Upregulation of 15 Antisense Long Non-Coding RNAs in Osteosarcoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Description
2.2. Total RNA Extraction and Sequencing
2.3. Data and Statistical Analyses
2.4. FANTOM-CAT Analysis
2.5. Real Time-Quantitative Polymerase Chain Reaction (RT-qPCR)
3. Results
3.1. Participants’ Characteristics
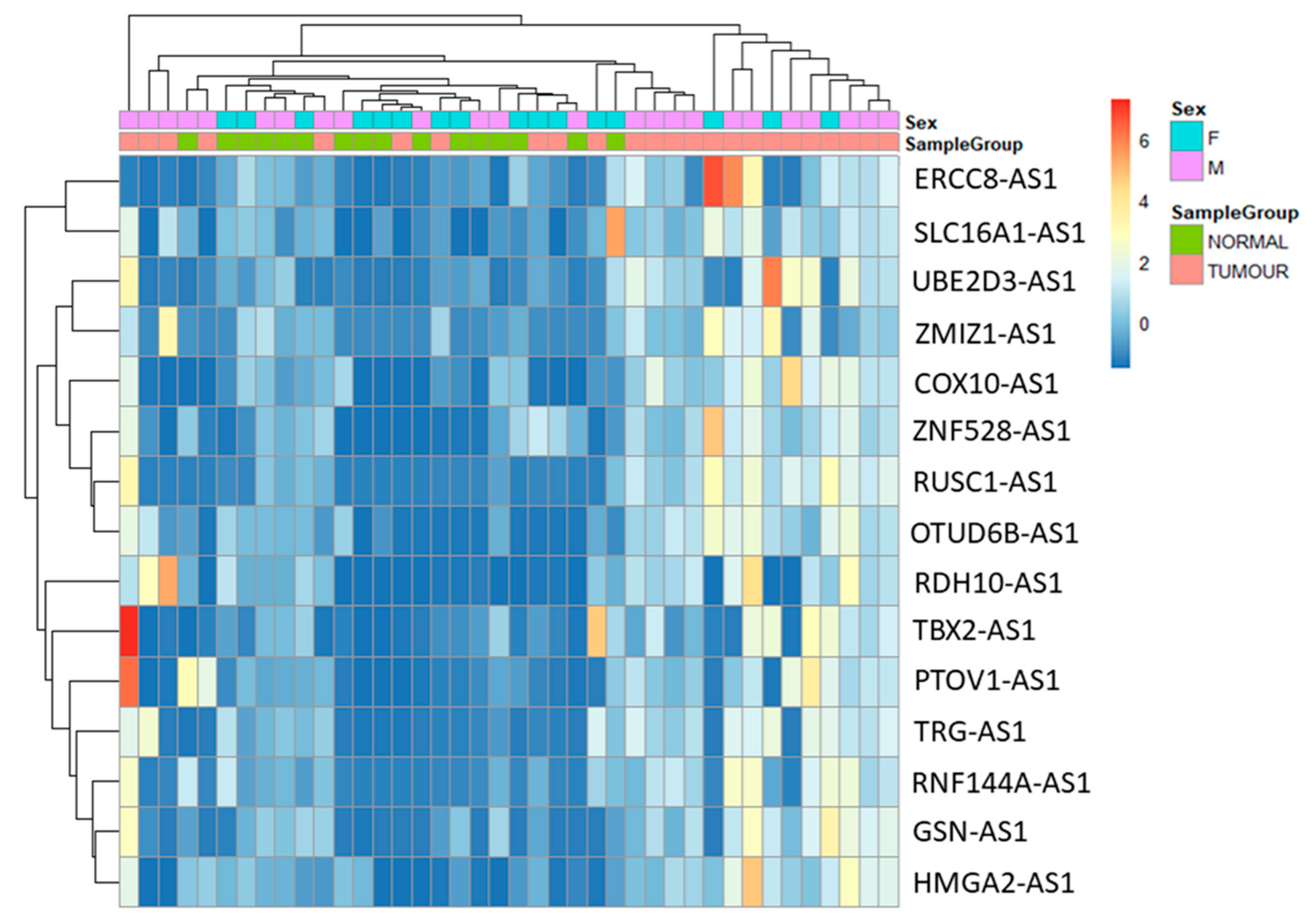
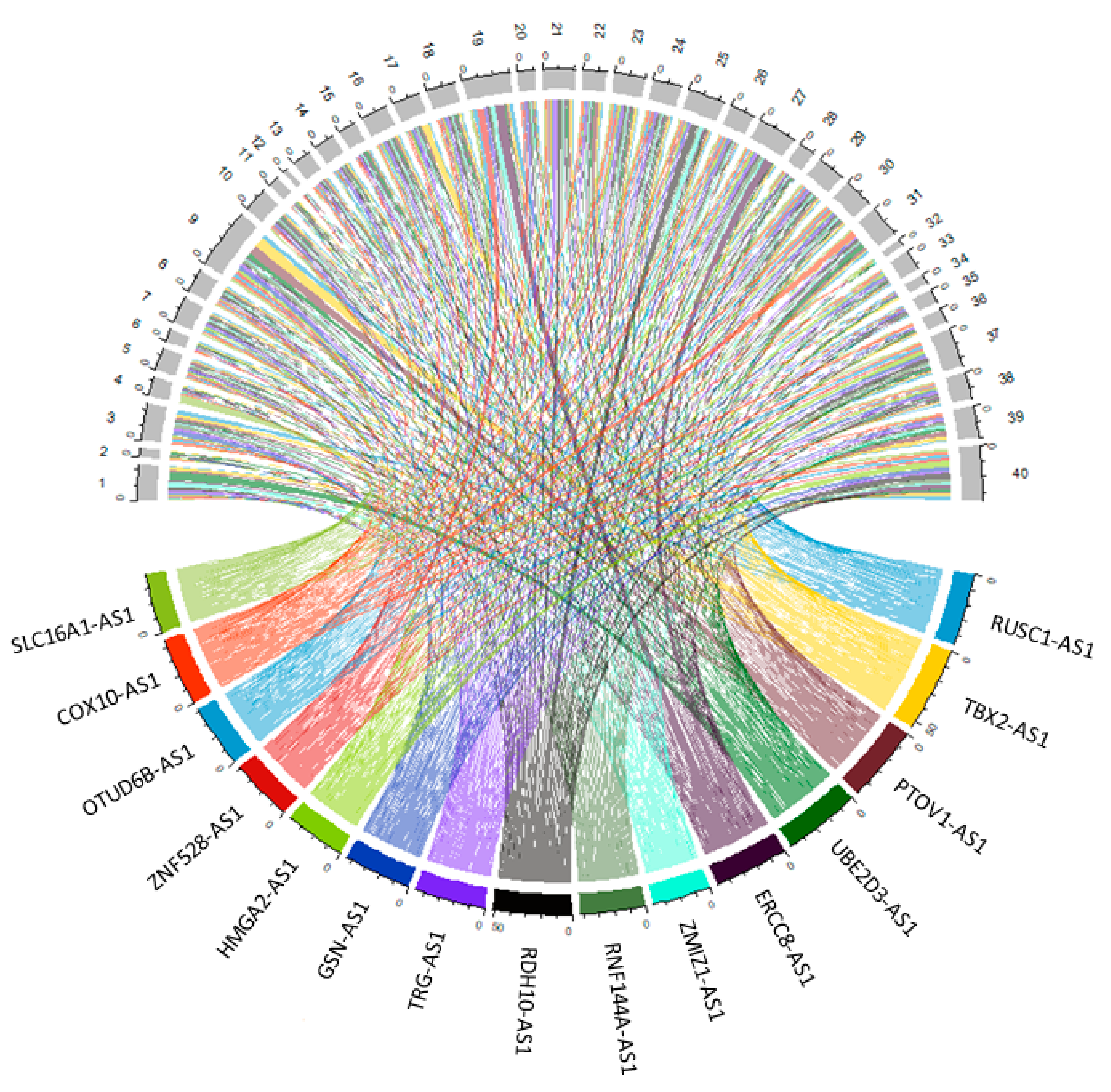
3.2. Differential Gene Expression Analysis between Tumour and Normal Samples
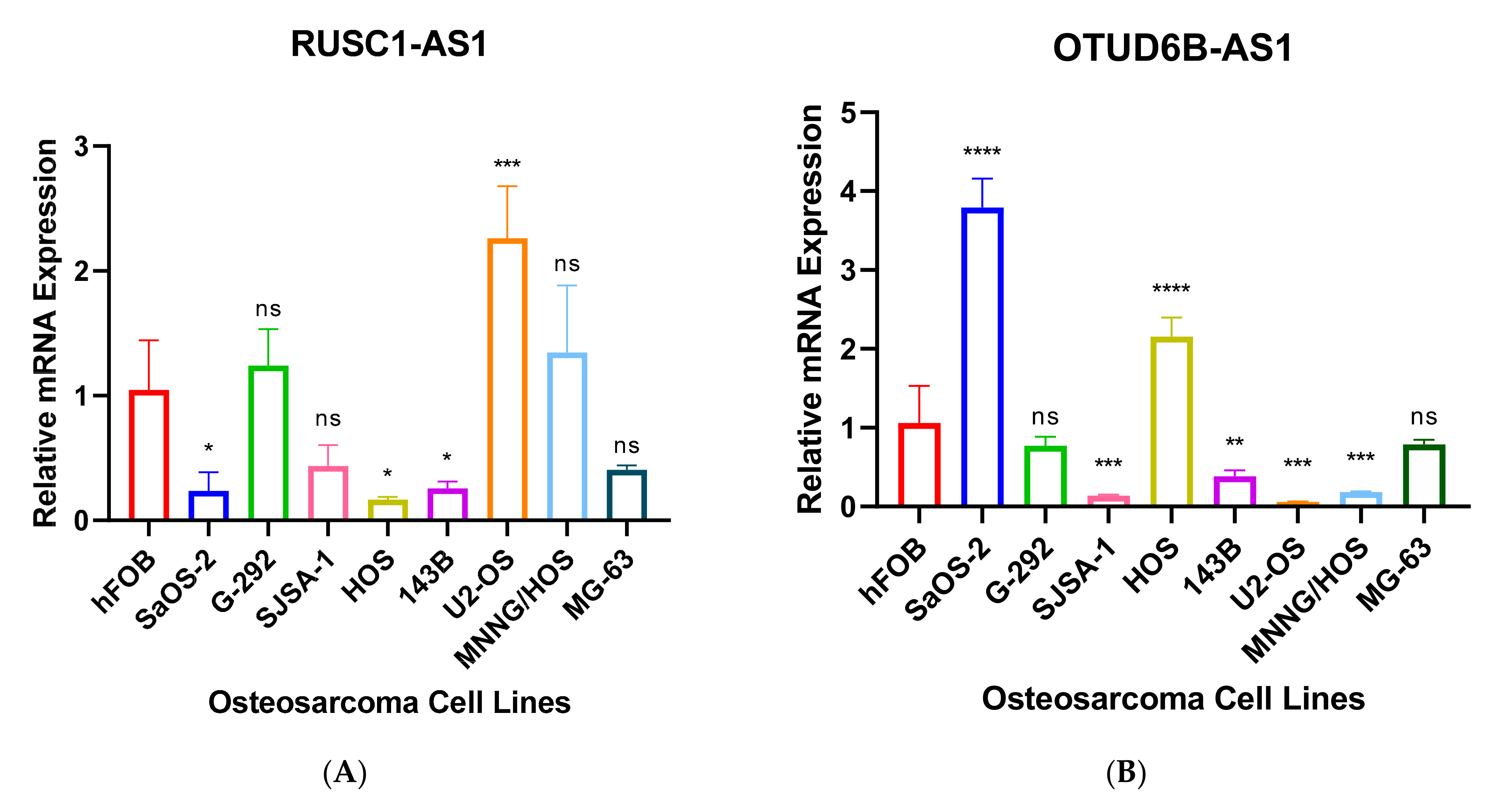
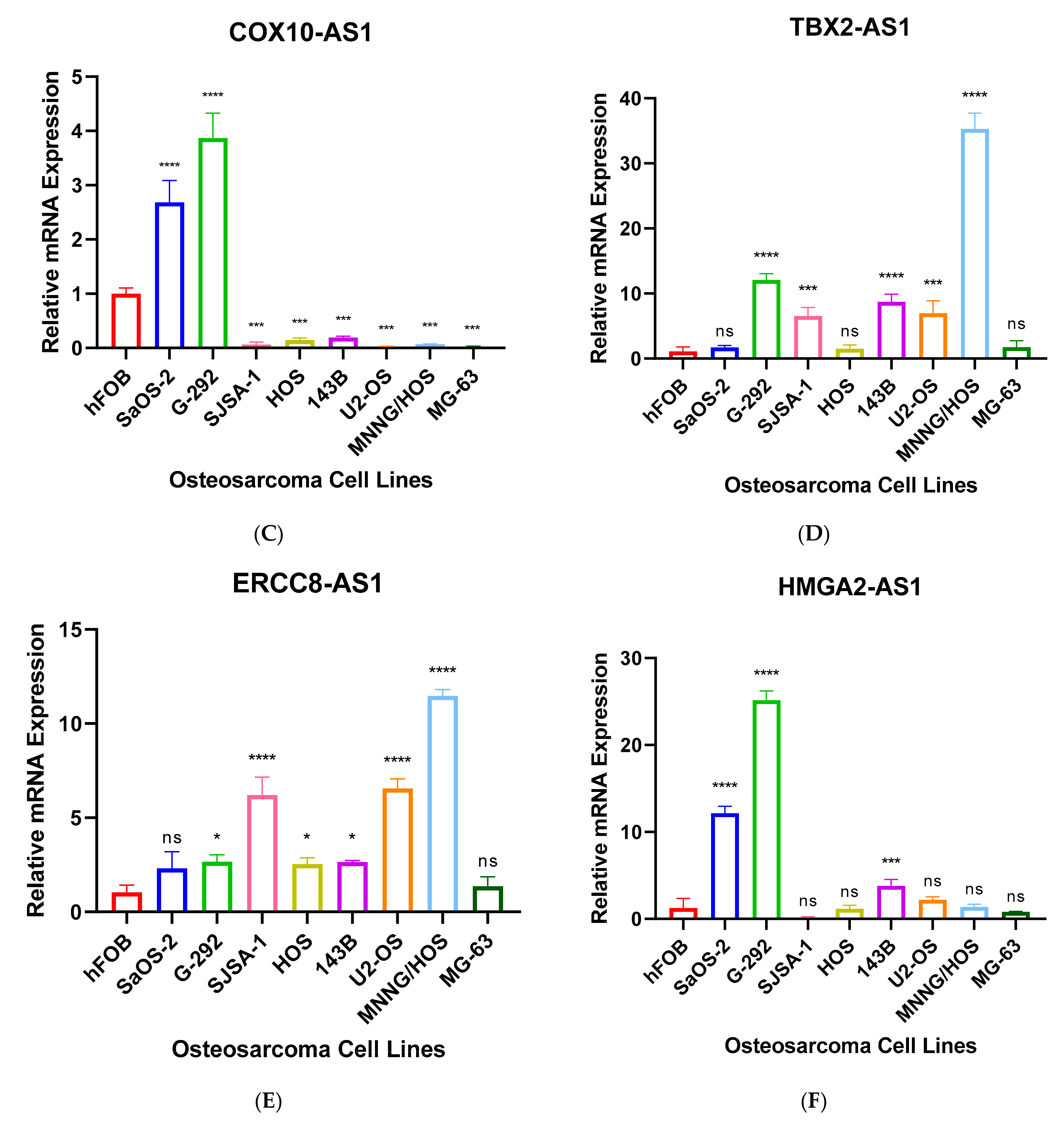
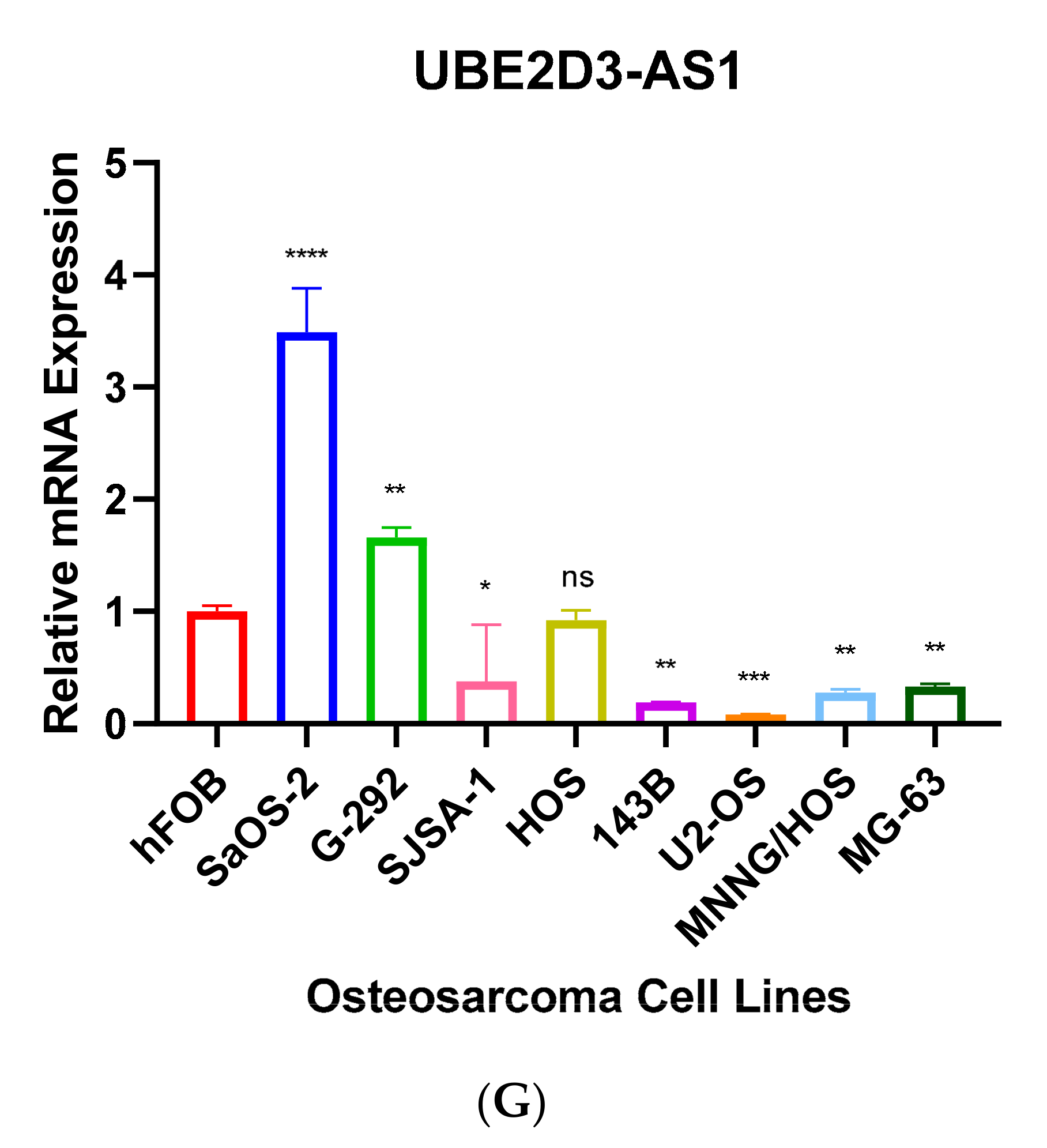
3.3. Validation of Seven Novel Candidate Transcripts Expression Profiles through RT-qPCR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, C.C.; Beird, H.C.; Andrew Livingston, J.; Advani, S.; Mitra, A.; Cao, S.; Reuben, A.; Ingram, D.; Wang, W.L.; Ju, Z.; et al. Immuno-genomic landscape of osteosarcoma. Nat. Commun. 2020, 11, 1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durfee, R.A.; Mohammed, M.; Luu, H.H. Review of Osteosarcoma and Current Management. Rheumatology 2016, 3, 221–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goryn, T.; Pienkowski, A.; Szostakowski, B.; Zdzienicki, M.; Lugowska, I.; Rutkowski, P. Functional outcome of surgical treatment of adults with extremity osteosarcoma after megaprosthetic reconstruction-single-center experience. J. Orthop. Surg. Res. 2019, 14, 346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindsey, B.A.; Markel, J.E.; Kleinerman, E.S. Osteosarcoma Overview. Rheumatology 2017, 4, 25–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothzerg, E.; Ingley, E.; Mullin, B.; Xue, W.; Wood, D.; Xu, J. The Hippo in the room: Targeting the Hippo signalling pathway for osteosarcoma therapies. J. Cell Physiol. 2021, 236, 1606–1615. [Google Scholar] [CrossRef] [PubMed]
- Lamplot, J.D.; Denduluri, S.; Qin, J.; Li, R.; Liu, X.; Zhang, H.; Chen, X.; Wang, N.; Pratt, A.; Shui, W.; et al. The Current and Future Therapies for Human Osteosarcoma. Curr. Cancer Rev. 2013, 9, 55–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothzerg, E.; Ho, X.D.; Xu, J.; Wood, D.; Martson, A.; Maasalu, K.; Koks, S. Alternative splicing of leptin receptor overlapping transcript in osteosarcoma. Exp. Biol. Med. 2020, 245, 1437–1443. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, K.; Yamamoto, N.; Shirai, T.; Nishida, H.; Hayashi, K.; Tanzawa, Y.; Kimura, H.; Takeuchi, A.; Miwa, S.; Inatani, H.; et al. Late recurrence of osteosarcoma: A report of two cases. J. Orthop. Surg. 2014, 22, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [Green Version]
- Lekka, E.; Hall, J. Noncoding RNAs in disease. FEBS Lett. 2018, 592, 2884–2900. [Google Scholar] [CrossRef]
- Hangauer, M.J.; Vaughn, I.W.; McManus, M.T. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013, 9, e1003569. [Google Scholar] [CrossRef]
