Glaucoma Heritability: Molecular Mechanisms of Disease
Abstract
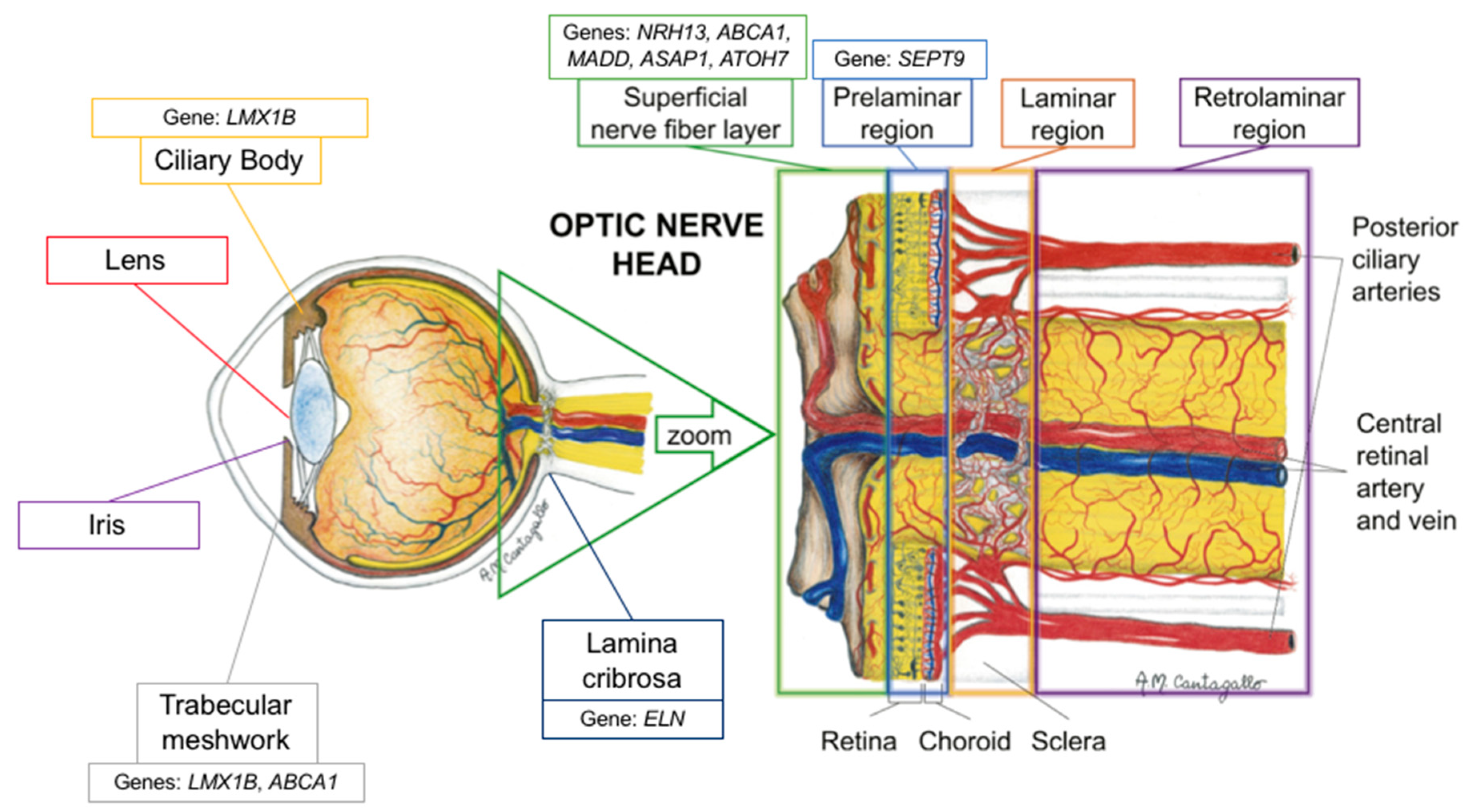
1. Introduction
2. Materials and Methods
3. Discussion
3.1. Intraocular Pressure
3.2. Vertical Cup-to-Disc Ratio
4. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Quigley, H.A.; Broman, A.T. The number of people with glaucoma worldwide in 2010 and 2020. Br. J. Ophthalmol. 2006, 90, 262–267. [Google Scholar] [CrossRef]
- Tham, Y.C.; Li, X.; Wong, T.Y.; Quigley, H.A.; Aung, T.; Cheng, C.Y. Global prevalence of glaucoma and projections of glaucoma burden through 2040: A systematic review and meta-analysis. Ophthalmology 2014, 121, 2081–2090. [Google Scholar] [CrossRef] [PubMed]
- Weinreb, R.N.; Aung, T.; Medeiros, F.A. The Pathophysiology and Treatment of Glaucoma: A Review. JAMA 2014, 311, 1901–1911. [Google Scholar] [CrossRef] [PubMed]
- Boland, M.V.; Ervin, A.M.; Friedman, D.S.; Jampel, H.D.; Hawkins, B.S.; Vollenweider, D.; Chelladurai, Y.; Ward, D.; Suarez-Cuervo, C.; Robinson, K.A. Comparative effectiveness of treatments for open-angle glaucoma: A systematic review for the U.S. Preventive Services Task Force. Ann. Intern. Med. 2013, 158, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Zukerman, R.; Harris, A.; Verticchio Vercellin, A.; Siesky, B.; Pasquale, L.R.; Ciulla, T.A. Molecular Genetics of Glaucoma: Subtype and Ethnicity Considerations. Genes 2021, 12, 55. [Google Scholar] [CrossRef] [PubMed]
- Tenesa, A.; Haley, C.S. The heritability of human disease: Estimation, uses and abuses. Nat. Rev. Genet. 2013, 14, 139–149. [Google Scholar] [CrossRef]
- Haworth, C.M.; Davis, O.S. From observational to dynamic genetics. Front. Genet. 2014, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Asefa, N.G.; Neustaeter, A.; Jansonius, N.M.; Snieder, H. Heritability of glaucoma and glaucoma-related endophenotypes: Systematic review and meta-analysis. Surv. Ophthalmol. 2019, 64, 835–851. [Google Scholar] [CrossRef]
- Charlesworth, J.; Kramer, P.L.; Dyer, T.; Diego, V.; Samples, J.R.; Craig, J.E.; Mackey, D.A.; Hewitt, A.W.; Blangero, J.; Wirtz, M.K. The path to open-angle glaucoma gene discovery: Endophenotypic status of intraocular pressure, cup-to-disc ratio, and central corneal thickness. Invest. Ophthalmol. Vis. Sci. 2010, 51, 3509–3514. [Google Scholar] [CrossRef]
- Bonnemaijer, P.W.M.; Leeuwen, E.M.V.; Iglesias, A.I.; Gharahkhani, P.; Vitart, V.; Khawaja, A.P.; Simcoe, M.; Höhn, R.; Cree, A.J.; Igo, R.P.; et al. Multi-trait genome-wide association study identifies new loci associated with optic disc parameters. Commun. Biol. 2019, 2, 435. [Google Scholar] [CrossRef]
