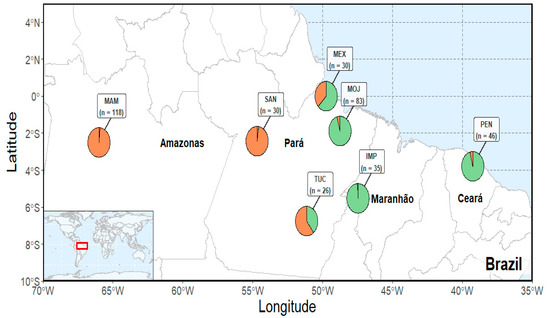
The samplings were preserved in 70% ethanol (Merck KGaA, Darmstadt, Germany) and posteriorly stored in −20 °C. Total genomic DNA was extracted from the digested tissue in proteinase K solution/Sodium Dodecyl Sulfate (SDS) (Thermo Scientific, Waltham, Massachusetts, United States) and purified in Phenol/Chloroform (Invitrogen, Waltham, Massachusetts, United States), followed by precipitation in Isopropanol (Merck KGaA, Darmstadt, Germany) [12]. The DNA concentration was measured in the NanoDrop ND1000 spectrophotometer (Thermo Scientific, Waltham, Massachusetts, United States).
2.3. Amplification and Genotyping of the Microsatellites
Twelve microsatellite loci were analyzed (Agig13519, Agig50571, Agig58115, Agig08356, Agig67103, Agig93614, Agig33291, Agig90836, Agig05001, Agig70664, Agig08912, and Agig06409), developed and selected as the most polymorphic loci for A. gigas by [13], and these loci were mixed in a multiplex-PCR system.
The multiplex PCR was run in a final volume of 9.5 µL, using a 5.0 µL 2X Qiagen Multiplex PCR Master Mix (Qiagen, Hilden, Germany), 1.0 µL of Q-Solution (Qiagen), 2.0 µL of H2O, 0.5 µL of primer mix, and 1.0 µL of genomic DNA, described by [13]. Amplification reactions were performed in a Veriti thermocycler (Applied Biosystems, Waltham, Massachusetts, United States). The thermocycling conditions were as follows: initial denaturation at 95 °C for 15 min, followed by 10 cycles at 94 °C for 30 s, 60 °C for 90 s, and 72 °C for 60 s; 20 cycles at 94 °C for 30 s, 58 °C for 90 s, and 72 °C for 60 s, and a final extension at 72 °C for 60 min, 10 °C for 5 min.
The samples were genotyped in an ABI 3130 Genetic Analyzer (Applied Biosystems Inc., Foster, CA, USA) using a mixture of 1 μL of the PCR product, 8.5 μL of formaldehyde and 0.5 μL of GeneScan 500 LIZ (Applied Biosystems). The determination of fragment size and allele designation was conducted with the GeneMapper 3.7 software (Applied Biosystems).
2.4. Data Analysis
2.4.1. Genetic Diversity, Inbreeding and Population Structure Analyses
The presence of the null allele in the markers was checked using the program Micro-Checker 2.2.3 [14]. The Polymorphic Information Content (PIC) of each marker was calculated in Cervus 3.0 [15], using [16]’s classification system, in which values of less than 0.25 indicate low polymorphism; those of 0.25–0.5, a moderate level of polymorphism, and those higher than 0.5, a highly polymorphic locus.
The genetic diversity indexes, the number of alleles per locus (NA) and the allelic richness (AR) were analyzed in Fstat 2.9.3.2 [17]. The observed heterozygosity (HO), the expected heterozygosity (HE) and Hardy–Weinberg Equilibrium (HWE) was determined in Arlequin 3.5.2.2 [18]. HWE p-values were adjusted by Bonferroni correction [19].
The effective population size (Ne) was estimated in COLONY v2.0.6.4 [20] assuming random mating and a 95% parametric confidence interval. The inbreeding coefficient was estimated as F = ½ NE.
The genetic differentiation among populations was estimated by pairwise FST [21] and RST, calculated in Arlequin 3.5 [18], according to the Hartl and Clark classification system [22]. The existence of genetic substructure and admixture between populations wild and farmed was determined using a Bayesian cluster analysis run in Structure 2.3.4 [23], adjusted by the method proposed by Evanno [24]. Besides, we also examined population structure using a model of multivariate ordination approach for discriminant analysis of principal components (DAPC), using the Adegenet package in R [25].