- Prensner, R.J.; Chinnaiyan, A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011, 1, 391–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dey, B.K.; Mueller, A.C.; Dutta, A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription 2014, 5, e944014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geisler, S.; Coller, J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013, 14, 699–712. [Google Scholar] [CrossRef] [Green Version]
- Ros, G.; Pegoraro, S.; De Angelis, P.; Sgarra, R.; Zucchelli, S.; Gustincich, S.; Manfioletti, G. HMGA2 Antisense Long Non-coding RNAs as New Players in the Regulation of HMGA2 Expression and Pancreatic Cancer Promotion. Front. Oncol. 2019, 9, 1526. [Google Scholar] [CrossRef]
- Faghihi, A.M.; Wahlestedt, C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 2009, 10, 637–643. [Google Scholar] [CrossRef]
- Liu, J.; Hu, J.; Corey, D.R. Expanding the action of duplex RNAs into the nucleus: Redirecting alternative splicing. Nucleic Acids Res. 2012, 40, 1240–1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallo, A.; Vukic, D.; Michalik, D.; O'Connell, M.A.; Keegan, L.P. ADAR RNA editing in human disease; more to it than meets the I. Hum. Genet. 2017, 136, 1265–1278. [Google Scholar]
- Slotkin, W.; Nishikura, K. Adenosine-to-inosine RNA editing and human disease. Genome Med. 2013, 5, 105. [Google Scholar] [CrossRef]
- Britto-Kido Sde, A.; Ferreira Neto, J.R.; Pandolfi, V.; Marcelino-Guimaraes, F.C.; Nepomuceno, A.L.; Vilela Abdelnoor, R.; Benko-Iseppon, A.M.; Kido, E.A. Natural antisense transcripts in plants: A review and identification in soybean infected with Phakopsora pachyrhizi SuperSAGE library. Sci. World J. 2013, 2013, 219798. [Google Scholar]
- Rosikiewicz, W.; Makalowska, I. Biological functions of natural antisense transcripts. Acta Biochim. Pol. 2016, 63, 665–673. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Sklutuis, R.; Groebner, J.L.; Romerio, F. HIV-1 Natural Antisense Transcription and Its Role in Viral Persistence. Viruses 2021, 13, 795. [Google Scholar] [CrossRef]
- Modi, A.; Vai, S.; Caramelli, D.; Lari, M. The Illumina Sequencing Protocol and the NovaSeq 6000 System. Methods Mol. Biol. 2021, 2242, 15–42. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Dai, Y.; Yu, H.; Zhao, S.; Samuels, D.C.; Shyr, Y. Improvements and impacts of GRCh38 human reference on high throughput sequencing data analysis. Genomics 2017, 109, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Severin, J.; Lizio, M.; Harshbarger, J.; Kawaji, H.; Daub, C.O.; Hayashizaki, Y.; Consortium, F.; Bertin, N.; Forrest, A.R. Interactive visualization and analysis of large-scale sequencing datasets using ZENBU. Nat. Biotechnol. 2014, 32, 217–219. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, X.; Chen, S.; Zhang, S. Natural antisense transcripts in the biological hallmarks of cancer: Powerful regulators hidden in the dark. J. Exp. Clin. Cancer Res. 2020, 39, 187. [Google Scholar] [CrossRef]