- Medeiros, F.A.; Alencar, L.M.; Zangwill, L.M.; Sample, P.A.; Weinreb, R.N. The Relationship between intraocular pressure and progressive retinal nerve fiber layer loss in glaucoma. Ophthalmology 2009, 116, 1125–1133. [Google Scholar] [CrossRef]
- Quigley, H.A.; Addicks, E.M.; Green, W.R.; Maumenee, A.E. Optic nerve damage in human glaucoma. II. The site of injury and susceptibility to damage. Arch. Ophthalmol. 1981, 99, 635–649. [Google Scholar] [CrossRef]
- Ju, W.K.; Kim, K.Y.; Lindsey, J.D.; Angert, M.; Duong-Polk, K.X.; Scott, R.T.; Kim, J.J.; Kukhmazov, I.; Ellisman, M.H.; Perkins, G.A.; et al. Intraocular pressure elevation induces mitochondrial fission and triggers OPA1 release in glaucomatous optic nerve. Invest. Ophthalmol. Vis. Sci. 2008, 49, 4903–4911. [Google Scholar] [CrossRef] [PubMed]
- Moore, D.; Harris, A.; Wudunn, D.; Kheradiya, N.; Siesky, B. Dysfunctional regulation of ocular blood flow: A risk factor for glaucoma? Clin. Ophthalmol. 2008, 2, 849–861. [Google Scholar] [PubMed]
- Aschard, H.; Kang, J.H.; Iglesias, A.I.; Hysi, P.; Cooke Bailey, J.N.; Khawaja, A.P.; Allingham, R.R.; Ashley-Koch, A.; Lee, R.K.; Moroi, S.E.; et al. Genetic correlations between intraocular pressure, blood pressure and primary open-angle glaucoma: A multi-cohort analysis. Eur. J. Hum. Genet. 2017, 25, 1261–1267. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.R.; Huang, H.; Nannini, D.R.; Fan, F.; Kim, H. Genome-wide association analyses identify new loci influencing intraocular pressure. Hum. Mol. Genet. 2018, 27, 2205–2213. [Google Scholar] [CrossRef]
- Pressman, C.L.; Chen, H.; Johnson, R.L. LMX1B, a LIM homeodomain class transcription factor, is necessary for normal development of multiple tissues in the anterior segment of the murine eye. Genesis 2000, 26, 15–25. [Google Scholar] [CrossRef]
- Tezel, G.; Wax, M.B. Increased production of tumor necrosis factor-α by glial cells exposed to simulated ischemia or elevated hydrostatic pressure induces apoptosis in cocultured retinal ganglion cells. J. Neurosci. 2000, 20, 8693–8700. [Google Scholar] [CrossRef]
- Hu, C.; Niu, L.; Li, L.; Song, M.; Zhang, Y.; Lei, Y.; Chen, Y.; Sun, X. ABCA1 Regulates IOP by Modulating Cav1/eNOS/NO Signaling Pathway. Invest. Ophthalmol. Vis. Sci. 2020, 61, 33. [Google Scholar] [CrossRef]
- Ou, B.; Ohno, S.; Tsukahara, S. Ultrastructural changes and immunocytochemical localization of microtubule-associated protein 1 in guinea pig optic nerves after acute increase in intraocular pressure. Invest. Ophthalmol. Vis. Sci. 1998, 39, 963–971. [Google Scholar]