2.4.2. Individual and Population Genetic Tracin
The capacity for the tracing of the A. gigas was evaluated for both individual and population identification methods. This analysis was based on 1000 genotypes from each population (total of 7000 genotypes), simulated by P-Loci [26] based on the first 10 pairs sampled at seven populations. In this simulation, sex was identified alternately as male or female.
The efficacy of the tracing by the identification of individuals was evaluated using the methods full likelihood, run in COLONY v2.0.6.4 [20]. To identify the population from each individual, we used the Bayesian method of Rannala and Mountain in Geneclass 2.0 [27]. The probability of assigning a specimen to a given population was calculated using the maximum likelihood estimation and expressed by scores.
The performance of each method used to allocate the specimens to their respective populations was evaluated based on the criteria adopted by Larrain et al., 2014 [28], i.e., sensitivity (S), calculated by the number of individuals assigned correctly to their respective population divided by the total number of individuals in this population; specificity (E), that is, the number of individuals correctly excluded from the population divided by the number of individuals that should be excluded from the population, and the average probability assignment score (AP), calculated by the mean likelihood of each successful identification for each population.
3. Results
3.1. Genetic Diversity, Inbreeding and Population Structure
The results of the Micro-Checker analysis indicated that there were null alleles in the markers Agig93614, Agig90836, Agig70664, Agig08912, and Agig06409. The 12-microsatellite system used in this research presented extremely informative PIC average rates for the MAM, SAN, MEX, and TUC populations. PIC moderate rates were observed for the populations of PEN, IMP, and MOJ (Supplementary Table S1).
The indices of genetic variability for each of the 12 microsatellites analyzed for each population are shown in complement Table S1. The 12-loci microsatellite system identified 102 alleles in the seven analyzed populations. The most polymorphic locus was the Agig90836, which presented 13 alleles, while the least polymorphic one was the Agig08912, presenting 5 alleles.
The average NA value varied from 2.83 to 7.08 and the average AR varied from 2.79 to 5.99. (Table 1). Wild populations of MAM (NA = 7.08, AR = 5.86) and SAN (NA = 6.08, AR = 5.99) presented high rates of genetic diversity, in contrast to the farmed population of IMP, which presented the lowest rates (NA = 2.83, AR = 2.79). The remaining populations presented intermediate NA and AR rates (Table 1).

Table 1.
Genetic diversity estimated by 12 microsatellite loci for three wild and four farmed populations of the A. gigas.
The population of SAN was the only one in Hardy–Weinberg Equilibrium. The other populations (MAM, MEX, TUC, PEN, IMP, and MOJ) presented at least one marker in disequilibrium, either for deficiency and/or heterozygosity (Supplementary Table S1). The lowest values of HO and HE were observed for the populations of MOJ (HO = 0.47) and PEN (HE = 0.51), respectively, while the highest values were observed in IMP (HO = 0.71) and TUC (HE = 0.69) (Table 1).
All farmed populations (TUC, PEN, IMP, and MOJ) presented a drastic reduction in NE, and endogamic depletion by F statistics. The wild population of MAM presented a higher value of NE = 168 and a low inbreeding rate, F = 0.003. Critical records of NE and F rates were observed for the farmed populations of IMP, NE = 4 and F = 0.125 (Table 2).

Table 2.
Sample size (N), effective population size (Ne), Confidence Interval (CI) and inbreeding coefficient (F) for the seven populations of the Arapaima gigas analyzed in the present study, based on 12 microsatellite loci.
RST and FST rates above 0.150 revealed high genetic differences among the farmed populations of PEN, IMP, and MOJ when compared to the wild populations of MAM, SAN and MEX. RST and FST rates varying from 0 to 0.050 indicate low differences between MAM and SAN populations, and that both populations are highly different from the MEX one. The comparison among the farmed populations (PEN × IMP × MOJ), revealed FST and RST rates varying from 0.050 to 0.150, indicating that they have a moderate differentiation. The population from TUC presented high differences when compared to the populations from MAM, MEX, PEN, IMP, and MOJ, and moderate difference when compared to SAN (Figure 2).