- Pelechano, V.; Steinmetz, L.M. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013, 14, 880–893. [Google Scholar] [CrossRef]
- Balbin, O.A.; Malik, R.; Dhanasekaran, S.M.; Prensner, J.R.; Cao, X.; Wu, Y.M.; Robinson, D.; Wang, R.; Chen, G.; Beer, D.G.; et al. The landscape of antisense gene expression in human cancers. Genome Res. 2015, 25, 1068–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hui, L.; Wang, J.; Zhang, J.; Long, J. lncRNA TMEM51-AS1 and RUSC1-AS1 function as ceRNAs for induction of laryngeal squamous cell carcinoma and prediction of prognosis. PeerJ 2019, 7, e7456. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Zhang, Q.; Lu, L.; Xu, Y. Long noncoding RNA RUSC1-AS1 promotes tumorigenesis in cervical cancer by acting as a competing endogenous RNA of microRNA-744 and consequently increasing Bcl-2 expression. Cell Cycle 2020, 19, 1222–1235. [Google Scholar] [CrossRef]
- Hu, C.C.; Liang, Y.W.; Hu, J.L.; Liu, L.F.; Liang, J.W.; Wang, R. LncRNA RUSC1-AS1 promotes the proliferation of breast cancer cells by epigenetic silence of KLF2 and CDKN1A. Eur. Rev. Med. Pharm. Sci. 2019, 23, 6602–6611. [Google Scholar]
- Chen, Y.A.; Cheng, L.; Zhang, Y.; Peng, L.; Yang, H.G. LncRNA RUSC1-AS1 promotes the proliferation of hepatocellular carcinoma cells through modulating NOTCH signaling. Neoplasma 2020, 67, 1204–1213. [Google Scholar] [CrossRef]
- Esquenet, M.; Swinnen, J.V.; Heyns, W.; Verhoeven, G. LNCaP prostatic adenocarcinoma cells derived from low and high passage numbers display divergent responses not only to androgens but also to retinoids. J. Steroid. Biochem. Mol. Biol. 1997, 62, 391–399. [Google Scholar] [CrossRef]
- Briske-Anderson, M.J.; Finley, J.W.; Newman, S.M. The influence of culture time and passage number on the morphological and physiological development of Caco-2 cells. Proc. Soc. Exp. Biol. Med. 1997, 214, 248–257. [Google Scholar] [CrossRef]
- Chang, L.; Woloschak, G.E. Effect of passage number on cellular response to DNA-damaging agents: Cell survival and gene expression. Cancer Lett. 1997, 113, 77–86. [Google Scholar] [CrossRef] [Green Version]
- Wenger, S.L.; Senft, J.R.; Sargent, L.M.; Bamezai, R.; Bairwa, N.; Grant, S.G. Comparison of established cell lines at different passages by karyotype and comparative genomic hybridization. Biosci. Rep. 2004, 24, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Sambuy, Y.; De Angelis, I.; Ranaldi, G.; Scarino, M.L.; Stammati, A.; Zucco, F. The Caco-2 cell line as a model of the intestinal barrier: Influence of cell and culture-related factors on Caco-2 cell functional characteristics. Cell Biol. Toxicol. 2005, 21, 1–26. [Google Scholar] [CrossRef]