- Ding, Y.Q.; Marklund, U.; Yuan, W.; Yin, J.; Wegman, L.; Ericson, J.; Deneris, E.; Johnson, R.L.; Chen, Z.F. Lmx1b is essential for the development of serotonergic neurons. Nat. Neurosci. 2003, 6, 933–938. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xu, L.; Chen, W.; Li, X.; Xia, Q.; Zheng, L.; Duan, Q.; Zhang, H.; Zhao, Y. Reduced Annexin A1 Secretion by ABCA1 Causes Retinal Inflammation and Ganglion Cell Apoptosis in a Murine Glaucoma Model. Front. Cell Neurosci. 2018, 12, 347. [Google Scholar] [CrossRef]
- Yeghiazaryan, K.; Flammer, J.; Wunderlich, K.; Schild, H.H.; Orgul, S.; Golubnitschaja, O. An enhanced expression of ABC 1 transporter in circulating leukocytes as a potential molecular marker for the diagnostics of glaucoma. Amino Acids 2005, 28, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Song, W.T.; Zhang, X.Y.; Xia, X.B. Atoh7 promotes the differentiation of Müller cells-derived retinal stem cells into retinal ganglion cells in a rat model of glaucoma. Exp. Biol. Med. 2015, 240, 682–690. [Google Scholar] [CrossRef]
- Miesfeld, J.B.; Ghiasvand, N.M.; Marsh-Armstrong, B.; Marsh-Armstrong, N.; Miller, E.B.; Zhang, P.; Manna, S.K.; Zawadzki, R.J.; Brown, N.L.; Glaser, T. The Atoh7 remote enhancer provides transcriptional robustness during retinal ganglion cell development. Proc. Natl. Acad. Sci. USA 2020, 117, 21690–21700. [Google Scholar] [CrossRef] [PubMed]
- Gelman, S.; Cone, F.E.; Pease, M.E.; Nguyen, T.D.; Myers, K.; Quigley, H.A. The presence and distribution of elastin in the posterior and retrobulbar regions of the mouse eye. Exp. Eye Res. 2010, 90, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Cross, S.H.; Macalinao, D.G.; McKie, L.; Rose, L.; Kearney, A.L.; Rainger, J.; Thaung, C.; Keighren, M.; Jadeja, S.; West, K.; et al. A Dominant-Negative Mutation of Mouse Lmx1b Causes Glaucoma and Is Semi-lethal via LBD1-Mediated Dimerisation. PLoS Genet 2014, 10, e1004359. [Google Scholar] [CrossRef] [PubMed]
- Schievella, A.R.; Chen, J.H.; Graham, J.R.; Lin, L.L. MADD, a novel death domain protein that interacts with the type 1 tumor necrosis factor receptor and activates mitogen-activated protein kinase. J. Biol. Chem. 1997, 272, 12069–12075. [Google Scholar] [CrossRef]
- Wang, L.; Schuster, G.U.; Hultenby, K.; Zhang, Q.; Andersson, S.; Gustafsson, J.A. Liver X receptors in the central nervous system: From lipid homeostasis to neuronal degeneration. Proc. Natl. Acad. Sci. USA 2002, 99, 13878–13883. [Google Scholar] [CrossRef]
- Yang, H.; Zheng, S.; Qiu, Y.; Yang, Y.; Wang, C.; Yang, P.; Li, Q.; Lei, B. Activation of liver X receptor alleviates ocular inflammation in experimental autoimmune uveitis. Invest. Ophthalmol. Vis. Sci. 2014, 55, 2795–2804. [Google Scholar] [CrossRef]
- Zheng, S.; Yang, H.; Chen, Z.; Zheng, C.; Lei, C.; Lei, B. Activation of Liver X Receptor Protects Inner Retinal Damage Induced by N-methyl-D-aspartate. Invest. Ophthalmol. Vis. Sci. 2015, 56, 1168–1180. [Google Scholar] [CrossRef]