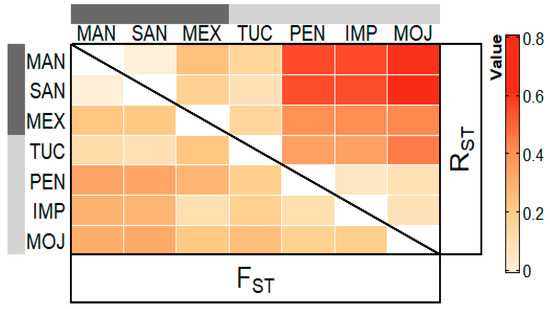
Figure 2.
Heat map of the pairwise FST values (above the diagonal) and the pairwise RST values (below the diagonal) for the three wild and four farmed populations of Arapaima gigas, estimated by microsatellite data.
The Bayesian clustering analysis in STRUCTURE was based on the criteria proposed by [12] and indicated that the seven populations are divided into two different subpopulations (K = 2). The first cluster is formed by the populations MAM, SAN, MEX and TUC. The second cluster is composed of the populations from PEN, IMP, and MOJ. It was not possible to analyze substructure in K = 3 and K = 4 (Figure 3 and Supplementary Figure S2).
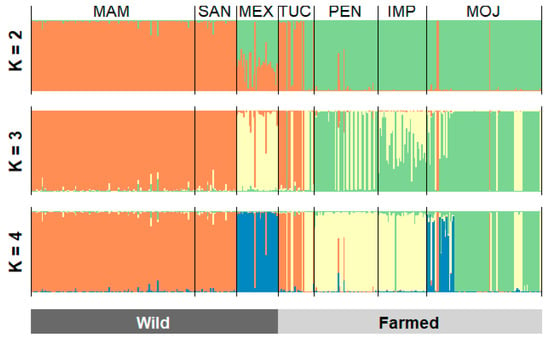
Figure 3.
Standard population structure of three wild and four farmed populations of Arapaima gigas. The analysis based on 12-loci microsatellite markers, supported by STRUCTURE, indicated the existence of two groups (K = 2). MAM—Mamirauá; SAN—Santarém; MEX—Mexiana; TUC—Tucumã; PEN—Pentecostes; IMP—Imperatriz; MOJ—Moju.
DAPC genetic structure analysis reveals that the seven populations are divided into four subpopulations. The first cluster is composed of the wild populations of MAN and SAN. The second group is formed by the farmed populations of IMP, MOJ, and PEN. The population from TUC formed the third cluster, which was genetically closer to the first and second clusters. The fourth group was composed of the wild population from MEX and was genetically more distant from the other ones.
3.2. Individual and Population Genetic Tracing
Individual identification based on the Full Likelihood method was able to correctly identify 100% of the simulated genotypes in populations from SAN and MOJ, by both parental probabilities (maternal and paternal). The other populations, MAM, MEX, TUC, IMP and PEN, had more than 90% of their genotype identified (Table 3).

Table 3.
Individual identification of simulated genotypes of Arapaima gigas from three wild and four farmed populations.
The identification at the population level, using the Bayesian method was able to distinguish 100% of the simulated genotypes for the populations of MEX, TUC and MOJ. The analysis for the populations of MAM, SAN, PEN, and IMP was able to correctly identify more than 99% of the genotypes. Maximum specificity was observed for all populations and maximum sensibility was seen for the populations of MAM, SAN, MEX, TUC, and MOJ. A lower sensibility was observed for the populations of PEN and IMP (Table 4).

Table 4.
Performance of identification of the simulated genotypes of Arapaima gigas from three wild and four farmed populations.
4. Discussion
4.1. Genetic Diversity, Inbreeding and Population Structure
The Amazon basin is home to the largest biodiversity of freshwater fish in the world, there are about 2300 recognized species, corresponding to more than 15% all of the freshwater fish species described on the planet [29], not counting the new ones that are described annually. However, all this aquatic biodiversity is under strong threat due to deforestation, construction of dams and navigable waterways, pollution and overfishing [30,31]. It is believed that anthropogenic impacts have a much greater effect on fish fauna in the Amazon than any predicted climate change [32]. Among the most endangered species, the A. gigas is one of the main sources of food, fishing and trade in riverside communities in the Amazon, and more recently has aroused the interests of aquaculture farming.