- Decaesteker, B. TBX2 is a neuroblastoma core regulatory circuitry component enhancing MYCN/FOXM1 reactivation of DREAM targets. Nat. Commun. 2018, 9, 4866. [Google Scholar] [CrossRef]
- Shin, C.H.; Ryu, S.; Kim, H.H. hnRNPK-regulated PTOV1-AS1 modulates heme oxygenase-1 expression via miR-1207-5p. BMB Rep. 2017, 50, 220–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, N.; Kim, K.; Shin, A.; Park, J.W.; Chang, H.J.; Shi, J.; Cai, Q.; Kim, D.Y.; Zheng, W.; Oh, J.H. Colorectal cancer susceptibility loci and influence on survival. Genes Chromosomes Cancer 2018, 57, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Shin, A.; Park, J.W.; Kim, J.; Oh, J.H. Common risk variants for colorectal cancer: An evaluation of associations with age at cancer onset. Sci. Rep. 2017, 7, 40644. [Google Scholar] [CrossRef] [Green Version]
- Alazzouzi, H.; Alhopuro, P.; Salovaara, R.; Sammalkorpi, H.; Jarvinen, H.; Mecklin, J.P.; Hemminki, A.; Schwartz, S., Jr.; Aaltonen, L.A.; Arango, D. SMAD4 as a prognostic marker in colorectal cancer. Clin. Cancer Res. 2005, 11, 2606–2611. [Google Scholar] [CrossRef] [Green Version]
- Rivlin, N.; Brosh, R.; Oren, M.; Rotter, V. Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes Cancer 2011, 2, 466–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, H.; Shang, Z.; Jia, C.; Wu, J.; Cui, B.; Wang, Q.; Ou, T. LncRNA RNF144A-AS1 Promotes Bladder Cancer Progression via RNF144A-AS1/miR-455-5p/SOX11 Axis. Oncol. Targets 2020, 13, 11277–11288. [Google Scholar] [CrossRef]
- Wang, Y.; Du, L.; Yang, X.; Li, J.; Li, P.; Zhao, Y.; Duan, W.; Chen, Y.; Wang, Y.; Mao, H.; et al. A nomogram combining long non-coding RNA expression profiles and clinical factors predicts survival in patients with bladder cancer. Aging 2020, 12, 2857–2879. [Google Scholar] [CrossRef] [PubMed]
- Huynh, N.P.; Gloss, C.C.; Lorentz, J.; Tang, R.; Brunger, J.M.; McAlinden, A.; Zhang, B.; Guilak, F. Long non-coding RNA GRASLND enhances chondrogenesis via suppression of the interferon type II signaling pathway. eLife 2020, 9, e49558. [Google Scholar] [CrossRef] [Green Version]
- He, S.; Wang, X.; Zhang, J.; Zhou, F.; Li, L.; Han, X. TRG-AS1 is a potent driver of oncogenicity of tongue squamous cell carcinoma through microRNA-543/Yes-associated protein 1 axis regulation. Cell Cycle 2020, 19, 1969–1982. [Google Scholar] [CrossRef]
- Sun, X.; Qian, Y.; Wang, X.; Cao, R.; Zhang, J.; Chen, W.; Fang, M. LncRNA TRG-AS1 stimulates hepatocellular carcinoma progression by sponging miR-4500 to modulate BACH1. Cancer Cell Int. 2020, 20, 367. [Google Scholar] [CrossRef]
- Xie, H.; Shi, S.; Chen, Q.; Chen, Z. LncRNA TRG-AS1 promotes glioblastoma cell proliferation by competitively binding with miR-877-5p to regulate SUZ12 expression. Pathol. Res. Pr. 2019, 215, 152476. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Lv, S.X.; Lv, L.; Liu, Y.H.; Dong, S.Y.; Yao, Z.H.; Dai, X.X.; Zhang, X.H.; Wang, O.C. Identification of lncRNA FAM83H-AS1 as a novel prognostic marker in luminal subtype breast cancer. Onco. Targets Ther. 2016, 9, 7039–7045. [Google Scholar] [CrossRef] [Green Version]