- Song, X.Y.; Wu, W.F.; Gabbi, C.; Dai, Y.B.; So, M.; Chaurasiya, S.P.; Wang, L.; Warner, M.; Gustafsson, J. Retinal and optic nerve degeneration in liver X receptor β knockout mice. Proc. Natl. Acad. Sci. USA 2019, 116, 16507–16512. [Google Scholar] [CrossRef]
- Ghossoub, R.; Hu, Q.; Failler, M.; Rouyez, M.C.; Spitzbarth, B.; Mostowy, S.; Wolfrum, U.; Saunier, S.; Cossart, P.; Jamesnelson, W.; et al. Septins 2, 7 and 9 and MAP4 colocalize along the axoneme in the primary cilium and control ciliary length. J. Cell Sci. 2013, 126, 2583–2594. [Google Scholar]
- Mimiwati, Z.; Mackey, D.A.; Craig, J.E.; Mackinnon, J.R.; Rait, J.L.; Liebelt, J.E.; Ayala-Lugo, R.; Vollrath, D.; Richards, J.E. Nail-patella syndrome and its association with glaucoma: A review of eight families. Br. J. Ophthalmol. 2006, 90, 1505–1509. [Google Scholar] [CrossRef]
- Vollrath, D.; Jaramillo-Babb, V.L.; Clough, M.V.; McIntosh, I.; Scott, K.M.; Lichter, P.R.; Richards, J.E. Loss-of-function mutations in the LIM-homeodomain gene, LMX1B, in nail-patella syndrome. Hum. Mol. Genet. 1998, 7, 1091–1098. [Google Scholar] [CrossRef]
- Park, S.; Jamshidi, Y.; Vaideanu, D.; Bitner-Glindzicz, M.; Fraser, S.; Sowden, J.C. Genetic risk for primary open-angle glaucoma determined by LMX1B haplotypes. Invest. Ophthalmol. Vis. Sci. 2009, 50, 1522–1530. [Google Scholar] [CrossRef] [PubMed]
- Choquet, H.; Paylakhi, S.; Kneeland, S.C.; Thai, K.K.; Hoffmann, T.J.; Yin, J.; Kvale, M.N.; Banda, Y.; Tolman, N.G.; Williams, P.A.; et al. A multiethnic genome-wide association study of primary open-angle glaucoma identifies novel risk loci. Nat. Commun. 2018, 9, 2278. [Google Scholar] [CrossRef] [PubMed]
- Gharahkhani, P.; Burdon, K.P.; Fogarty, R.; Sharma, S.; Hewitt, A.W.; Martin, S.; Law, M.H.; Cremin, K.; Bailey, J.N.C.; Loomis, S.J.; et al. Common variants near ABCA1, AFAP1 and GMDS confer risk of primary open-angle glaucoma. Nat. Genet. 2014, 46, 1120–1125. [Google Scholar] [CrossRef] [PubMed]
- Hysi, P.G.; Cheng, C.Y.; Springelkamp, H.; Macgregor, S.; Bailey, J.N.C.; Wojciechowski, R.; Vitart, V.; Nag, A.; Hewitt, A.W.; Höhn, R.; et al. Genome-wide analysis of multi-ancestry cohorts identifies new loci influencing intraocular pressure and susceptibility to glaucoma. Nat. Genet. 2014, 46, 1126–1130. [Google Scholar] [CrossRef]
- Wang, S.; Smith, J.D. ABCA1 and nascent HDL biogenesis. Biofactors 2014, 40, 547–554. [Google Scholar] [CrossRef]
- Hozoji, M.; Munehira, Y.; Ikeda, Y.; Makishima, M.; Matsuo, M.; Kioka, N.; Ueda, K. Direct interaction of nuclear liver X receptor-β with ABCA1 modulates cholesterol efflux. J. Biol. Chem. 2008, 283, 30057–30063. [Google Scholar] [CrossRef]
