Next-Generation Sequencing Analysis of the Tineola bisselliella Larval Gut Transcriptome Reveals Candidate Enzymes for Keratin Digestion
Abstract
1. Introduction
2. Materials and Methods
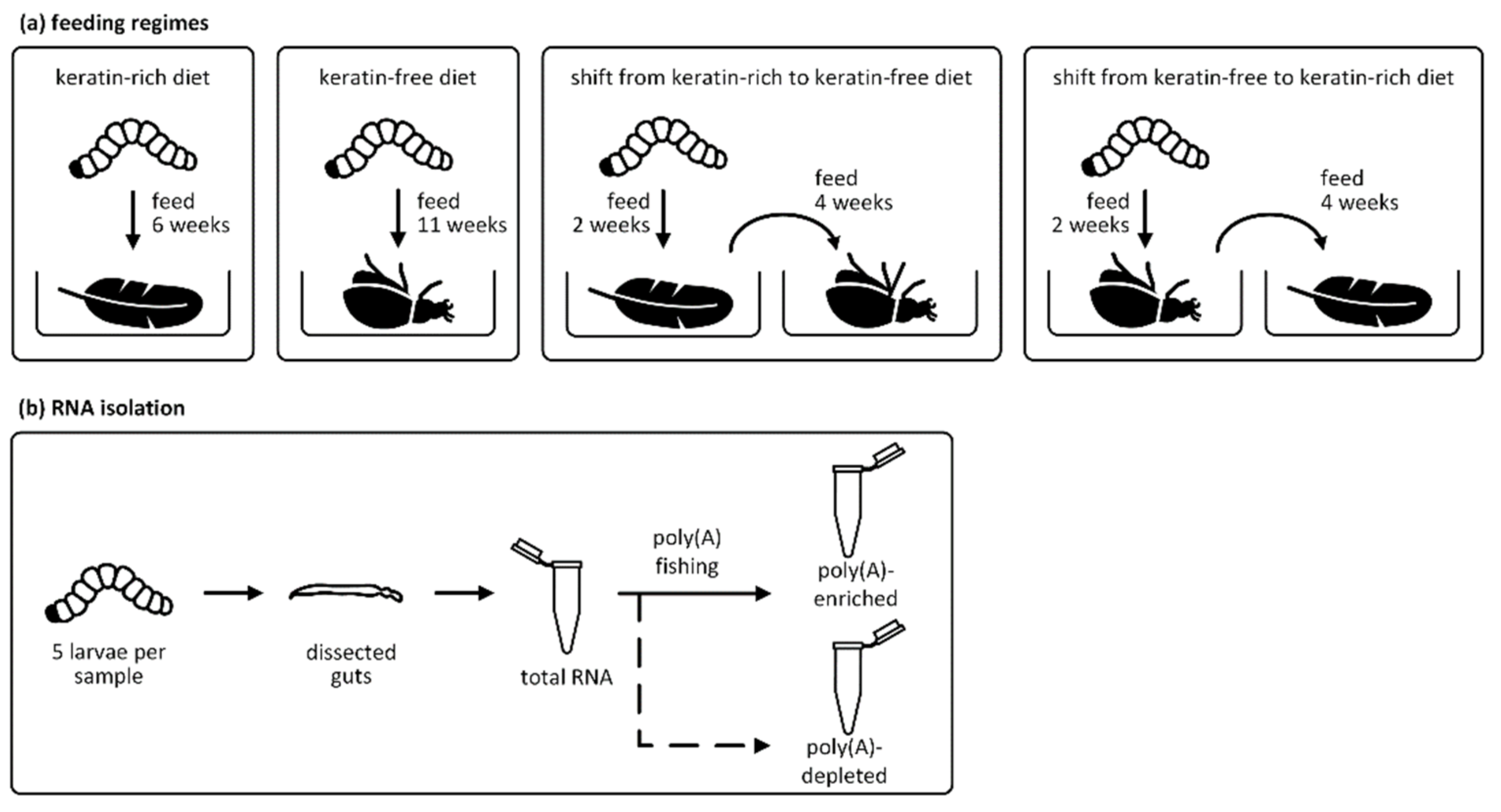
2.1. Animal Rearing and Sample Preparation
2.2. RNA Isolation and Sequencing
2.3. Sequence Data Quality Control and Transcriptome Assembly