Unfortunately, since 1975 A. gigas has been on the IUCN list of endangered species, within the “data deficient” category, meaning there is a lack of data to assess its population status. Thus, tools such as the multiplex microsatellite panel described by [13] are critical for obtaining genetic data and trade monitoring. In the present study, this panel was applied in the analysis of wild and farmed populations of the A. gigas, being efficient in answering questions on genetic diversity and tracing. The PIC values of the 12 combined markers allowed the obtention of accurate data t for statistical analysis.
The seven populations of A. gigas investigated in this study showed low rates of genetic diversity, NA, AR, HO, HE, PIC, NE and F—especially in the NA and AR rates (Table 1), compared to the Colossoma macropomum [33], an Amazonian species that, like the A. gigas, has great economic importance for fishing and aquaculture. Several studies, such as the ones performed by several authors before [34,35,36,37,38,39,40,41], also studied the genetic diversity of wild stocks of the A. gigas in different locations throughout the Amazon basin, presenting different results that range from low to high genetic variability.
A common point among the populations is the record of higher rates of genetic diversity in populations belonging to the Mamirauá reserve, thus reinforcing the importance of the participatory management model used in this region.
Our results warn the loss of genetic diversity in the farmed populations from TUC, IMP, PEN, and MOJ, when compared to the wild ones from MAM, SAN, and MEX (Table 1). We assume that the loss of genetic variation is due to genetic bottlenecks, caused by the domestication process and the founding effect. We reinforce that without sufficient genetic variability, there is always a risk that the population will not be able to respond well to new selective pressures caused by environmental changes [42]. We highlight that in farmed populations, this is an issue for the genetic management.
To decrease the high level of inbreeding recorded in the farmed populations of the A. gigas (Table 2), measures such as the implantation of reproductive genetic management and recruiting additional founders, aiming to increase the genetic variability and minimize inbreeding, must be adopted.
As in all farmed populations, the reduction of NE is observed in the wild populations of MEX (considering the 50 as the minimum NE) [43], meaning they deserve more attention from the competent organizations and entities. This index is seen by conservationists as a measure of the "genetic status" of a population [44], which helps in understanding the genetic health, evaluating the risk of inbreeding and inbreeding depression, thus, also the risk of extinction [45]. For wild populations, we advise the inclusion of NE with the census size for monitoring the effectiveness of species protection actions.
We believe that actions commonly used in fish farming environments, such as consanguineous mating and the low number of breeders, contributed to the high genetic differentiation of the farmed populations from PEN, IMP, and MOJ in comparison to the wild ones from MAM, SAN and MEX (Figure 3). The intense employment of these practices added to the length of time of activity, and the founding effect may justify the moderate differentiation registered among the cultivated populations.
The low differentiation between the wild populations of SAN and MAM (Figure 3), supported by the structure pattern revealed by STRUCUTURE and DAPC analysis (Figure 1 and Figure 2), may reflect a historical gene flow between these areas, even though A. gigas is considered a sedentary species in the literature. Ref. [34], upon finding high gene flow rates among populations of the A. gigas that were up to 1500 km distant, pointed out the important role for juvenile dispersal as a means of conveying gene flow between subpopulations, requiring further studies.
The high differentiation of the wild population of MEX from the MAM and SAN populations (Figure 3) and the structuring analysis by DAPC (Figure 4) reveal a lower gene flow among these wild populations. This is perhaps indicative of a possible process of isolation by distance, due to environmental factors hindering the high rates of migration, and also distinct anthropogenic factors associated with overexploitation [39].
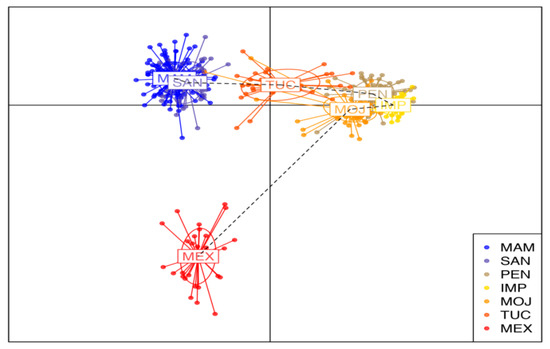
Figure 4.