- Yang, X.; Luo, C.; Cai, J.; Powell, D.W.; Yu, D.; Kuehn, M.H.; Tezel, G. Neurodegenerative and inflammatory pathway components linked to TNF-α/TNFR1 signaling in the glaucomatous human retina. Invest. Ophthalmol. Vis. Sci. 2011, 52, 8442–8454. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Tezel, G.; Wax, M.B.; Edward, D.P. Matrix metalloproteinases and tumor necrosis factor α in glaucomatous optic nerve head. Arch. Ophthalmol. 2000, 118, 666–673. [Google Scholar] [CrossRef]
- Tezel, G.; Li, L.Y.; Patil, R.V.; Wax, M.B. TNF-α and TNF-α receptor-1 in the retina of normal and glaucomatous eyes. Invest. Ophthalmol. Vis. Sci. 2001, 42, 1787–1794. [Google Scholar] [PubMed]
- Yang, Z.; Quigley, H.A.; Pease, M.E.; Yang, Y.; Qian, J.; Valenta, D.; Zack, D.J. Changes in gene expression in experimental glaucoma and optic nerve transection: The equilibrium between protective and detrimental mechanisms. Investig. Ophthalmol. Vis. Sci. 2007, 48, 5539–5548. [Google Scholar] [CrossRef]
- Yuan, L.; Neufeld, A.H. Activated microglia in the human glaucomatous optic nerve head. J. Neurosci. Res. 2001, 64, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, T.; Nakazawa, C.; Matsubara, A.; Noda, K.; Hisatomi, T.; She, H.; Michaud, N.; Hafezi-Moghadam, A.; Miller, J.W.; Benowitz, L.I. Tumor necrosis factor-α mediates oligodendrocyte death and delayed retinal ganglion cell loss in a mouse model of glaucoma. J. Neurosci. 2006, 26, 12633–12641. [Google Scholar] [CrossRef]
- Agarwal, R.; Agarwal, P. Glaucomatous neurodegeneration: An eye on tumor necrosis factor-α. Indian J. Ophthalmol. 2012, 60, 255–261. [Google Scholar] [CrossRef]
- Sawada, H.; Fukuchi, T.; Tanaka, T.; Abe, H. Tumor Necrosis Factor-α Concentrations in the Aqueous Humor of Patients with Glaucoma. Invest. Ophthalmol. Vis. Sci. 2010, 51, 903–906. [Google Scholar] [CrossRef] [PubMed]
- Bridges, A.A.; Gladfelter, A.S. Septin Form and Function at the Cell Cortex. J. Biol. Chem. 2015, 290, 17173–17180. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.R. Normal-tension glaucoma (Low-tension glaucoma). Indian J. Ophthalmol. 2011, 59 (Suppl. 1), S97–S101. [Google Scholar] [CrossRef]
- Weisman, R.L.; Asseff, C.F.; Phelps, C.D.; Podos, S.M.; Becker, B. Vertical elongation of the optic cup in glaucoma. Trans. Am. Acad. Ophthalmol. Otolaryngol. 1973, 77, OP157–OP161. [Google Scholar]
- Tatham, A.J.; Weinreb, R.N.; Zangwill, L.M.; Liebmann, J.M.; Girkin, C.A.; Medeiros, F.A. The relationship between cup-to-disc ratio and estimated number of retinal ganglion cells. Invest. Ophthalmol. Vis. Sci. 2013, 54, 3205–3214. [Google Scholar] [CrossRef]
- Hopley, C.R.; Crowston, J.; Healey, P.R.; Lee, A.; Rochtchina, E.; Mitchell, P.; Study, B.M.E. Vertical Cup: Disc Ratio in a Non-glaucomatous Population-based Sample: The Blue Mountains Eye Study. Invest. Ophthalmol. Vis. Sci. 2003, 44, 3408. [Google Scholar]