2.4. Gene Finding and Functional Annotation
2.5. Identification of Proteins with Putative Keratinolytic Activity
2.6. Differential Gene Expression
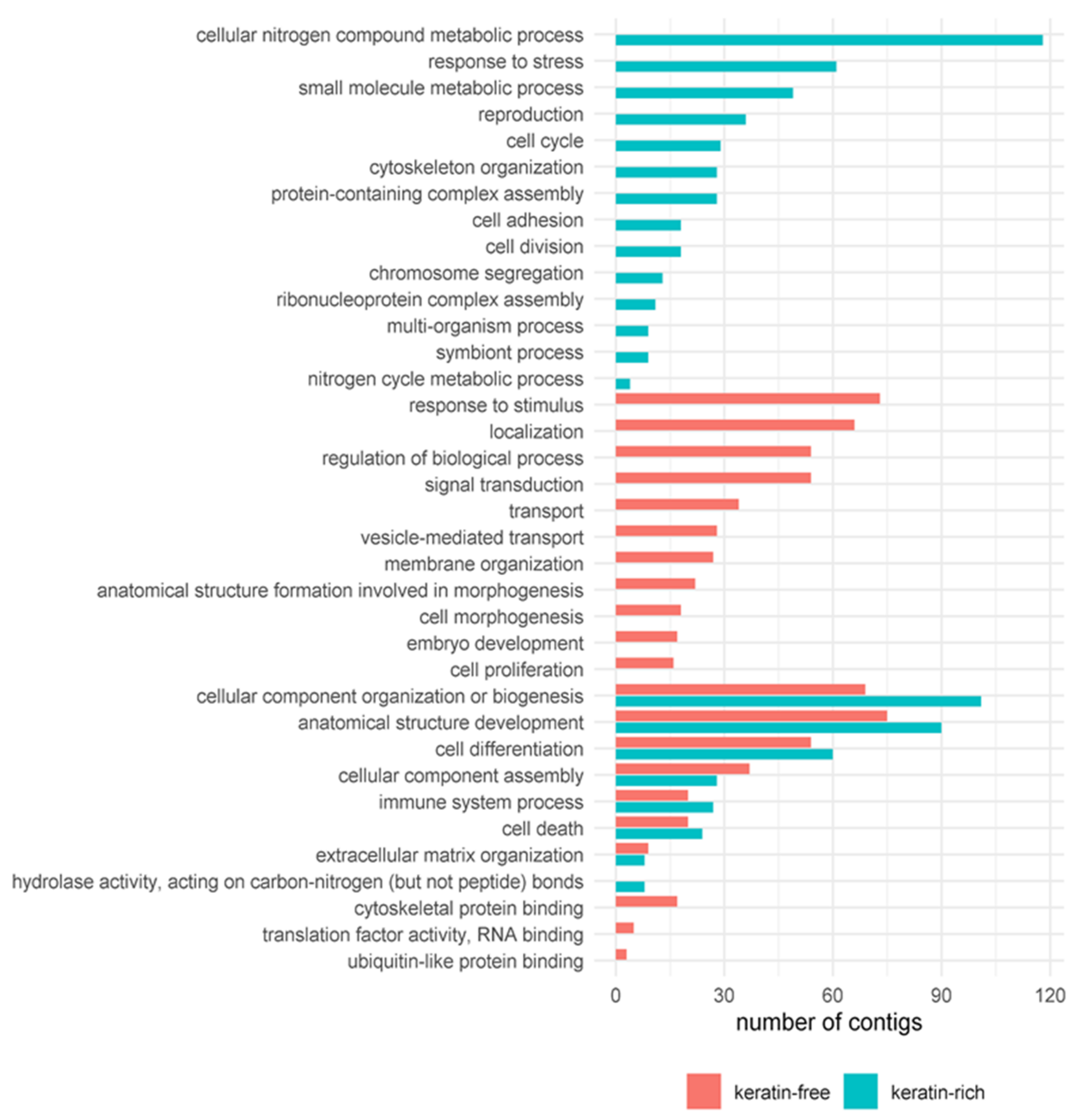
2.7. GO Enrichment Analysis
2.8. Clustering of Contigs Containing a Trypsin Domain
3. Results
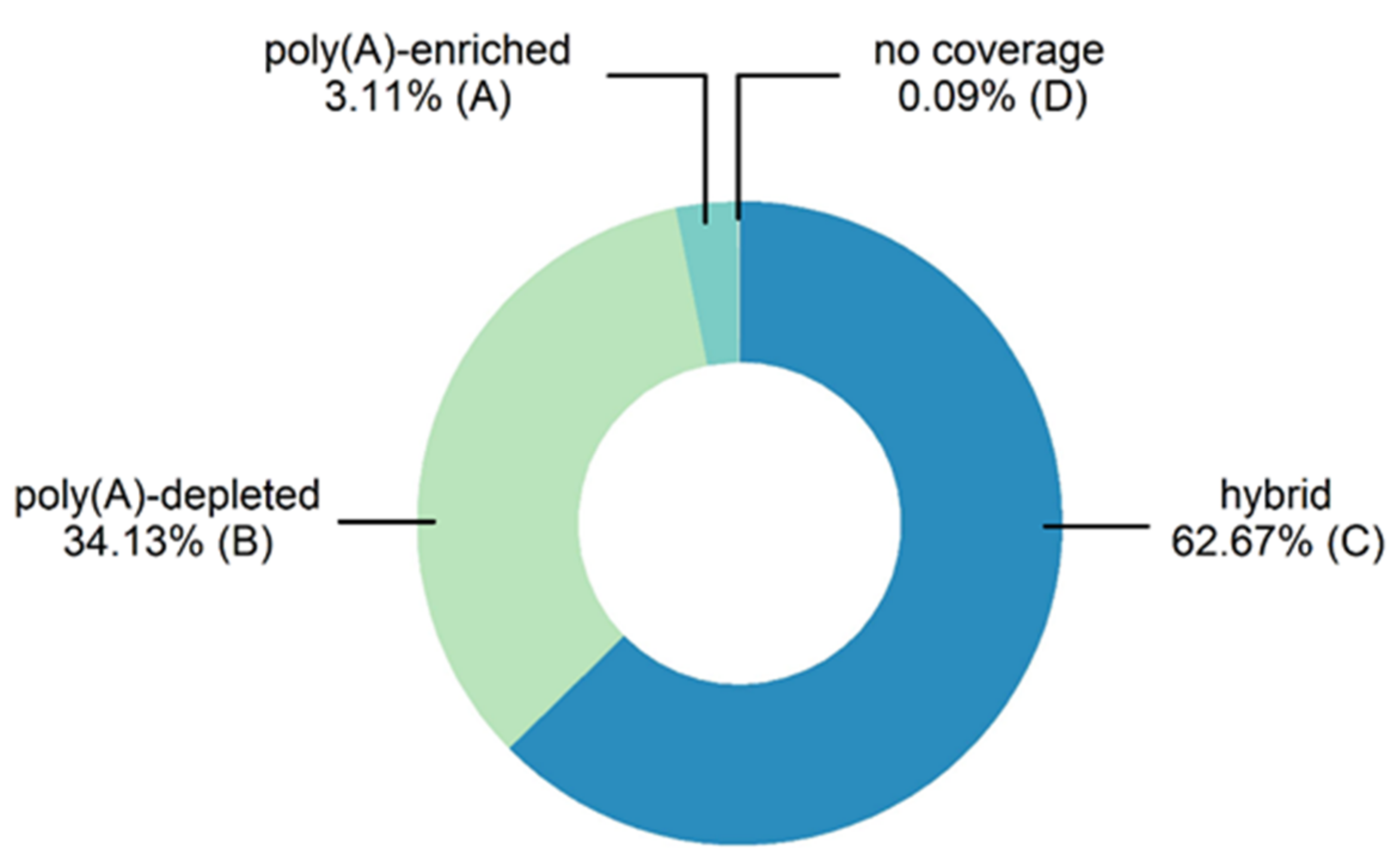
3.1. Generation of a High-Quality Larval Gut Transcriptome from T. bisselliella
- A.
- poly(A)-enriched contigs with no coverage from poly(A)-depleted samples
- B.
- poly(A)-depleted contigs with no coverage from poly(A)-enriched samples
- C.
- contigs covered by reads from poly(A)-enriched and poly(A)-depleted samples
- D.