Scatterplot of Discriminant Analysis of Principle Components (DAPC) of three wild and four farmed populations of Arapaima gigas. Clusters are shown by different colors and inertia ellipses, while dots represent individuals. Wild population: MAM—Mamirauá; SAN—Santarém; MEX—Mexiana. Farmed population: TUC—Tucumã; PEN—Pentecostes; IMP—Imperatriz; MOJ—Moju.
The formation of two clusters (K = 2) in the Bayesian analysis reinforces the loss of genetic diversity in the farmed populations from PEN, IMP, and MOJ, which is reflected in the pattern of distribution of allele frequencies of these populations in a different cluster apart from the wild populations and also from the population from TUC (Figure 1 and Figure 4). We believe the population from TUC, despite being a farmed population, was structured with the wild ones for containing a high representation of wild individuals in its base population.
When analyzing the structuring of wild populations of the A. gigas along the Amazon basin, [34] conclude it is possible to observe changes in the structure of the MEX population, although we have only one species different from what was described by Stewart, 2003 [46]. Our data corroborate these findings (Figure 1), but it is possible to observe changes in the structure of the MEX population (Figure 2 and Figure 4).
In view of the low genetic diversity of the wild populations of A. gigas, we reiterate the importance of measures to control the A. gigas fishing in the state of Amazonas. We also emphasize the need for the authorities responsible for fisheries management of developing a national standard for species management throughout the entire Amazon basin.
4.2. Individual and Population Genetic Tracing
Food traceability is becoming increasingly important for ensuring international food-safety and assisting in the control and sustainability of commercial activities. For this, tools based on DNA analysis have been highlighted, as they present many advantages for DNA being found in all cells—requiring a small amount of material for analysis, and the possibility of the collection at any stage of the production chain [47].
The individual and population discrimination tests used for species tracing can be used in different contexts. For instance, in conservation circumstances, they can be used to assess the genetic impact of domesticated specimens on wild populations. Moreover, the genetic tracing can be used in the combat against illegal fishing through the identification of the origin of seized materials, among others. In aquaculture, these tests are commonly used to track farm fleeting, create genetic relationships maps among breeders, and to identify the farm of origin of contaminated stocks, in cases of detection of diseases or toxins in commercialized fish.
For the A. gigas, identification at the population level had better performance than the individual discrimination, which corroborates the findings of [11,28]. A simulation study reported that at least 15 microsatellites are necessary to correctly reach 95% of confidence in attribution decisions [48]. Based on the populations used in this study, we demonstrated that with 12 microsatellites our results were more than 99% accurate in correctly identifying the genotype of seven populations, at the population level. Furthermore, at the individual level, only one population presented results under 95%. However, the number of markers required for the genetic tracing is different for each species, and it depends on various factors, such as the genetic diversity and population structure [49].
The 12-loci microsatellite panel tested for the A. gigas comes to meet the growing need for reliable, fast and low-cost molecular tools for direct application to improving legal aspects of fish trading, such as the discrimination of farmed individuals from wild ones in/from different locations. For further studies, we recommend an increase in the number of markers on the tested panel, aiming to reach greater accuracy in the analyses, especially in populations that have a lower genetic diversity.
5. Conclusions
Genetic tracing using the 12 microsatellite markers was effective and presented a better performance in identifying samples at the population level. The 12-microsatellite panel is appliable in the legal aspects of the trade of the A. gigas, such as origin discrimination, reproductive genetic management by DNA profiling, and evaluation and monitoring of genetic diversity.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes12091324/s1, Figure S1: Delta K values for STRUCTURE analysis of seven populations of the Arapaima gigas—Delta K, calculated according to Evanno et al. [12], is plotted against the number of modeled gene pools (K). Table S1: Genetic diversity estimated by 12 microsatellite loci for three populations wild of the A. gigas and four farmed.