- Jing, Z.; Guo, S.; Zhang, P.; Liang, Z. LncRNA-Associated ceRNA Network Reveals Novel Potential Biomarkers of Laryngeal Squamous Cell Carcinoma. Technol. Cancer Res. Treat. 2020, 19, 1533033820985787. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, X.; Xu, F.; Zhang, L.; Wang, T.; Fu, X.; Jin, T.; Zhang, W.; Ye, L. The regulation of acetylation and stability of HMGA2 via the HBXIP-activated Akt-PCAF pathway in promotion of esophageal squamous cell carcinoma growth. Nucleic Acids Res. 2020, 48, 4858–4876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Mo, Q.; Wang, X. Oncological role of HMGA2 (Review). Int. J. Oncol. 2019, 55, 775–788. [Google Scholar] [CrossRef]
- Xi, X.; Teng, M.; Zhang, L.; Xia, L.; Chen, J.; Cui, Z. MicroRNA-204-3p represses colon cancer cells proliferation, migration, and invasion by targeting HMGA2. J. Cell Physiol. 2020, 235, 1330–1338. [Google Scholar] [CrossRef]
- Li, Y.; Qiang, W.; Griffin, B.B.; Gao, T.; Chakravarti, D.; Bulun, S.; Kim, J.J.; Wei, J.J. HMGA2-mediated tumorigenesis through angiogenesis in leiomyoma. Fertil. Steril. 2020, 114, 1085–1096. [Google Scholar] [CrossRef]
- Dong, J.; Wang, R.; Ren, G.; Li, X.; Wang, J.; Sun, Y.; Liang, J.; Nie, Y.; Wu, K.; Feng, B.; et al. HMGA2-FOXL2 Axis Regulates Metastases and Epithelial-to-Mesenchymal Transition of Chemoresistant Gastric Cancer. Clin. Cancer Res. 2017, 23, 3461–3473. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Dai, M.; Li, Q.; Wang, Z.; Lu, Y.; Song, Z. HMGA2 regulates lung cancer proliferation and metastasis. Thorac. Cancer 2017, 8, 501–510. [Google Scholar] [CrossRef]
- Wang, X.; Wang, J.; Wu, J. Emerging roles for HMGA2 in colorectal cancer. Transl. Oncol. 2021, 14, 100894. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Li, Y.; Shi, C.; Zhang, Z.; Xu, J.; Sun, B. LncRNA OTUD6B-AS1 inhibits many cellular processes in colorectal cancer by sponging miR-21-5p and regulating PNRC2. Hum. Exp. Toxicol. 2021. [Google Scholar] [CrossRef]
- Wang, W.; Cheng, X.; Zhu, J. Long non-coding RNA OTUD6B-AS1 overexpression inhibits the proliferation, invasion and migration of colorectal cancer cells via downregulation of microRNA-3171. Oncol. Lett. 2021, 21, 193. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhang, Z.J.; Jian, W.G.; Liu, P.H.; Xue, W.; Wang, T.D.; Meng, Y.Y.; Yuan, C.; Li, H.M.; Yu, Y.P.; et al. Novel long noncoding RNA OTUD6B-AS1 indicates poor prognosis and inhibits clear cell renal cell carcinoma proliferation via the Wnt/beta-catenin signaling pathway. Mol. Cancer 2019, 18, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Xia, F.; Feng, T.; Jiang, B.; Wang, W.; Li, X. OTUD6B-AS1 Inhibits Viability, Migration, and Invasion of Thyroid Carcinoma by Targeting miR-183-5p and miR-21. Front. Endocrinol. 