- Hollands, H.; Johnson, D.; Hollands, S.; Simel, D.L.; Jinapriya, D.; Sharma, S. Do findings on routine examination identify patients at risk for primary open-angle glaucoma? The rational clinical examination systematic review. JAMA 2013, 309, 2035–2042. [Google Scholar] [CrossRef] [PubMed]
- Nannini, D.R.; Torres, M.; Chen, Y.I.; Taylor, K.D.; Rotter, J.I.; Varma, R.; Gao, X. A Genome-Wide Association Study of Vertical Cup-Disc Ratio in a Latino Population. Invest. Ophthalmol. Vis. Sci. 2017, 58, 87–95. [Google Scholar] [CrossRef]
- Ramdas, W.D.; van Koolwijk, L.M.; Ikram, M.K.; Jansonius, N.M.; de Jong, P.T.; Bergen, A.A.; Isaacs, A.; Amin, N.; Aulchenko, Y.S.; Wolfs, R.C.; et al. A genome-wide association study of optic disc parameters. PLoS Genet. 2010, 6, e1000978. [Google Scholar] [CrossRef]
- Springelkamp, H.; Höhn, R.; Mishra, A.; Hysi, P.G.; Khor, C.-C.; Loomis, S.J.; Bailey, J.N.C.; Gibson, J.; Thorleifsson, G.; Janssen, S.F.; et al. Meta-analysis of genome-wide association studies identifies novel loci that influence cupping and the glaucomatous process. Nat. Commun. 2014, 5, 4883. [Google Scholar] [CrossRef]
- Springelkamp, H.; Iglesias, A.I.; Mishra, A.; Höhn, R.; Wojciechowski, R.; Khawaja, A.P.; Nag, A.; Wang, Y.X.; Wang, J.J.; Cuellar-Partida, G.; et al. New insights into the genetics of primary open-angle glaucoma based on meta-analyses of intraocular pressure and optic disc characteristics. Hum. Mol. Genet. 2017, 26, 438–453. [Google Scholar] [CrossRef] [PubMed]
- Nannini, D.R.; Kim, H.; Fan, F.; Gao, X. Genetic Risk Score Is Associated with Vertical Cup-to-Disc Ratio and Improves Prediction of Primary Open-Angle Glaucoma in Latinos. Ophthalmology 2018, 125, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Steven, K.; Qassim, A.; Marshall, H.N.; Bean, C.; Tremeer, M.; An, J.; Siggs, O.M.; Gharahkhani, P.; Craig, J.E.; et al. Automated AI labeling of optic nerve head enables insights into cross-ancestry glaucoma risk and genetic discovery in >280,000 images from UKB and CLSA. Am. J. Hum. Genet. 2021, 108, 1204–1216. [Google Scholar] [CrossRef]
- Han, X.; Qassim, A.; An, J.; Marshall, H.; Zhou, T.; Ong, J.S.; Hassall, M.M.; Hysi, P.G.; Foster, P.J.; Khaw, P.T.; et al. Genome-wide association analysis of 95 549 individuals identifies novel loci and genes influencing optic disc morphology. Hum. Mol. Genet. 2019, 28, 3680–3690. [Google Scholar] [CrossRef]
- Pers, T.H.; Karjalainen, J.M.; Chan, Y.; Westra, H.J.; Wood, A.R.; Yang, J.; Lui, J.C.; Vedantam, S.; Gustafsson, S.; Esko, T.; et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 2015, 6, 5890. [Google Scholar] [CrossRef]
