Exploring Viral Diversity in a Gypsum Karst Lake Ecosystem Using Targeted Single-Cell Genomics
Abstract
1. Introduction
2. Materials and Methods
3. Results
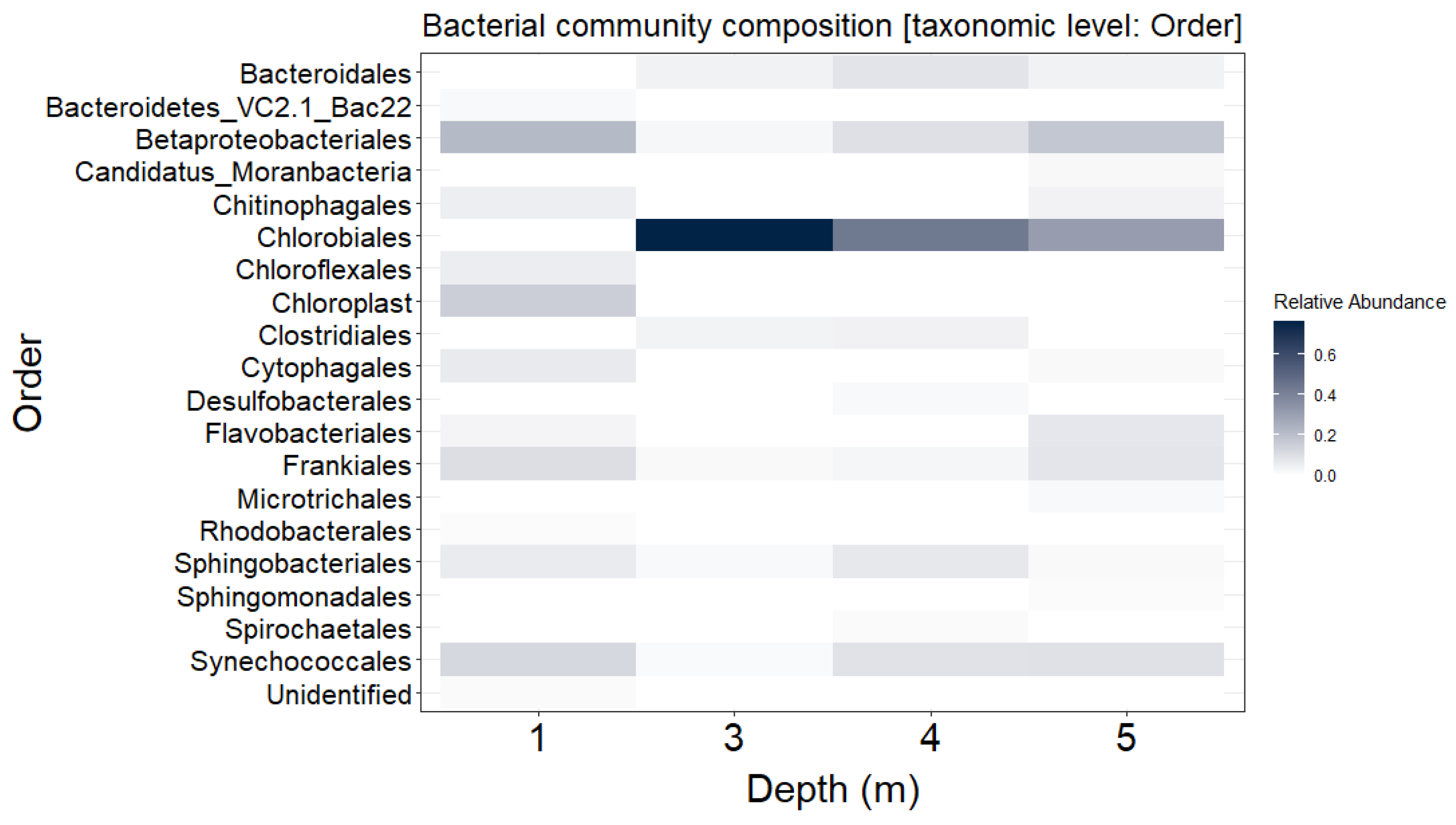
3.1. Distribution of Bacterial Community throughout the Water Column of Lake Kirkilai
3.2. Bacterial Diversity within Single Amplified Genomes
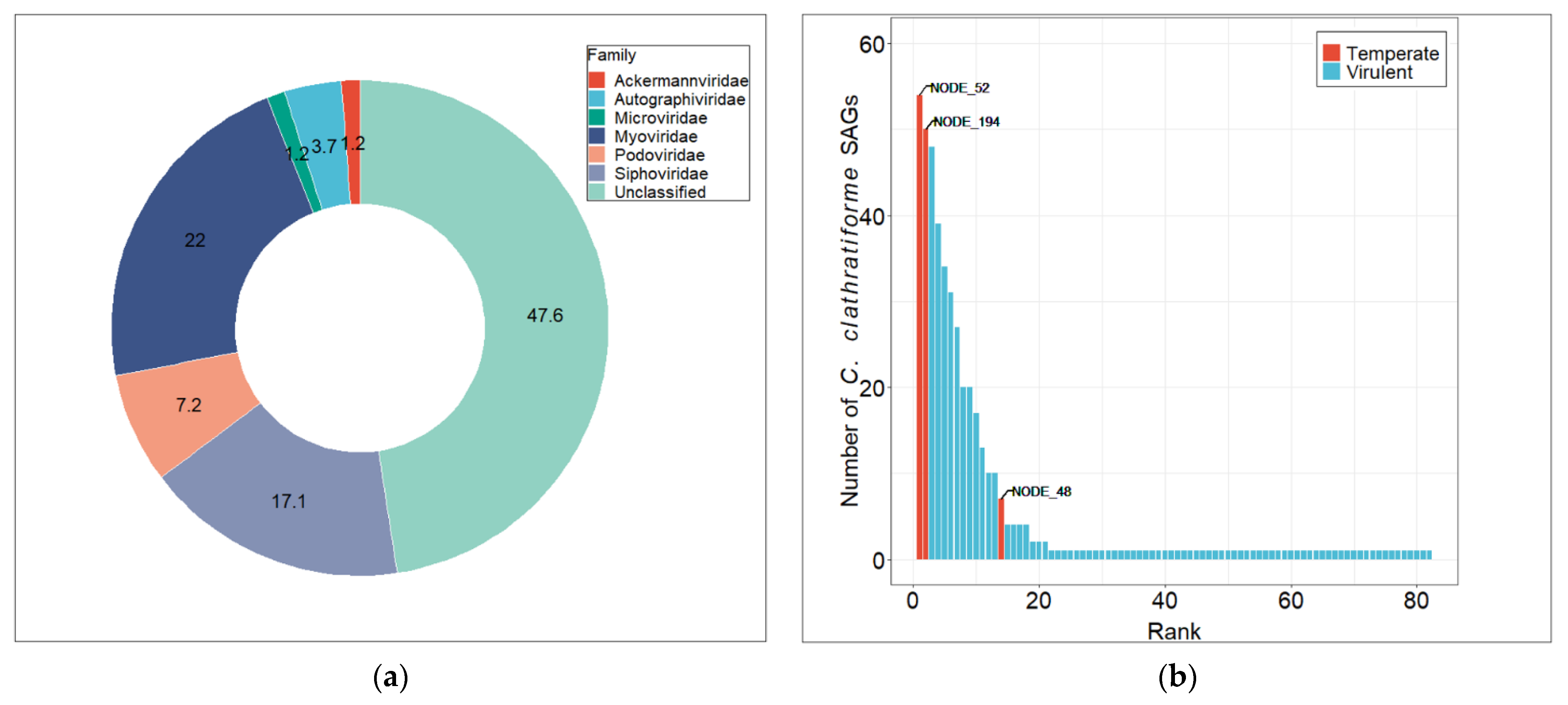
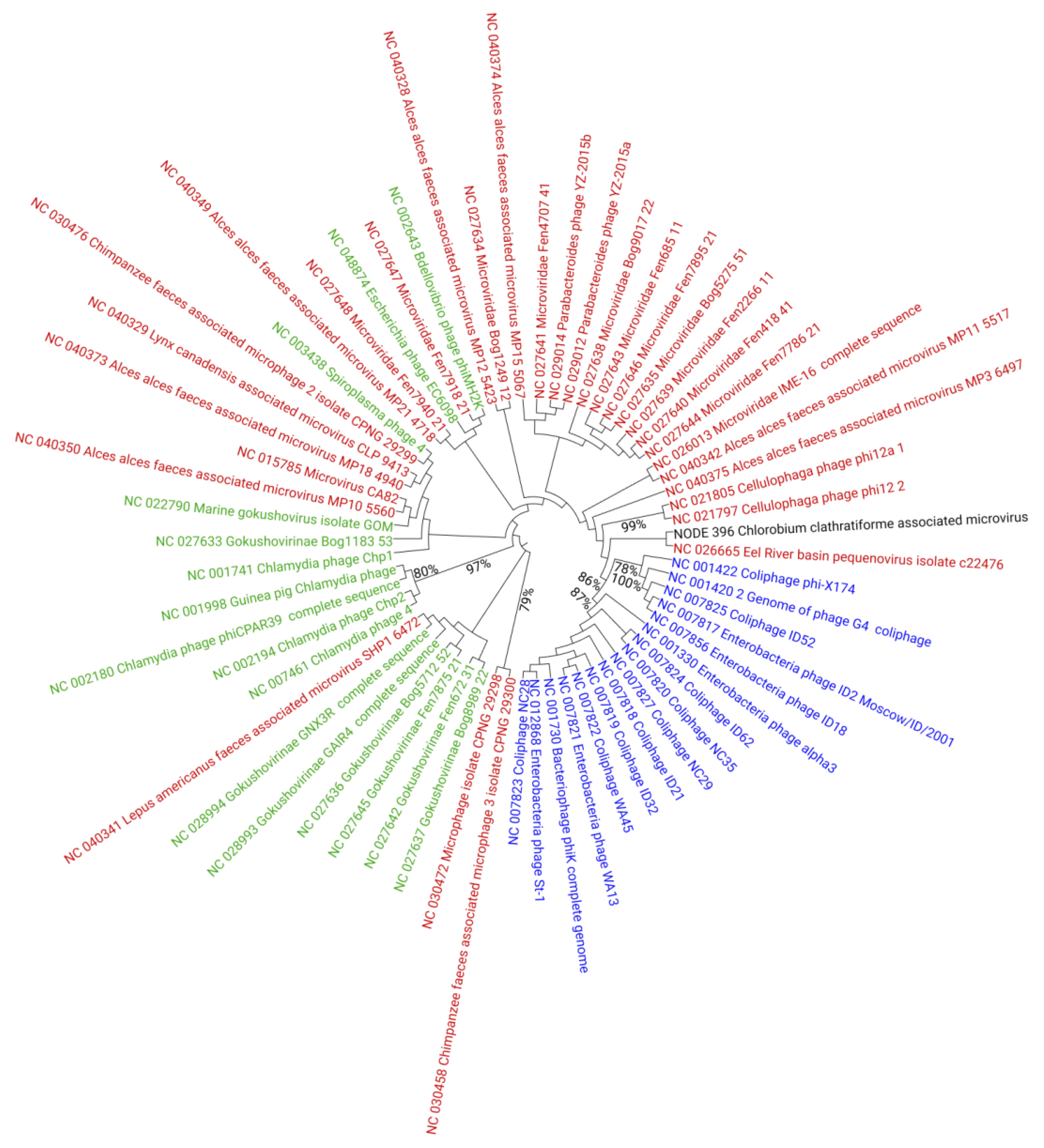
3.3. Diversity and Distribution of Chlorobium clathratiforme-Associated Phage Sequences
3.4. Gene Content Analysis of Chlorobium clathratiforme-Associated Phage Sequences
3.5. Analysis of CRISPR-Cas Loci in the Chlorobium clathratiforme Genome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Green, J.; Paget, M. Bacterial redox sensors. Nat. Rev. Microbiol. 2004, 2, 954–966. [Google Scholar] [CrossRef]
- Keshri, J.; Pradeep Ram, A.S.; Nana, P.A.; Sime-Ngando, T. Taxonomical resolution and distribution of bacterioplankton along the vertical gradient reveals pronounced spatiotemporal patterns in contrasted temperate freshwater lakes. Microb. Ecol. 2018, 76, 372–386. [Google Scholar] [CrossRef] [PubMed]
- İnceoğlu, Ö.; Llirós, M.; Crowe, S.A.; Morana, C.; Darchambeau, F.; Borges, A.V.; Descy, J.P.; Servais, P. Vertical Distribution of functional potential and active microbial communities in meromictic Lake Kivu. Microb. Ecol. 2015, 70, 596–611. [Google Scholar] [CrossRef] [PubMed]
- Baatar, B.; Chiang, P.W.; Rogozin, D.Y.; Wu, Y.T.; Tseng, C.H.; Yang, C.Y.; Chiu, H.H.; Oyuntsetseg, B.; Degermendzhy, A.G.; Tang, S.L. Bacterial communities of three saline meromictic lakes in Central Asia. PLoS ONE 2016, 11, e0150847. [Google Scholar] [CrossRef] [PubMed]
- Diao, M.; Huisman, J.; Muyzer, G. Spatio-temporal dynamics of sulfur bacteria during oxic-anoxic regime shifts in a seasonally stratified lake. FEMS Microbiol. Ecol. 2018, 94, fiy040. [Google Scholar] [CrossRef]
- Tonolla, M.; Peduzzi, S.; Hahn, D.; Peduzzi, R. Spatio-temporal distribution of phototrophic sulfur bacteria in the chemocline of meromictic Lake Cadagno (Switzerland). FEMS Microbiol. Ecol. 2003, 43, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, L.H.; Habicht, K.S.; Peduzzi, S.; Tonolla, M.; Canfield, D.E.; Miller, M.; Cox, R.P.; Frigaard, N.-U. Dominance of a clonal green sulfur bacterial population in a stratified lake. FEMS Microbiol. Ecol. 2009, 70, 30–41. [Google Scholar] [CrossRef]