2020, 11, 136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Zhan, X. Identification of clinical trait-related lncRNA and mRNA biomarkers with weighted gene co-expression network analysis as useful tool for personalized medicine in ovarian cancer. EPMA J. 2019, 10, 273–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, C.; Jiang, X.; Liang, A.; Zhu, R.; Yang, Y.; Zhong, L.; Wan, D. COX10-AS1 Facilitates Cell Proliferation and Inhibits Cell Apoptosis in Glioblastoma Cells at Post-Transcription Level. Neurochem. Res. 2020, 45, 2196–2203. [Google Scholar] [CrossRef]
- Tian, J.; Hu, D. LncRNA SLC16A1-AS1 is upregulated in hepatocellular carcinoma and predicts poor survival. Clin. Res. Hepatol. Gastroenterol. 2021, 45, 101490. [Google Scholar] [CrossRef]
- Long, Y.; Li, H.; Jin, Z.; Zhang, X. LncRNA SLC16A1-AS1 is Upregulated in Glioblastoma and Promotes Cancer Cell Proliferation by Regulating miR-149 Methylation. Cancer Manag. Res. 2021, 13, 1215–1223. [Google Scholar] [CrossRef]
- Liu, H.Y.; Lu, S.R.; Guo, Z.H.; Zhang, Z.S.; Ye, X.; Du, Q.; Li, H.; Wu, Q.; Yu, B.; Zhai, Q.; et al. lncRNA SLC16A1-AS1 as a novel prognostic biomarker in non-small cell lung cancer. J. Investig. Med. 2020, 68, 52–59. [Google Scholar] [CrossRef] [Green Version]
- Van der Pluijm, I.; Garinis, G.A.; Brandt, R.M.; Gorgels, T.G.; Wijnhoven, S.W.; Diderich, K.E.; de Wit, J.; Mitchell, J.R.; van Oostrom, C.; Beems, R.; et al. Impaired genome maintenance suppresses the growth hormone--insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol. 2007, 5, e2. [Google Scholar]
- Karikkineth, A.C.; Scheibye-Knudsen, M.; Fivenson, E.; Croteau, D.L.; Bohr, V.A. Cockayne syndrome: Clinical features, model systems and pathways. Ageing Res. Rev. 2017, 33, 3–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manandhar, M.; Boulware, K.S.; Wood, R.D. The ERCC1 and ERCC4 (XPF) genes and gene products. Gene 2015, 569, 153–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashiyama, K. Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes Cockayne syndrome, xeroderma pigmentosum, and Fanconi anemia. Am. J. Hum. Genet. 2013, 92, 807–819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenberg, P.S.; Greene, M.H.; Alter, B.P. Cancer incidence in persons with Fanconi anemia. Blood 2003, 101, 822–826. [Google Scholar] [CrossRef] [PubMed]
- Rothzerg, E.; Xu, J.; Wood, D.; Koks, S. 12 Survival-related differentially expressed genes based on the TARGET-osteosarcoma database. Exp. Biol. Med. 2021. [Google Scholar] [CrossRef]



| Transcript | Forward Primer | Reverse Primer |
|---|---|---|
| UBE2D3-AS1 | TGAATGCTTATGCCGGTGGT | CGGCCCGAGCTA-GACTAAAG |
| OTUD6B-AS1 | GACATATCCGGGTGACGTTTTT | TTGTTCCACTGTCTTCTGGCATT |
| COX10-AS1 | TACCTCTGGGAAGTAC-GGGG | CACTTGCCACTGAAAGCACC |