- no-coverage contigs with no coverage from either of the samples
3.2. Taxonomic Analysis of the Larval Gut Transcriptome
3.3. General Functional Annotation of the Larval Gut Transcriptome
3.4. Diet-Dependent Differentially Expressed Contigs
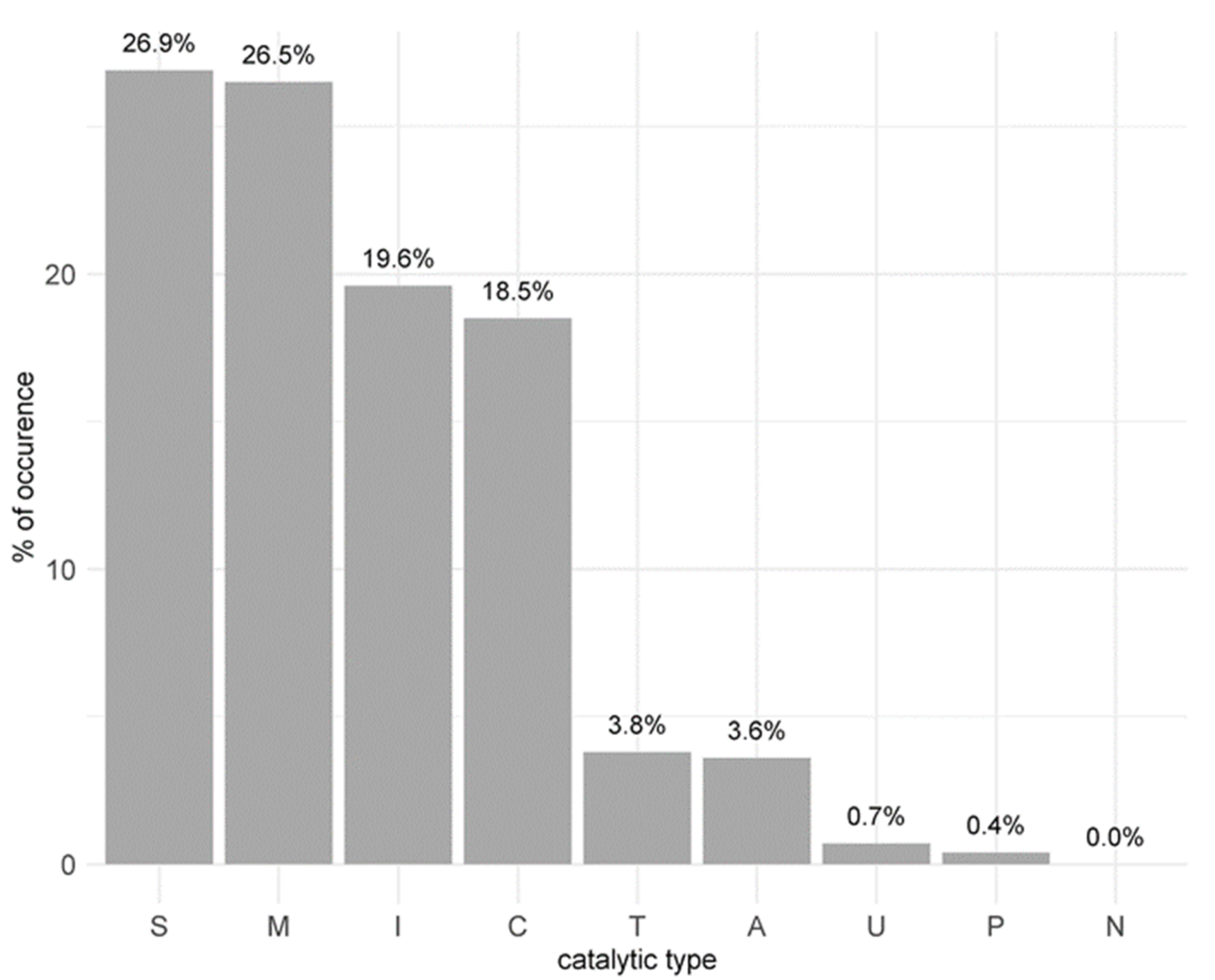
3.5. The Peptidase Landscape of the Larval Gut Transcriptome
3.6. Detailed Screening for Potential Keratinases in the Larval Gut
3.7. Comparison of the Transcriptome with Reported Keratinase Related Enzymes of T. bisselliella
3.8. Identification of Additional Differentially Expressed Genes Potentially Involved in Keratin Degradation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gerard, P.J. The Digestive System of the Keratin-feeding Larvae of Hofmannophila pseudospretella (Lepidoptera: Oecophoridae). N. Z. J. Zool. 2002, 29, 15–22. [Google Scholar] [CrossRef]
- Hughes, J.; Vogler, A.P. Gene Expression in the Gut of Keratin-Feeding Clothes Moths (Tineola) and Keratin Beetles (Trox) Revealed by Subtracted CDNA Libraries. Insect Biochem. Mol. Biol. 2006, 36, 584–592. [Google Scholar] [CrossRef]
- Plarre, R.; Krüger-Carstensen, B. An Attempt to Reconstruct the Natural and Cultural History of the Webbing Clothes Moth Tineola bisselliella Hummel (Lepidoptera: Tineidae). J. Entomol. Acarol. Res. 2011, 43, 83–93. [Google Scholar] [CrossRef]
- Sinkiewicz, I.; Śliwińska, A.; Staroszczyk, H.; Kołodziejska, I. Alternative Methods of Preparation of Soluble Keratin from Chicken Feathers. Waste Biomass Valoriz. 2017, 8, 1043–1048. [Google Scholar] [CrossRef]
- Zhao, W.; Yang, R.; Zhang, Y.; Wu, L. Sustainable and Practical Utilization of Feather Keratin by an Innovative Physicochemical Pretreatment: High Density Steam Flash-Explosion. Green Chem. 2012, 14, 3352–3360. [Google Scholar] [CrossRef]
- Kumawat, T.K.; Sharma, A.; Chandra, V.S.; Keratin Waste, S. The Biodegradable Polymers. Keratin 2018. [Google Scholar] [CrossRef]
- Navone, L.; Speight, R. Understanding the Dynamics of Keratin Weakening and Hydrolysis by Proteases. PLoS ONE 2018, 13, e0202608. [Google Scholar] [CrossRef]
- Wang, B.; Yang, W.; McKittrick, J.; Meyers, M.A. Keratin: Structure, Mechanical Properties, Occurrence in Biological Organisms, and Efforts at Bioinspiration. Prog. Mater. Sci. 2016, 76, 229–318. [Google Scholar] [CrossRef]
- Martinez-Hernandez, A.L.; Velasco-Santos, C.; De Icaza, M.; Castano, V.M. Microstructural Characterisation of Keratin Fibres from Chicken Feathers. Int. J. Environ. Pollut. 2005, 23, 162–178. [Google Scholar] [CrossRef]
- Fraenkel, G.; Blewett, M. The Dietetics of the Clothes Moth, Tinelola bisselliella Hum. J. Exp. Biol. 1946, 22, 156–161. [Google Scholar] [CrossRef]
- Linderstrøm-Lang, K.; Duspiva, F. Beiträge Zur Enzymatischen Histochemie. XVI. Die Verdauung von Keratin durch die Larven der Kleidermotte (Tineola biselliella Humm.). Biol. Chem. 1935, 237, 131–158. [Google Scholar] [CrossRef]
- Day, M.F. Studies on the Digestion of Wool by Insects. I. Microscopy of Digestion of Wool by Clothes Moth Larvae (Tineola bisselliella Humm). Aust. J. Sci. Res. Ser. B Biol. Sci. 1951, 4, 42–48. [Google Scholar] [CrossRef]