- Llorens-Marès, T.; Liu, Z.; Allen, L.Z.; Rusch, D.B.; Craig, M.T.; Dupont, C.L.; Bryant, D.A.; Casamayor, E.O. Speciation and ecological success in dimly lit waters: Horizontal gene transfer in a green sulfur bacteria bloom unveiled by metagenomic assembly. ISME J. 2017, 11, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Danza, F.; Ravasi, D.; Storelli, N.; Roman, S.; Lüdin, S.; Bueche, M.; Tonolla, M. Bacterial diversity in the water column of meromictic Lake Cadagno and evidence for seasonal dynamics. PLoS ONE 2018, 13, e0209743. [Google Scholar] [CrossRef] [PubMed]
- Weitz, J.S.; Stock, C.A.; Wilhelm, S.W.; Bourouiba, L.; Coleman, M.L.; Buchan, A.; Follows, M.J.; Fuhrman, J.A.; Jover, L.F.; Lennon, J.T.; et al. A multitrophic model to quantify the effects of marine viruses on microbial food webs and ecosystem processes. ISME J. 2015, 9, 1352–1364. [Google Scholar] [CrossRef]
- Zimmerman, A.E.; Howard-Varona, C.; Needham, D.M.; John, S.G.; Worden, A.Z.; Sullivan, M.B.; Waldbauer, J.R.; Coleman, M.L. Metabolic and biogeochemical consequences of viral infection in aquatic ecosystems. Nat. Rev. Microbiol. 2020, 18, 21–34. [Google Scholar] [CrossRef]
- Sullivan, M.B. Viromes, not gene markers, for studying double-stranded DNA virus communities. J. Virol. 2015, 89, 2459–2461. [Google Scholar] [CrossRef]
- Simmonds, P.; Adams, M.J.; Benk, M.; Breitbart, M.; Brister, J.R.; Carstens, E.B.; Davison, A.J.; Delwart, E.; Gorbalenya, A.E.; Harrach, B.; et al. Consensus statement: Virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 2017, 15, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Wilson, W.H.; Gilg, I.C.; Moniruzzaman, M.; Field, E.K.; Koren, S.; Lecleir, G.R.; Martínez Martínez, J.; Poulton, N.J.; Swan, B.K.; Stepanauskas, R.; et al. Genomic exploration of individual giant ocean viruses. ISME J. 2017, 11, 1736–1745. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Hernandez, F.; Fornas, O.; Lluesma Gomez, M.; Bolduc, B.; De La Cruz Peña, M.J.; Martínez, J.M.; Anton, J.; Gasol, J.M.; Rosselli, R.; Rodriguez-Valera, F.; et al. Single-virus genomics reveals hidden cosmopolitan and abundant viruses. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Martínez Martínez, J.; Martinez-Hernandez, F.; Martinez-Garcia, M. Single-virus genomics and beyond. Nat. Rev. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, C.M.; Rodriguez-Valera, F.; Garcia-Heredia, I.; Martin-Cuadrado, A.B.; Ghai, R. Reconstruction of novel cyanobacterial siphovirus genomes from Mediterranean metagenomic fosmids. Appl. Environ. Microbiol. 2013, 79, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, C.M.; Ghai, R.; Saghaï, A.; López-García, P.; Rodriguez-Valeraa, F. Genomes of abundant and widespread viruses from the deep ocean. MBio 2016, 7, e00805-16. [Google Scholar] [CrossRef]