- Chen, Y.; Lin, Y.; Vithana, E.N.; Jia, L.; Zuo, X.; Wong, T.Y.; Chen, L.J.; Zhu, X.; Tam, P.O.S.; Gong, B.; et al. Common variants near ABCA1 and in PMM2 are associated with primary open-angle glaucoma. Nat. Genet. 2014, 46, 1115–1119. [Google Scholar] [CrossRef] [PubMed]
- Alipanahi, B.; Hormozdiari, F.; Behsaz, B.; Cosentino, J.; McCaw, Z.R.; Schorsch, E.; Sculley, D.; Dorfman, E.H.; Foster, P.J.; Peng, L.H.; et al. Large-scale machine learning-based phenotyping significantly improves genomic discovery for optic nerve head morphology. Am. J. Hum. Genet. 2021, 108, 1217–1230. [Google Scholar] [CrossRef]
- García-Bermúdez, M.Y.; Freude, K.K.; Mouhammad, Z.A.; van Wijngaarden, P.; Martin, K.K.; Kolko, M. Glial Cells in Glaucoma: Friends, Foes, and Potential Therapeutic Targets. Front. Neurol. 2021, 12, 624983. [Google Scholar] [CrossRef]
- Mithieux, S.M.; Weiss, A.S. Elastin. Adv. Protein Chem. 2005, 70, 437–461. [Google Scholar]
- Melin, B.S.; Barnholtz-Sloan, J.S.; Wrensch, M.R.; Johansen, C.; Il’yasova, D.; Kinnersley, B.; Ostrom, Q.T.; Labreche, K.; Chen, Y.; Armstrong, G.; et al. Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nat. Genet. 2017, 49, 789–794. [Google Scholar] [CrossRef]
- Fan, B.J.; Wang, D.Y.; Pasquale, L.R.; Haines, J.L.; Wiggs, J.L. Genetic variants associated with optic nerve vertical cup-to-disc ratio are risk factors for primary open angle glaucoma in a US Caucasian population. Invest. Ophthalmol. Vis. Sci. 2011, 52, 1788–1792. [Google Scholar] [CrossRef]
- Newman, E.; Reichenbach, A. The Müller cell: A functional element of the retina. Trends Neurosci. 1996, 19, 307–312. [Google Scholar] [CrossRef]
- Wu, J.; Bell, O.H.; Copland, D.A.; Young, A.; Pooley, J.R.; Maswood, R.; Evans, R.S.; Khaw, P.T.; Ali, R.R.; Dick, A.D.; et al. Gene Therapy for Glaucoma by Ciliary Body Aquaporin 1 Disruption Using CRISPR-Cas9. Mol. Ther. 2020, 28, 820–829. [Google Scholar] [CrossRef]
- Chu, C.J.; Wu, J.; Bell, O.H.; Copland, D.; Maswood, R.; Young, A.; Pooley, J.; Khaw, P.T.; Ali, R.R.; Dick, A.D. Gene therapy for Glaucoma by CRISPR-Cas9 mediated disruption of Aquaporin 1 in the Ciliary Body. Invest. Ophthalmol. Vis. Sci. 2019, 60, 5120. [Google Scholar]
- Rhee, J.; Shih, K.C. Use of Gene Therapy in Retinal Ganglion Cell Neuroprotection: Current Concepts and Future Directions. Biomolecules 2021, 11, 581. [Google Scholar] [CrossRef] [PubMed]
| Gene | GWAS Identified | Suggested Mechanism | Reference |
|---|---|---|---|
| LMX1B | Gao et al., 2018 [16] | Altered Anterior Segment Development; Altered Aqueous Humor Dynamics | Pressman et al., 2000 [17] |
| MADD | Gao et al., 2018 [16] | TNF-a-Mediated Microglial Activation | Tezel et al., 2000 [18] |