- Yoshimura, T.; Tabata, H.; Nishio, M.; Ide, E.; Yamaoka, R.; Hayashiya, K. L-Cysteine Lyase of the Webbing Clothes Moth, Tineola bisselliella. Insect Biochem. 1988, 18, 771–777. [Google Scholar] [CrossRef]
- Robinson, C.P.; Ciccotosto, S.; Sparrow, L.G. Identification of a Key Enzyme in the Digestion of Wool by Larvae of the Webbing Clothes Moth, Tineola bisselliella. J. Text. Inst. 1993, 84, 39–48. [Google Scholar] [CrossRef]
- Waterhouse, D. Studies on the Digestion of Wool by Insects IV. Absorption and Elimination of Metals by Lepidopterous Larvae, with Special Reference to the Clothes Moth, Tineola bisselliella (Humm.). Aust. J. Biol. Sci. 1952, 5, 143. [Google Scholar] [CrossRef]
- Powning, R.F. A Study of Cysteine Desulphydrase in Certain Insects. Aust. J. Biol. Sci. 1954, 7, 308–318. [Google Scholar] [CrossRef] [PubMed]
- Powning, R.F.; Irzykiewicz, H. Cystine and Glutathione Reductases in the Clothes Moth Tineola bisselliella. Aust. J. Biol. Sci. 1960, 13, 59–68. [Google Scholar] [CrossRef]
- Robinson, C.P.; Ciccotosto, S.; Gaal, A.M.; Sparrow, L.G. Purification of Cysteine Lyase from Larvae of the Webbing Clothes Moth, Tineola bisselliella. Insect Biochem. Mol. Biol. 1993, 23, 491–498. [Google Scholar] [CrossRef]
- Waterhouse, D.F. Studies on the Digestion of Wool by Insects VII. Some Features of Digestion in Three Species of Dermestid Larvae and a Comparison with Tineola Larvae. Aust. J. Biol. Sci. 1952, 5, 444–459. [Google Scholar] [CrossRef]
- Crewther, W.G.; McQuade, A.B. The Intestinal Microflora of the Clothes Moth Larva Tineola bisselliella in Relation to Wool Digestion. J. Gen. Microbiol. 1955, 12, 311–313. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hammer, T.J.; Janzen, D.H.; Hallwachs, W.; Jaffe, S.P.; Fierer, N. Caterpillars Lack a Resident Gut Microbiome. Proc. Natl. Acad. Sci. USA 2017, 114, 9641–9646. [Google Scholar] [CrossRef] [PubMed]
- Vilcinskas, A.; Schwabe, M.; Brinkrolf, K.; Plarre, R.; Wielsch, N.; Vogel, H. Larvae of the webbing clothes moth Tineola bisselliella maintain gut bacteria that secrete enzyme cocktails to facilitate the digestion of keratin. Microorganisms 2020, 8, 1415. [Google Scholar] [CrossRef]
- Lange, L.; Huang, Y.; Busk, P.K. Microbial Decomposition of Keratin in Nature—a New Hypothesis of Industrial Relevance. Appl. Microbiol. Biotechnol. 2016, 100, 2083–2096. [Google Scholar] [CrossRef] [PubMed]
- Vidmar, B.; Vodovnik, M. Microbial Keratinases: Enzymes with Promising Biotechnological Applications. Food Technol. Biotechnol. 2018, 56, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 30 August 2017).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a Full-Length Transcriptome without a Genome from RNA-Seq Data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO Applications from Quality Assessments to Gene Prediction and Phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De Novo Transcript Sequence Reconstruction from RNA-Seq Using the Trinity Platform for Reference Generation and Analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS Database of Proteolytic Enzymes, Their Substrates and Inhibitors in 2017 and a Comparison with Peptidases in the PANTHER Database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef] [PubMed]
- UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [CrossRef]
- Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016, 44, D7–D19. [CrossRef]
- Tian, W.; Skolnick, J. How Well Is Enzyme Function Conserved as a Function of Pairwise Sequence Identity? J. Mol. Biol. 2003, 333, 863–882. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in Homology Search: HMMER3 and Convergent Evolution of Coiled-Coil Regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam Protein Families Database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Owlcollab/Owltools; Owl Collaboratory. 2020. Available online: https://github.com/owlcollab/owltools (accessed on 25 September 2020).
- Morgan, M.; Falcon, S.; Gentleman, R. GSEABase: Gene Set Enrichment Data Structures and Methods; R Package Version 1.48.0; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Falcon, S.; Gentleman, R. Using GOstats to Test Gene Lists for GO Term Association. Bioinformatics 2007, 23, 257–258. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for Clustering the next-Generation Sequencing Data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-Hit: A Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Ward, C.W. Resolution of Proteases in the Keratinolytic Larvae of the Webbing Clothes Moth. Aust. J. Biol. Sci. 1975, 28, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Ward, C.W. Diversity of Proteases in the Keratinolytic Larvae of the Webbing Clothes Moth, Tineola bisselliella. Comp. Biochem. Physiol. Part. B Comp. Biochem. 1972, 42, 131–135. [Google Scholar] [CrossRef]
- Ward, C.W. Aminopeptidases in Webbing Clothes Moth Larvae. Properties and Specificities of Enzymes of Highest Electrophoretic Mobility. Aust. J. Biol. Sci. 1975, 28, 447–456. [Google Scholar] [CrossRef]
- Ward, C.W. Properties and Specificity of the Major Anionic Trypsin-like Enzyme in the Keratinolytic Larvae of the Webbing Clothes Moth. Biochim. Biophys. Acta 1975, 391, 201–211. [Google Scholar] [CrossRef]
- Powning, R.F.; Irzykiewicz, H. Studies on the Digestive Proteinase of Clothes Moth Larvae (Tineola bisselliella)—II: Digestion of Wool and Other Substrates by Tineola Proteinase and Comparison with Trypsin. J. Insect Physiol. 1962, 8, 275–284. [Google Scholar] [CrossRef]
- Timmermans, M.J.T.N.; Lees, D.C.; Simonsen, T.J. Towards a Mitogenomic Phylogeny of Lepidoptera. Mol. Phylogenet. Evol. 2014, 79, 169–178. [Google Scholar] [CrossRef]
- Qi, L.; Fang, Q.; Zhao, L.; Xia, H.; Zhou, Y.; Xiao, J.; Li, K.; Ye, G. De Novo Assembly and Developmental Transcriptome Analysis of the Small White Butterfly Pieris rapae. PLoS ONE 2016, 11, e0159258. [Google Scholar] [CrossRef]
- Xie, W.; Lei, Y.; Fu, W.; Yang, Z.; Zhu, X.; Guo, Z.; Wu, Q.; Wang, S.; Xu, B.; Zhou, X.; et al. Tissue-Specific Transcriptome Profiling of Plutella xylostella Third Instar Larval Midgut. Int. J. Biol. Sci. 2012, 8, 1142–1155. [Google Scholar] [CrossRef]