- Danhorn, T.; Young, C.R.; Delong, E.F. Comparison of large-insert, small-insert and pyrosequencing libraries for metagenomic analysis. ISME J. 2012, 6, 2056–2066. [Google Scholar] [CrossRef]
- Labonté, J.M.; Swan, B.K.; Poulos, B.; Luo, H.; Koren, S.; Hallam, S.J.; Sullivan, M.B.; Woyke, T.; Eric Wommack, K.; Stepanauskas, R. Single-cell genomics-based analysis of virus–host interactions in marine surface bacterioplankton. ISME J. 2015, 9, 2386–2399. [Google Scholar] [CrossRef]
- Munson-McGee, J.H.; Peng, S.; Dewerff, S.; Stepanauskas, R.; Whitaker, R.J.; Weitz, J.S.; Young, M.J. A virus or more in (nearly) every cell: Ubiquitous networks of virus–host interactions in extreme environments. ISME J. 2018, 12, 1706–1714. [Google Scholar] [CrossRef] [PubMed]
- Castillo, Y.M.; Sebastián, M.; Forn, I.; Grimsley, N.; Yau, S.; Moraru, C.; Vaqué, D. Visualization of viral infection dynamics in a unicellular eukaryote and quantification of viral production using VirusFISH. bioRxiv 2019, 37–49. [Google Scholar] [CrossRef]
- Berg, M.; Goudeau, D.; Olmsted, C.; McMahon, K.D.; Yitbarek, S.; Thweatt, J.L.; Bryant, D.A.; Eloe-Fadrosh, E.A.; Malmstrom, R.R.; Roux, S. Host population diversity as a driver of viral infection cycle in wild populations of green sulfur bacteria with long standing virus-host interactions. ISME J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Hawley, A.K.; Torres Beltran, M.; Scofield, M.; Schwientek, P.; Stepanauskas, R.; Woyke, T.; Hallam, S.J.; Sullivan, M.B. Ecology and evolution of viruses infecting uncultivated SUP05 bacteria as revealed by single-cell- and meta-genomics. eLife 2014, 3, e03125. [Google Scholar] [CrossRef] [PubMed]
- Humphries, P.; Baldwin, D.S. Drought and aquatic ecosystems: An introduction. Freshw. Biol. 2003, 48, 1141–1146. [Google Scholar] [CrossRef]
- Leibold, M.A.; Holyoak, M.; Mouquet, N.; Amarasekare, P.; Chase, J.M.; Hoopes, M.F.; Holt, R.D.; Shurin, J.B.; Law, R.; Tilman, D.; et al. The metacommunity concept: A framework for multi-scale community ecology. Ecol. Lett. 2004, 7, 601–613. [Google Scholar] [CrossRef]
- Taminskas, J.; Marcinkevičius, V. Karst geoindicators of environmental change: The case of Lithuania. Environ. Geol. 2002, 42, 757–766. [Google Scholar] [CrossRef]
- Paškauskas, R.; Kučinskienė, A.; Žvikas, A. Sulfate-reducing bacteria in gypsum karst lakes of northern Lithuania. Mikrobiologiya 2005, 74, 823–830. [Google Scholar] [CrossRef]