| RUSC1-AS1 | GAAAAGGATGGAGCAGCCGTCA | GGCTGAACGATGGAGACGAATG |
| HMGA2-AS1 | GCAGCTTGTTTTCTGGGTGG | ACTTTGGGGGCAAAGTGTCA |
| ERCC8-AS1 | GCCAAACCGAGATCACATGC | CACACAGTGGGAGCCTGAAT |
| TBX2-AS1 | AACATCCAGGGCAATCTGGG | GTGCCGAGAGAATCGGTAGG |
| PTOV1-AS1 | AGGCGATCCTCAGGAATGTG | AATAAGCAAGCCCCGGTTCA |
| RNF144A-AS1 | CACACAGCAAGCTAGGA | ACTTTCCTTGCGAGGGTTGG |
| RDH10-AS1 | TGACTACAGCGAGCAACAGC | TCCACTGAGACGGAAACTGC |
| TRG-AS1 | CTCCTTCATTCCCTATTC | TTATGATGGCTACGATGT |
| ZMIZ1-AS1 | TCTCAAGGCTCCGCTAGTCT | TCACCTGCATCCCCCAATTC |
| GSN-AS1 | CCCATCAGCGGCTATCCAAA | TGGACATCGAGGAGGTCACT |
| ZNF528-AS1 | ACACTGGCCTTAG-TCCTCCA | CTGCGCTTGTTTTCAGGGTT |
| SLC16A1-AS1 | CCCTGGGAGGTAGGCCTTAT | TCTACCACCCTATGGGGCTC |
| GAPDH | GAAGGTGAAGGTCGGAGTC | GAAGATGGTGATGGGATTTC |
| Patient ID | Tumour | Normal | Gender | Age of Diagnosis | Site of Tumour | Chemotherapy | Vital Status |
|---|---|---|---|---|---|---|---|
| Q17B029593M | A7 | A23 | Female | 26 | Femur | Yes | Died from disease at the age of 26 |
| Q17B045995J | A5 | A29 | Female | 78 | Femur | No | Died from disease at the age of 80 |
| Q18B006524D | A28 | A8 | Female | 74 | Illium | No | Died from disease at the age of 74 |
| Q18B009680H ’ | A1 | - | Male | 17 | Humerus | Yes | Died from disease at the age of 18 |
| Q18B009680H ’ | A12 | - | Male | 17 | Humerus | Yes | - |
| Q18B014955A | A15 | A23 | Female | 17 | Femur | Yes | Died from disease at the age of 18 |
| Q18B015603E * | A5 | - | Male | 26 | Femur | Yes | Alive no evidence of disease |
| Q18B015603E * | A52 | - | Male | 26 | Femur | Yes | - |
| Q18B018266Y | A1 | E1 | Male | 58 | T9-10 vertebra | Yes | Died from disease at the age of 58 |
| Q18B028621H | A4 | A1 | Male | 26 | Femur | Yes | Alive no evidence of disease |
| Q18B034715Y | A6 | A11 | Female | 19 | Femur | Yes | Alive with metastatic disease |
| Q18B051017F | A1 | A16 | Male | 69 | Femur | Yes | Died from disease at the age of 71 |
| Q19B001229R | A30 | A22 | Female | 13 | Femur | Yes | Alive no evidence of disease |
| Q19B005830Y | A2 | - | Male | 17 | Tibia | Yes | Alive no evidence of disease |
| Q19B007088F | B10 | B22 | Female | 17 | Tibia | Yes | Alive no evidence of disease |
| Q19B013567K ^ | A1 | - | Male | 63 | Femur | No | Died from disease at the age of 64 |
| Q19B013567K ^ | A12 | - | Male | 63 | Femur | No | - |
| Q19B021879L | A19 | A21 | Male | 17 | Tibia | Yes | Alive no evidence of disease |
| Q19B035672T | A1 | - | Male | 33 | Femur | Yes | Alive no evidence of disease |
| Q19B051495P | B19 | A2 | Female | 14 | Humerus | Yes | Alive no evidence of disease |
| Q19B052024A | B2 | B6 | Male | 36 | Femur | No | Alive no evidence of disease |
| Q17B018941H | A12 | B1 | Male | 36 | Tibia | Yes | Died from disease at the age of 37 |
| Q16B040208X | A33 | A25 | Male | 17 | Femur | Yes | Died from disease at the age of 18 |