| NR1H3 | Gao et al., 2018 [16] | Alteration of IOP via-ABCA1-Regulated Aqueous Humor Dynamic Alterations | Hu et al., 2020 [19] |
| SEPT9 | Gao et al., 2018 [16] | Cytoskeletal Alterations | Ou et al., 1998 [20] |
| Gene | Study | Environment |
|---|---|---|
| ABCA1 | Hu et al., 2020 [19] | Ex vivo Human Model |
| ABCA1 | Li et al., 2018 [22] | In vivo Mouse Model |
| ABCA1 | Yeghiazaryan et al., 2005 [23] | In vivo Human Model |
| ATOH7 | Song et al., 2015 [24] | In vivo Rat Model |
| ATOH7 | Miesfeld et al., 2020 [25] | In vivo Mouse Model |
| ELN | Gelman et al., 2010 [26] | Ex vivo Mouse Model |
| LMX1B | Cross et al., 2014 [27] | In vivo Mouse Model |
| LMX1B | Pressman et al., 2000 [17] | In vivo Mouse Model |
| MADD | Schievella et al., 1997 [28] | In vitro Yeast Interaction Trap |
| NR1H3 | Wang et al., 2002 [29] | In vivo Mouse Model |
| NR1H3 | Yang et al., 2014 [30] | In vivo Mouse Model |
| NR1H3 | Zheng et al., 2015 [31] | In vivo Mouse Model |
| NR1H3 | Song et al., 2019 [32] | Ex vivo Mouse Model |
| SEPT9 | Ghossoub et al., 2013 [33] | Ex vivo Human Model |
| SEPT9 | Ou et al., 1998 [20] | In vivo Guinea Pig Model |
| Gene | GWAS Identified | Suggested Mechanism | Reference |
|---|---|---|---|
| ABCA1 | Springelkamp et al., 2017 [59] | Normal Function and Cell Death of Retinal Ganglion Cells | Chen et al., 2014 [64] |
| ELN | Han et al., 2021 [61] | Alteration to Normal Function of Elastin, Leading to Optic Nerve Head Degeneration | Gelman et al., 2010 [26] |
| ASAP1 | Alipanahi et al., 2021 [65] | Glial Cell-Mediated Retinal Ganglion Cell Loss | García-Bermúdez et al., 2021 [66] |
| ATOH7 | Nannini et al., 2018 [60] | Alteration to Müller Cel Differentiation and Retinal Ganglion Cell Genesis | Miesfeld et al., 2020 [25] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zukerman, R.; Harris, A.; Oddone, F.; Siesky, B.; Verticchio Vercellin, A.; Ciulla, T.A. Glaucoma Heritability: Molecular Mechanisms of Disease. Genes 2021, 12, 1135. https://doi.org/10.3390/genes12081135
Zukerman R, Harris A, Oddone F, Siesky B, Verticchio Vercellin A, Ciulla TA. Glaucoma Heritability: Molecular Mechanisms of Disease. Genes. 2021; 12(8):1135. https://doi.org/10.3390/genes12081135
Chicago/Turabian StyleZukerman, Ryan, Alon Harris, Francesco Oddone, Brent Siesky, Alice Verticchio Vercellin, and Thomas A. Ciulla. 2021. "Glaucoma Heritability: Molecular Mechanisms of Disease" Genes 12, no. 8: 1135. https://doi.org/10.3390/genes12081135
APA StyleZukerman, R., Harris, A., Oddone, F., Siesky, B., Verticchio Vercellin, A., & Ciulla, T. A. (2021). Glaucoma Heritability: Molecular Mechanisms of Disease. Genes, 12(8), 1135. https://doi.org/10.3390/genes12081135