- Krevš, A.; Kučinskienė, A. Vertical distribution of bacteria and intensity of microbiological processes in two stratified gypsum Karst Lakes in Lithuania. Knowl. Manag. Aquat. Ecosyst. 2011, 2. [Google Scholar] [CrossRef]
- Krevš, A.; Kučinskienė, A.; Kuisienė, N. Anoxygenic phototrophic bacteria from gypsum karst lakes of Lithuania. Inland Water Biol. 2014, 7, 25–33. [Google Scholar] [CrossRef]
- Imhoff, J.F. The Family Chlorobiaceae. In The Prokaryotes: Other Major Lineages of Bacteria and the Archaea; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 501–514. ISBN 978-3-642-38954-2. [Google Scholar]
- Riemann, L.; Leitet, C.; Pommier, T.; Simu, K.; Holmfeldt, K.; Larsson, U.; Hagström, A. The native bacterioplankton community in the central Baltic Sea is influenced by freshwater bacterial species. Appl. Environ. Microbiol. 2008, 74, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Primers for marine microbiome studies. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Mahé, F.; Rognes, T.; Quince, C.; de Vargas, C.; Dunthorn, M. Swarm: Robust and fast clustering method for amplicon-based studies. PeerJ 2014, 2, e593. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.O.N.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Kieft, K.; Zhou, Z.; Anantharaman, K. VIBRANT: Automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome 2020, 8, 90. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Menzel, P.; Ng, K.L.; Krogh, A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 2016, 7, 11257. [Google Scholar] [CrossRef]
- Uritskiy, G.V.; DiRuggiero, J.; Taylor, J. MetaWRAP—A flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 2018, 6, 158. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-W.; Simmons, B.A.; Singer, S.W. MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 2016, 32, 605–607. [Google Scholar] [CrossRef] [PubMed]
- Alneberg, J.; Bjarnason, B.S.; de Bruijn, I.; Schirmer, M.; Quick, J.; Ijaz, U.Z.; Lahti, L.; Loman, N.J.; Andersson, A.F.; Quince, C. Binning metagenomic contigs by coverage and composition. Nat. Methods 2014, 11, 1144–1146. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.D.; Froula, J.; Egan, R.; Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 2015, 3, e1165. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Russel, J.; Pinilla-Redondo, R.; Mayo-Muñoz, D.; Shah, S.A.; Sørensen, S.J. CRISPRCasTyper: Automated Identification, Annotation, and Classification of CRISPR-Cas Loci. CRISPR J. 2020, 3, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Mitrofanov, A.; Alkhnbashi, O.S.; Shmakov, S.A.; Makarova, K.S.; Koonin, E.V.; Backofen, R. CRISPRidentify: Identification of CRISPR arrays using machine learning approach. Nucleic Acids Res. 2021, 49, e20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Mirdita, M.; Levy Karin, E.; Norroy, C.; Galiez, C.; Söding, J. SpacePHARER: Sensitive identification of phages from CRISPR spacers in prokaryotic hosts. Bioinformatics 2021. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The viral proteomic tree server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef]
- Roux, S.; Adriaenssens, E.M.; Dutilh, B.E.; Koonin, E.V.; Kropinski, A.M.; Krupovic, M.; Kuhn, J.H.; Lavigne, R.; Brister, J.R.; Varsani, A.; et al. Minimum information about an uncultivated virus genome (MIUVIG). Nat. Biotechnol. 2019, 37, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M.; Rohwer, F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005, 13, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Overmann, J.; Pfennig, N. Pelodictyon phaeoclathratiforme sp. nov.; a new brown-colored member of the Chlorobiaceae forming net-like colonies. Arch. Microbiol. 1989, 152, 401–406. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC—A novel tool to calculate the intergenomic similarities of prokaryote-infecting viruses. Viruses 2020, 12, 1268. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.; Brister, J.R. How to Name and Classify Your Phage: An Informal Guide. Viruses 2017, 9, 70. [Google Scholar] [CrossRef] [PubMed]
- Crummett, L.T.; Puxty, R.J.; Weihe, C.; Marston, M.F.; Martiny, J.B.H. The genomic content and context of auxiliary metabolic genes in marine cyanomyoviruses. Virology 2016, 499, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, C.M.; Guyomar, C.; Roux, S.; Lavigne, R.; Rodriguez-Valera, F.; Sullivan, M.B.; Gillet, R.; Forterre, P.; Krupovic, M. Numerous cultivated and uncultivated viruses encode ribosomal proteins. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Tisza, M.J.; Pastrana, D.V.; Welch, N.L.; Stewart, B.; Peretti, A.; Starrett, G.J.; Pang, Y.-Y.S.; Krishnamurthy, S.R.; Pesavento, P.A.; McDermott, D.H.; et al. Discovery of several thousand highly diverse circular DNA viruses. eLife 2020, 9, e51971. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Brum, J.R.; Dutilh, B.E.; Sunagawa, S.; Duhaime, M.B.; Loy, A.; Poulos, B.T.; Solonenko, N.; Lara, E.; Poulain, J.; et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 2016, 537, 689–693. [Google Scholar] [CrossRef]
- Roux, S.; Hallam, S.J.; Woyke, T.; Sullivan, M.B. Viral dark matter and virus–host interactions resolved from publicly available microbial genomes. eLife 2015, 4, e08490. [Google Scholar] [CrossRef]