- Zhang, T.; Coates, B.S.; Wang, Y.; Wang, Y.; Bai, S.; Wang, Z.; He, K. Down-Regulation of Aminopeptidase N and ABC Transporter Subfamily G Transcripts in Cry1Ab and Cry1Ac Resistant Asian Corn Borer, Ostrinia furnacalis (Lepidoptera: Crambidae). Int. J. Biol. Sci. 2017, 13, 835–851. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Song, Y.; Li, W.; Shi, J.; Wang, Z. Identification of Putative Chemosensory Receptor Genes from the Athetis dissimilis Antennal Transcriptome. PLoS ONE 2016, 11, e0147768. [Google Scholar] [CrossRef]
- Yadav, C.; Smith, M.L.; Yack, J.E. Transcriptome Analysis of a Social Caterpillar, Drepana arcuata: De Novo Assembly, Functional Annotation and Developmental Analysis. PLoS ONE 2020, 15, e0234903. [Google Scholar] [CrossRef]
- Rahmat, N.L.; Zifruddin, A.N.; Zainal Abidin, C.M.R.; Nor Muhammad, N.-A.; Hassan, M. The Developmental Transcriptome of Bagworm, Metisa plana (Lepidoptera: Psychidae) and Insights into Chitin Biosynthesis Genes. Genes 2021, 12, 7. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J. Transcriptome Assembly Quality Assessment. 2019. Available online: https://github.com/trinityrnaseq/trinityrnaseq/wiki/Transcriptome-Assembly-Quality-Assessment (accessed on 23 February 2020).
- Breeschoten, T.; Ros, V.I.D.; Schranz, M.E.; Simon, S. An Influential Meal: Host Plant Dependent Transcriptional Variation in the Beet Armyworm, Spodoptera exigua (Lepidoptera: Noctuidae). BMC Genom. 2019, 20, 845. [Google Scholar] [CrossRef] [PubMed]
- Tassone, E.E.; Zastrow-Hayes, G.; Mathis, J.; Nelson, M.E.; Wu, G.; Flexner, J.L.; Carrière, Y.; Tabashnik, B.E.; Fabrick, J.A. Sequencing, de Novo Assembly and Annotation of a Pink Bollworm Larval Midgut Transcriptome. GigaScience 2016, 5, 28. [Google Scholar] [CrossRef]
- Terra, W.R.; Ferreira, C.; Jordão, B.P.; Dillon, R.J. Digestive enzymes. In Biology of the Insect Midgut; Lehane, M.J., Billingsley, P.F., Eds.; Springer: Dordrecht, The Netherlands, 1996. [Google Scholar] [CrossRef]
- Srinivasan, A.; Giri, A.P.; Gupta, V.S. Structural and functional diversities in lepidopteran serine proteases. Cell. Mol. Biol. Lett. 2006, 11, 132. [Google Scholar] [CrossRef]
- Powning, R.F.; Day, M.F.; Irzykiewicz, H. Studies on the Digestion of Wool by Insects. II. The Properties of Some Insect Proteinases. Aust. J. Sci. Res. Ser. B Biol. Sci. 1951, 4, 49–63. [Google Scholar] [CrossRef]
- Macedo, M.L.R.; Freire, M.G.M. Insect Digestive Enzymes as a Target for Pest Control. Invertebr. Surviv. J. 2011, 8, 190–198. [Google Scholar]
- De Oliveira Martinez, J.P.; Cai, G.; Nachtschatt, M.; Navone, L.; Zhang, Z.; Robins, K.; Speight, R. Challenges and Opportunities in Identifying and Characterising Keratinases for Value-Added Peptide Production. Catalysts 2020, 10, 184. [Google Scholar] [CrossRef]
- Tesfaye, T.; Sithole, B.; Ramjugernath, D. Valorisation of Chicken Feathers: A Review on Recycling and Recovery Route—Current Status and Future Prospects. Clean Technol. Environ. Policy 2017, 19, 2363–2378. [Google Scholar] [CrossRef]
- Wang, L.; Cheng, G.; Ren, Y.; Dai, Z.; Zhao, Z.-S.; Liu, F.; Li, S.; Wei, Y.; Xiong, J.; Tang, X.-F.; et al. Degradation of Intact Chicken Feathers by Thermoactinomyces sp. CDF and Characterization of Its Keratinolytic Protease. Appl. Microbiol. Biotechnol. 2015, 99, 3949–3959. [Google Scholar] [CrossRef]