| Q15B001034Y | A15 | B1 | Male | 19 | Femur | Yes | Died from disease at the age of 20 |
| ENSEMBL | Symbol | Log2FC | p-Value | padj | Transcripts Name |
|---|---|---|---|---|---|
| ENSG00000225855.7 | RUSC1-AS1 | 4.556139 | 3.83E-05 | 0.008835 | RUSC1 antisense RNA 1 |
| ENSG00000267280.5 | TBX2-AS1 | 3.98406 | 0.001648 | 0.035197 | TBX2 antisense RNA 1 |
| ENSG00000268006.1 | PTOV1-AS1 | 3.833228 | 0.000187 | 0.017564 | PTOV1 antisense RNA 1 |
| ENSG00000246560.2 | UBE2D3-AS1 | 3.770542 | 0.003651 | 0.045301 | UBE2D3 antisense RNA 1 |
| ENSG00000233847.1 | ERCC8-AS1 | 3.385903 | 0.004691 | 0.048827 | ERCC8 antisense RNA 1 |
| ENSG00000224596.8 | ZMIZ1-AS1 | 3.366849 | 0.001591 | 0.035104 | ZMIZ1 antisense RNA 1 |
| ENSG00000228203.7 | RNF144A-AS1 | 3.300567 | 0.002698 | 0.041347 | RNF144A antisense RNA 1 |
| ENSG00000250295.6 | RDH10-AS1 | 3.227144 | 0.003145 | 0.043071 | RDH10 antisense RNA 1 |
| ENSG00000281103.2 | TRG-AS1 | 3.160751 | 0.001404 | 0.033432 | T cell receptor γ locus antisense RNA 1 |
| ENSG00000235865.2 | GSN-AS1 | 3.103048 | 0.002449 | 0.040134 | GSN antisense RNA 1 |
| ENSG00000197301.7 | HMGA2-AS1 | 2.912661 | 0.000946 | 0.030737 | HMGA2 antisense RNA 1 |
| ENSG00000269834.6 | ZNF528-AS1 | 2.774884 | 0.001737 | 0.03563 | ZNF528 antisense RNA 1 |
| ENSG00000253738.2 | OTUD6B-AS1 | 2.764901 | 0.001943 | 0.03686 | OTUD6B antisense RNA 1 |
| ENSG00000236088.10 | COX10-AS1 | 2.722786 | 0.004343 | 0.04782 | COX10 antisense RNA 1 |
| ENSG00000226419.8 | SLC16A1-AS1 | 2.427554 | 0.001752 | 0.03577 | SLC16A1 antisense RNA 1 |
| Transcripts | Type of Overlap |
|---|---|
| RUSC1-AS1 | Head-to-head |
| TBX2-AS1 | Head-to-head |
| PTOV1-AS1 | Head-to-head |
| UBE2D3-AS1 | Embedded |
| ERCC8-AS1 | Embedded |
| ZMIZ1-AS1 | Head-to-head |
| RNF144A-AS1 | Head-to-head |
| RDH10-AS1 | Tail-to-tail |
| TRG-AS1 | Head-to-head |
| GSN-AS1 | Embedded |
| HMGA2-AS1 | Embedded |
| ZNF528-AS1 | Head-to-head |
| OTUD6B-AS1 | Head-to-head |
| COX10-AS1 | Head-to-head |
| SLC16A1-AS1 | Head-to-head |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rothzerg, E.; Ho, X.D.; Xu, J.; Wood, D.; Märtson, A.; Kõks, S. Upregulation of 15 Antisense Long Non-Coding RNAs in Osteosarcoma. Genes 2021, 12, 1132. https://doi.org/10.3390/genes12081132
Rothzerg E, Ho XD, Xu J, Wood D, Märtson A, Kõks S. Upregulation of 15 Antisense Long Non-Coding RNAs in Osteosarcoma. Genes. 2021; 12(8):1132. https://doi.org/10.3390/genes12081132
Chicago/Turabian StyleRothzerg, Emel, Xuan Dung Ho, Jiake Xu, David Wood, Aare Märtson, and Sulev Kõks. 2021. "Upregulation of 15 Antisense Long Non-Coding RNAs in Osteosarcoma" Genes 12, no. 8: 1132. https://doi.org/10.3390/genes12081132
APA StyleRothzerg, E., Ho, X. D., Xu, J., Wood, D., Märtson, A., & Kõks, S. (2021). Upregulation of 15 Antisense Long Non-Coding RNAs in Osteosarcoma. Genes, 12(8), 1132. https://doi.org/10.3390/genes12081132