- Mavrich, T.N.; Hatfull, G.F. Bacteriophage evolution differs by host, lifestyle and genome. Nat. Microbiol. 2017, 2, 17112. [Google Scholar] [CrossRef]
- Luque, A.; Silveira, C.B. Quantification of lysogeny caused by phage coinfections in microbial communities from biophysical principles. mSystems 2020, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Trinh, J.T.; Székely, T.; Shao, Q.; Balázsi, G.; Zeng, L. Cell fate decisions emerge as phages cooperate or compete inside their host. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Erez, Z.; Steinberger-Levy, I.; Shamir, M.; Doron, S.; Stokar-Avihail, A.; Peleg, Y.; Melamed, S.; Leavitt, A.; Savidor, A.; Albeck, S.; et al. Communication between viruses guides lysis-lysogeny decisions. Nature 2017, 541, 488–493. [Google Scholar] [CrossRef] [PubMed]
- Knowles, B.; Silveira, C.B.; Bailey, B.A.; Barott, K.; Cantu, V.A.; Cobián-Güemes, A.G.; Coutinho, F.H.; Dinsdale, E.A.; Felts, B.; Furby, K.A.; et al. Lytic to temperate switching of viral communities. Nature 2016, 531, 466–470. [Google Scholar] [CrossRef] [PubMed]
- Howard-Varona, C.; Hargreaves, K.R.; Abedon, S.T.; Sullivan, M.B. Lysogeny in nature: Mechanisms, impact and ecology of temperate phages. ISME J. 2017, 11, 1511–1520. [Google Scholar] [CrossRef]
- Angly, F.E.; Felts, B.; Breitbart, M.; Salamon, P.; Edwards, R.A.; Carlson, C.; Chan, A.M.; Haynes, M.; Kelley, S.; Liu, H.; et al. The marine viromes of four oceanic regions. PLoS Biol. 2006, 4, 2121–2131. [Google Scholar] [CrossRef]
- Hu, X.; Liu, J.; Liu, H.; Zhuang, G.; Xun, L. Sulfur metabolism by marine heterotrophic bacteria involved in sulfur cycling in the ocean. Sci. China Earth Sci. 2018, 61, 1369–1378. [Google Scholar] [CrossRef]
- Wasmund, K.; Mußmann, M.; Loy, A. The life sulfuric: Microbial ecology of sulfur cycling in marine sediments. Environ. Microbiol. Rep. 2017, 9, 323–344. [Google Scholar] [CrossRef] [PubMed]
- Harada, M.; Yoshida, T.; Kuwahara, H.; Shimamura, S.; Takaki, Y.; Kato, C.; Miwa, T.; Miyake, H.; Maruyama, T. Expression of genes for sulfur oxidation in the intracellular chemoautotrophic symbiont of the deep-sea bivalve calyptogena okutanii. Extremophiles 2009, 13, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Grein, F.; Ramos, A.R.; Venceslau, S.S.; Pereira, I.A.C. Unifying concepts in anaerobic respiration: Insights from dissimilatory sulfur metabolism. Biochim. Biophys. Acta-Bioenerg. 2013, 1827, 145–160. [Google Scholar] [CrossRef]