- Day, M. Studies on the Digestion of Wool by Insects III. A Comparison Between the Tracheation of the Midgut of Tineola Larvae and That of Other Insect Tissues. Aust. J. Biol. Sci. 1951, 4, 64. [Google Scholar] [CrossRef]
| Category | Number of Contigs |
|---|---|
| Complete | 76,936 |
| Partial | 7999 |
| Fragment | 51,978 |
| Comparison | Differentially Expressed Contigs | Downregulated on kr Diet | Upregulated on kr Diet | |
|---|---|---|---|---|
| poly(A)-enriched | kf↔kr | 625 | 265 | 360 |
| kf↔kf2kr | 1622 | 793 | 829 | |
| kr↔kr2kf | 808 | 366 | 442 |
| Contig | ORF pred. | Database Hit ID | Database Hit Species Name | Subject Coverage | Identity | Pfam Domain | DE |
|---|---|---|---|---|---|---|---|
| DN159021_c0_g1_i1 | n.d. | EU362730.1 | B. subtilis | 19.46% | 98.66% (nt) | - | no |
| DN300764_c0_g1_i1 | n.d. | EU362730.1 | B. subtilis | 19.37% | 99.10% (nt) | - | no |
| DN123288_c0_g1_i1 | yes | A0A0H3YE38 | B. tequilensis | 36.55% | 41.38% (aa) | peptidase S8 (f) | no |
| DN94215_c0_g1_i1 | yes | A0A2H4A2Y5 | M. taiwanensis | 91.64% | 42.29% (aa) | peptidase S8 (c) inhibitor I9 (c) | no |
| Comparison of Differential Gene Expression | |||||
|---|---|---|---|---|---|
| ORF (Contig Name) | kf↔kr | kf↔kf2kr | kr↔kr2kf | UniProt Entry | Pfam Domain |
| DN25_c0_g1_i12 | + | + | + | collagenase | trypsin (c) |
| DN25_c0_g1_i18 | + | + | n.d. | collagenase | trypsin (p) |
| DN3115_c0_g1_i8 | + | + | n.d. | collagenase | trypsin (c) |
| DN2565_c0_g1_i9 | n.d. | + | + | collagenase | trypsin (f) |
| DN2565_c0_g1_i10 | n.d. | + | + | collagenase | trypsin (c) |
| DN25_c0_g1_i3 | n.d. | + | n.d. | collagenase | trypsin (c) |
| DN1221_c0_g2_i2 | n.d. | + | n.d. | collagenase | trypsin (c) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schwabe, M.; Griep, S.; Schmidtberg, H.; Plarre, R.; Goesmann, A.; Vilcinskas, A.; Vogel, H.; Brinkrolf, K. Next-Generation Sequencing Analysis of the Tineola bisselliella Larval Gut Transcriptome Reveals Candidate Enzymes for Keratin Digestion. Genes 2021, 12, 1113. https://doi.org/10.3390/genes12081113
Schwabe M, Griep S, Schmidtberg H, Plarre R, Goesmann A, Vilcinskas A, Vogel H, Brinkrolf K. Next-Generation Sequencing Analysis of the Tineola bisselliella Larval Gut Transcriptome Reveals Candidate Enzymes for Keratin Digestion. Genes. 2021; 12(8):1113. https://doi.org/10.3390/genes12081113
Chicago/Turabian StyleSchwabe, Michael, Sven Griep, Henrike Schmidtberg, Rudy Plarre, Alexander Goesmann, Andreas Vilcinskas, Heiko Vogel, and Karina Brinkrolf. 2021. "Next-Generation Sequencing Analysis of the Tineola bisselliella Larval Gut Transcriptome Reveals Candidate Enzymes for Keratin Digestion" Genes 12, no. 8: 1113. https://doi.org/10.3390/genes12081113
APA StyleSchwabe, M., Griep, S., Schmidtberg, H., Plarre, R., Goesmann, A., Vilcinskas, A., Vogel, H., & Brinkrolf, K. (2021). Next-Generation Sequencing Analysis of the Tineola bisselliella Larval Gut Transcriptome Reveals Candidate Enzymes for Keratin Digestion. Genes, 12(8), 1113. https://doi.org/10.3390/genes12081113