Author Contributions
Conceptualization, P.F.F.-G. and J.d.P.A.; methodology, P.F.F.-G., J.d.P.A. and C.S.S.; software, D.M., F.C.M.; validation, P.F.F.-G., C.S.S. and J.d.P.A.; formal analysis, P.F.F.-G., D.M. and F.C.M.; investigation, P.F.F.-G.; resources, M.D.N.R., I.H. and S.S.; data curation, P.F.F.-G., D.M, J.d.P.A. and G.F.C.; writing—original draft preparation, P.F.F.-G.; writing—review and editing, P.F.F.-G. and G.F.C.; visualization, S.S., I.H. and M.D.N.R.; supervision, I.H. and S.S.; project administration, S.S. and I.H.; funding acquisition, M.D.N.R., I.H and S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), grant number 165498/2015-3 and The L’Oréal Brazil—2017/01, Edital Mulheres na Ciência 2017.
Institutional Review Board Statement
This research was approved by the Ethics Committee on Animal Use (CEUA) of the Federal Rural University of Amazon (UFRA), protocol number 055/2017 (CEUA)–23084.017501/2017-02 (UFRA).
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Universidade Federal do Pará and Universidade Federal Rural da Amazônia for all the support.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Imbiriba, E.P. potencial de criação de pirarucu, Arapaima gigas, em cativeiro. Acta. Amaz. 2001, 31, 18. [Google Scholar] [CrossRef]
- Gonçalves, A.C.T. O Gigante Amazônico: Manejo Sustentável de Pirarucu = The Amazing Giant: Sustainable Management of Arapaima (Pirarucu); Instituto de Desenvolvimento Sustentável Mamirauá: Tefé, Brasil, 2018; ISBN 978-85-88758-76-6. [Google Scholar]
- ABP—Anuário Brasileiro da Piscicultura PEIXE BR 2020. Available online: https://www.peixebr.com.br/anuario-2020/ (accessed on 15 February 2020).
- Brandão, F.R.; Gomes, L.D.C.; Crescêncio, R.; Carvalho, E.D.S. Uso de sal durante o transporte de juvenis (1 kg) de pirarucu (Arapaima gigas). Acta. Amaz. 2008, 38, 767–771. [Google Scholar] [CrossRef]
- FAO. Fisheries & Aquaculture: Cultured Aquatic Species Information Programme—Arapaima Gigas (Schinz, 1822). Available online: http://www.fao.org/fishery/culturedspecies/Arapaima_gigas/en (accessed on 26 November 2019).
- Heinrich, S.; Ross, J.V.; Cassey, P. Of Cowboys, fish, and pangolins: US trade in exotic leather. Conserv. Sci. Pr. 2019, 1, 75. [Google Scholar] [CrossRef]
- Väli, U.; Einarsson, A.; Waits, L.; Ellegren, H. to what extent do microsatellite markers reflect genome-wide genetic diversity in natural populations? Mol. Ecol. 2008, 17, 3808–3817. [Google Scholar] [CrossRef] [PubMed]
- Groeneveld, L.F.; Lenstra, J.A.; Eding, H.; Toro, M.A.; Scherf, B.; Pilling, D.; Negrini, R.; Finlay, E.; Jianlin, H.; Groeneveld, E.; et al. Genetic diversity in farm animals—A review. Anim. Genet. 2010, 41, 6–31. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, E.S. Manejo Comunitário do Pirarucu Arapaima gigas na Reserva de Desenvolvimento Sustentável Mamirauá Amazonas, Brasil; IBAMA: Bonn, Germany, 2007; pp. 239–261. [Google Scholar]
- Kanthaswamy, S. Review: Domestic animal forensic genetics—biological evidence, genetic markers, analytical approaches and challenges. Anim. Genet. 2015, 46, 473–484. [Google Scholar] [CrossRef]
- Aguiar, J.D.P.; Fazzi-Gomes, P.F.; Hamoy, I.G.; Dos Santos, S.E.; Sampaio, I.; Dos Santos, S.E.B. Tracing individuals and populations of the tambaqui, Colossoma macropomum (Cuvier, 1818), from Brazilian hatcheries using microsatellite markers. J. Sci. Food Agric. 2019, 99, 2998–3004. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.F.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989; Available online: https://www.sigmaaldrich.com/catalog/product/sigma/m8265 (accessed on 21 October 2019).