- Ding, H.; Clark, R.J.; Ding, B. IscA mediates iron delivery for assembly of iron-sulfur clusters in IscU under the limited accessible free iron conditions. J. Biol. Chem. 2004, 279, 37499–37504. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Zheng, C.; Liu, J. Characterization of iron-sulfur cluster assembly protein isca from Acidithiobacillus ferrooxidans. Biochemistry 2013, 78, 244–251. [Google Scholar] [CrossRef]
- López-Bueno, A.; Tamames, J.; Velázquez, D.; Moya, A.; Quesada, A.; Alcamí, A. High diversity of the viral community from an Antarctic lake. Science 2009, 326, 858–861. [Google Scholar] [CrossRef] [PubMed]
- Tucker, K.P.; Parsons, R.; Symonds, E.M.; Breitbart, M. Diversity and distribution of single-stranded DNA phages in the North Atlantic Ocean. ISME J. 2011, 5, 822–830. [Google Scholar] [CrossRef]
| Metabolism | Pathway | Total AMGs | AMG KO a | AMG KO Name |
|---|---|---|---|---|
| Carbohydrate metabolism | Pentose phosphate pathway | 1 | K07404 | pgl; 6-phosphogluconolactonase |
| Fructose and mannose metabolism | 4 | K01711 | Gmd; GDPmannose 4,6-dehydratase | |
| K02377 | fcl; GDP-L-fucose synthase | |||
| Galactose metabolism | 1 | K01784 | gale; UDP-glucose 4-epimerase | |
| Amino sugar and nucleotide sugar metabolism | 7 | K01709 | rfbG; CDP-glucose 4,6-dehydratase | |
| K01711 | Gmd; GDPmannose 4,6-dehydratase | |||
| K02377 | fcl; GDP-L-fucose synthase | |||
| K13010 | rfbE; perosamine synthetase | |||
| C5-Branched dibasic acid and Butanoate metabolism | 1 | K01652 | ilvB, ilvG, ilvI; acetolactate synthase I/II/III large subunit | |
| Energy metabolism | Sulfur metabolism | 2 | K00390 | cysH; thioredoxin-dependent phosphoadenosine phosphosulfate (PAPS) reductase |
| 2 | K02439 | TST; thiosulfate sulfurtransferase (rhodanese) | ||
| Amino acid metabolism | Alanine, aspartate and glutamate metabolism | 1 | K01953 | asnB; asparagine synthase (glutamine-hydrolyzing) |
| Glycine, serine and threonine metabolism | 1 | K00613 | Glycine amidinotransferase | |
| Cysteine and methionine metabolism | 3 | K00558 | dcm; DNA (cytosine-5)-methyltransferase | |
| K00789 | metK; S-adenosylmethionine synthetase | |||
| Valine, leucine, and isoleucine biosynthesis | 1 | K01652 | ilvB, ilvG, ilvI; acetolactate synthase I/II/III large subunit | |
| Arginine and proline metabolism | 1 | K00613 | Glycine amidinotransferase | |
| Cysteine and methionine metabolism | K00558 | dcm; DNA (cytosine-5)-methyltransferase | ||
| Metabolism of cofactors and vitamins | Pantothenate and CoA biosynthesis | 1 | K01652 | ilvB, ilvG, ilvI; acetolactate synthase I/II/III large subunit |
| Porphyrin and chlorophyll metabolism | 1 | K04034 | bchE; anaerobic magnesium-protoporphyrin IX monomethyl ester cyclase | |
| Ubiquinone and other terpenoid-quinone biosynthesis | 2 | K03183 | ubiE; demethylmenaquinone methyltransferase/2-methoxy-6-polyprenyl-1,4-benzoquinol methylase | |
| Unclassified | – | 1 | K02039 | phoU; phosphate transport system protein |
| CRISPR No. (Subfamily) | Array Length (bp) | Repeat Sequence (5′→3′) | Cas Proteins | Number of Spacers | Spacer No. | Viral Contig Match | ViralDB Match (e ≤ 10−4) | NCBI Accession No. |
|---|---|---|---|---|---|---|---|---|
| CRISPR1 (subfamily III-A) | 2983 | ATTATCTCCGACCTGACATATCAAAAGGGATTACGAC | Cas1, Cas2, Cas6, Cas10, Csm2, Csm3, Csm4, Csm5, RT, DExK | 40 | 13 | NODE_31 | – | – |
| 35 | NODE_77 | – | – | |||||
| 37 | NODE_31 | – | – | |||||
| CRISPR2 (subfamily I-C) | 2078 | GTCGCGCCCCCTGCGGGCGCGTGGATTGAAAC | – | 31 | 1 | NODE_31 | – | – |
| 4 | NODE_113 | – | – | |||||
| 5 | NODE_396 | Gokushovirus WZ-2015a | KT264813.1 | |||||
| 16 | – | Microviridae ctec913 | MH617588.1 | |||||
| 18 | NODE_113 | – | – | |||||
| 19 | NODE_113 | – | – | |||||
| CRISPR3 (subfamily Unknown) | 5829 | GTTTCAATCCGCTATGCGTGCAATAAGATATGATG | Cas 1, Cas2 | 81 | 52 | NODE_31 | – | – |
| 54 | NODE_48 | Erythrobacter phage vB_EliS_R6L | KY006853.1 | |||||
| 62 | NODE_395 | – | – | |||||
| 66 | NODE_31 | – | – | |||||
| 73 | NODE_48 | – | – |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šulčius, S.; Alzbutas, G.; Juknevičiūtė, V.; Šimoliūnas, E.; Venckus, P.; Šimoliūnienė, M.; Paškauskas, R. Exploring Viral Diversity in a Gypsum Karst Lake Ecosystem Using Targeted Single-Cell Genomics. Genes 2021, 12, 886. https://doi.org/10.3390/genes12060886
Šulčius S, Alzbutas G, Juknevičiūtė V, Šimoliūnas E, Venckus P, Šimoliūnienė M, Paškauskas R. Exploring Viral Diversity in a Gypsum Karst Lake Ecosystem Using Targeted Single-Cell Genomics. Genes. 2021; 12(6):886. https://doi.org/10.3390/genes12060886
Chicago/Turabian StyleŠulčius, Sigitas, Gediminas Alzbutas, Viktorija Juknevičiūtė, Eugenijus Šimoliūnas, Petras Venckus, Monika Šimoliūnienė, and Ričardas Paškauskas. 2021. "Exploring Viral Diversity in a Gypsum Karst Lake Ecosystem Using Targeted Single-Cell Genomics" Genes 12, no. 6: 886. https://doi.org/10.3390/genes12060886
APA StyleŠulčius, S., Alzbutas, G., Juknevičiūtė, V., Šimoliūnas, E., Venckus, P., Šimoliūnienė, M., & Paškauskas, R. (2021). Exploring Viral Diversity in a Gypsum Karst Lake Ecosystem Using Targeted Single-Cell Genomics. Genes, 12(6), 886. https://doi.org/10.3390/genes12060886