- Fazzi-Gomes, P.; Aguiar, J.; Cabral, G.F.; Marques, D.; Palheta, H.; Moreira, F.; Rodrigues, M.; Cavalcante, R.; Souza, J.; Silva, C.; et al. Genomic approach for conservation and the sustainable management of endangered species of the Amazon. PLoS ONE 2021, 16, e0240002. [Google Scholar] [CrossRef]
- Oosterhout, C.V.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. micro-checker: Software for identifying and correcting geno-typing errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program cervus accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Goudet, J.; Goudet, J. FSTAT, a Program to Estimate and Test Gene Diversities and Fixation Indices (Version 2.9.3). J. Goudet. 2001. [Google Scholar]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Rice, W.R. Analyzing Tables of Statistical Tests. Evolution 1989, 43, 223–225. [Google Scholar] [CrossRef]
- Jones, O.R.; Wang, J. COLONY: A program for parentage and sibship inference from multilocus genotype data. Mol. Ecol. Resour. 2010, 10, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the Analysis of Population Structure. Evolution 1984, 38, 1358. [Google Scholar] [PubMed]
- Hartl, D.L.; Clark, A.G. Princípios de Genética de Populações; Artmed Editora: Marnate, Varese, Italy, 2010; ISBN 978-85-363-2374-9. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of ge-netically structured populations. BMC Genet 2010, 11, 94. [Google Scholar] [CrossRef]
- Matson, S.E.; Camara, M.D.; Eichert, W.; Banks, M.A. P-LOCI: A computer program for choosing the most efficient set of loci for parentage assignment. Mol. Ecol. Resour. 2016, 16, 599. [Google Scholar] [CrossRef]
- Piry, S.; Alapetite, A.; Cornuet, J.-M.; Paetkau, D.; Baudouin, L.; Estoup, A. GENECLASS2: A Software for Genetic Assignment and First-Generation Migrant Detection. J. Hered. 2004, 95, 536–539. [Google Scholar] [CrossRef] [PubMed]
- Larraín, M.A.; Díaz, N.F.; Lamas, C.; Uribe, C.; Araneda, C. Traceability of mussel (Mytilus chilensis) in southern Chile using microsatellite molecular markers and assignment algorithms. Exploratory survey. Food Res. Int. 2014, 62, 104–110. [Google Scholar] [CrossRef]
- Darias, M.J.; Amadio, S.A.; Rosenthal, H. The Research Network on Amazonian Ichthyofauna. J. Appl. Ichthyol. 2015, 31, 1–3. [Google Scholar] [CrossRef]
- Castello, L.; Stewart, D.J.; Arantes, C.C. O que Sabemos e Precisamos Fazer a Respeito da Conservação do Pirarucu (Arapaima spp.) na Amazônia in Figueiredo, Ellen Sílvia Amaral (Org.) Biologia, Conservação e Manejo Participativo de Pirarucus na Pan-Amazônia; Organizado por Ellen Amaral: Tefé, Brazil, 2013; p. 278. [Google Scholar]
- Hurt, C.; Hedrick, P. Conservation genetics in aquatic species: General approaches and case studies in fishes and springsnails of arid lands. Aquat. Sci. 2004, 66, 402–413. [Google Scholar] [CrossRef]
- Oberdorff, T.; Jézéquel, C.; Campero, M.; Carvajal-Vallejos, F.; Cornu, J.F.; Dias, M.; Duponchelle, F.; Ocampo, J.A.M.; Ortega, H.; Renno, J.F.; et al. Opinion Paper: How vulnerable are Amazonian freshwater fishes to ongoing climate change? J. Appl. Ichthyol. 2015, 31, 4–9. [Google Scholar] [CrossRef]
- Fazzi-Gomes, P.; Guerreiro, S.; Palheta, G.D.A.; Melo, N.F.A.C.; de Santos, S.; Hamoy, I. High genetic diversity and connectivity in Colossoma macropomum in the Amazon basin revealed by microsatellite markers. Genet. Mol. Biol. 2017, 40, 142–146. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Farias, I.P.; Willis, S.; Leão, A.; Verba, J.T.; Crossa, M.; Foresti, F.; Porto-Foresti, F.; Sampaio, I.; Hrbek, T. The largest fish in the world’s biggest river: Genetic connectivity and conservation of Arapaima gigas in the Amazon and Araguaia-Tocantins drainages. PLoS ONE 2019, 14, e0220882. [Google Scholar] [CrossRef]
- Fazzi-Gomes, P.; Melo, N.; Palheta, G.; Guerreiro, S.; Amador, M.; Ribeiro-Dos-Santos, A.; Santos, S.; Hamoy, I. Rapid-communication Genetic diversity and differentiation in natural populations of Arapaima gigas from lower Amazon revealed by microsatellites. Genet. Mol. Res. 2017, 16. [Google Scholar] [CrossRef]
- Vitorino, C.A.; Nogueira, F.; Souza, I.L.; Araripe, J.; Venere, P.C. Low Genetic Diversity and Structuring of the Arapaima (Osteoglossiformes, Arapaimidae) Population of the Araguaia-Tocantins Basin. Front. Genet. 2017, 8, 159. [Google Scholar] [CrossRef] [PubMed]
- Araripe, J.; Rêgo, P.S.D.; Queiroz, H.; Sampaio, I.; Schneider, H. Dispersal Capacity and Genetic Structure of Arapaima gigas on Different Geographic Scales Using Microsatellite Markers. PLoS ONE 2013, 8, e54470. [Google Scholar] [CrossRef] [PubMed]
- Hamoy, I.G.; Santos, E.J.M.; Santos, S.E.B. Rapid and inexpensive analysis of genetic variability in Arapaima gigas by PCR multiplex panel of eight microsatellites. Genet. Mol. Res. 2008, 7, 29–32. [Google Scholar] [CrossRef]
- Hrbek, T.; Crossa, M.; Farias, I. Conservation strategies for Arapaima gigas (Schinz, 1822) and the Amazonian várzea ecosystem. Braz. J. Biol. 2007, 67, 909–917. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hrbek, T.; Farias, I.; Crossa, M.; Sampaio, I.; Porto, J.; Meyer, A. Population genetic analysis of Arapaima gigas, one of the largest freshwater fishes of the Amazon basin: Implications for its conservation. Anim. Conserv. 2005, 8, 297–308. [Google Scholar] [CrossRef]
- Farias, I.P.; Hrbek, T.; Brinkmann, H.; Sampaio, I.; Meyer, A. Characterization and isolation of DNA microsatellite primers for Arapaima gigas, an economically important but severely over-exploited fish species of the Amazon basin. Mol. Ecol. Notes 2003, 3, 128–130. [Google Scholar] [CrossRef]
- Pray, L. Genetic drift: Bottleneck effect and the case of the bearded vulture. Nat. Educ. 2008, 1, 61. [Google Scholar]
- Gjedrem, T.; Baranski, M. Selective Breeding in Aquaculture: An Introduction; Springer: Berlin, Germany, 2009; p. 221. [Google Scholar]
- Hamblin, M.T.; Buckler, E.; Jannink, J.-L. Population genetics of genomics-based crop improvement methods. Trends Genet. 2011, 27, 98–106. [Google Scholar] [CrossRef]
- Wang, Y.-Z.; Cao, L.-J.; Zhu, J.-Y.; Wei, S.-J. Development and Characterization of Novel Microsatellite Markers for the Peach Fruit Moth Carposina sasakii (Lepidoptera: Carposinidae) Using Next-Generation Sequencing. Int. J. Mol. Sci. 2016, 17, 362. [Google Scholar] [CrossRef]
- Stewart, D.J. A New Species ofArapaima(Osteoglossomorpha: Osteoglossidae) from the Solimões River, Amazonas State, Brazil. Copeia 2013, 2013, 470–476. [Google Scholar] [CrossRef]
- Nielsen, R.; Paul, J.S.; Albrechtsen, A.; Song, Y.S. Genotype and SNP calling from next-generation sequencing data. Nat. Rev. Genet. 2011, 12, 443–451. [Google Scholar] [CrossRef]
- Hayes, B.; Sonesson, A.K.; Gjerde, B. Evaluation of three strategies using DNA markers for traceability in aquaculture species. Aquaculture 2005, 250, 70–81. [Google Scholar] [CrossRef]
- Hansen, M.M.; Kenchington, E.; Nielsen, E.E. Assigning individual fish to populations using microsatellite DNA markers. Fish Fish. 2001, 2, 93–112. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).