Genotyping-by-Sequencing-Based Genome-Wide Association Studies of Fusarium Wilt Resistance in Radishes (Raphanus sativus L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Pathogen Inoculation
2.3. Preparation of Genotyping-by-Sequencing (GBS) Libraries
2.4. Sequencing and Genotyping
2.5. Imputation and SNP Filtering
2.6. Population Structure, Linkage Disequilibrium, and Genetic Diversity
2.7. Genome-Wide Association Analysis
2.8. Linkage Map Construction & QTL Mapping
3. Results
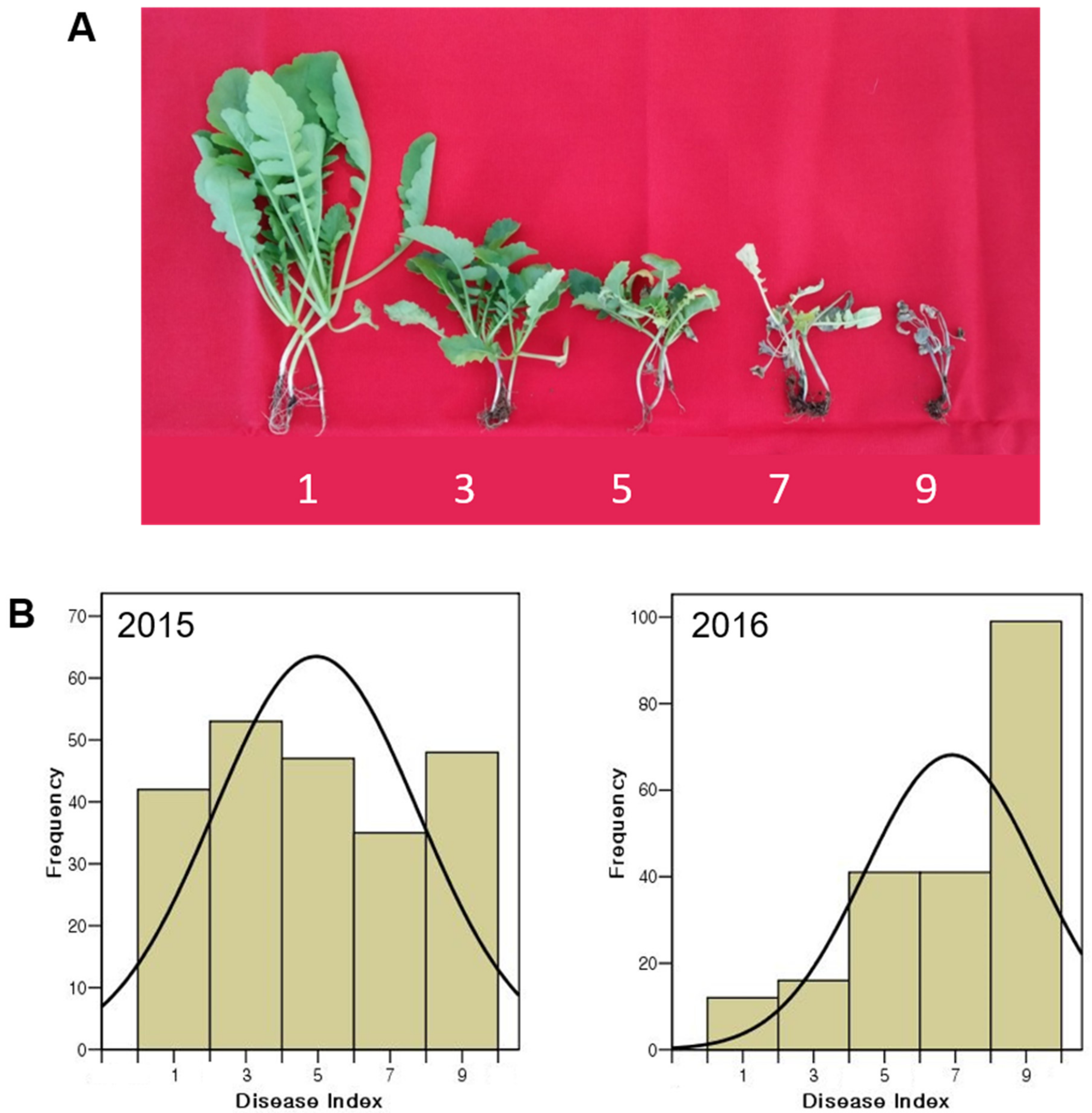
3.1. Phenotypic Variation
3.2. Characterization and Distribution of Markers in the Radish Genome
3.3. Population Structure and Linkage Disequilibrium with Genotypic Data
3.4. Genome-Wide Association Study (GWAS)
3.5. QTL Mapping for Fusarium Wilt Resistance
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, O.N.; Park, H.Y. Assessment of genetic diversity in cultivated radishes (Raphanus sativus) by agronomic traits and SSR markers. Sci. Hortic. 2017, 223, 19–30. [Google Scholar] [CrossRef]
- Lim, S.-H.; Song, J.-H.; Kim, D.-H.; Kim, J.K.; Lee, J.-Y.; Kim, Y.-M.; Ha, S.-H. Activation of anthocyanin biosynthesis by ex-pression of the radish R2R3-MYB transcription factor gene RsMYB1. Plant Cell Rep. 2016, 35, 641–653. [Google Scholar] [CrossRef] [PubMed]
- Niikura, S.; Nishio, T.; Kitashiba, H. F1 Hybrid Breeding Using Genome Information. In The Radish Genome; Springer Science and Business Media LLC: Cham, Switzerland, 2017; pp. 199–216. [Google Scholar]
- Kopta, T.; Pokluda, R. Yields, Quality and nutritional parameters of radish (Raphanus sativus) cultivars when grown in organically in Czech Republic. Hort. Sci. 2013, 40, 16–21. [Google Scholar] [CrossRef]
- Kaneko, Y.; Kimizuka-Takagi, C.; Bang, S.W.; Matsuzawa, Y. Radish. In Vegetables; Springer: Berlin/Heidelberg, Germany, 2007; pp. 141–160. [Google Scholar]
- KOSTAT. Agricultural Research Report. Available online: http://kostat.go.kr/ (accessed on 1 March 2015).
- Kitamura, S. Varieties and transitions of radish. Japanese Radish. Jpn. Sci. Soc. Tokyo 1958, 1–19. [Google Scholar]
- Li, S. The origin and resources of vegetable crops in China. In Proceedings of the International Symposium on Horticultural Germplasm, Cultivated and Wild, Beijing, China, 5–9 September 1988; pp. 197–202. [Google Scholar]
- Park, H.G.; Kwon, O.H.; Kim, H.T.; Na, J.H.; Park, Y.; Park, J.Y.; Park, C.S.; Song, K.H.; Yang, D.H.; Om, Y.H.; et al. The Recent History of Vegetable Seed Industry in KOREA; Seoul National Univ Press: Seoul, Korea, 2008. [Google Scholar]
- Yamane, K.; Lü, N.; Ohnishi, O. Chloroplast DNA variations of cultivated radish and its wild relatives. Plant Sci. 2005, 168, 627–634. [Google Scholar] [CrossRef]
- Lü, N.; Yamane, K.; Ohnishi, O. Genetic diversity of cultivated and wild radish and phylogenetic relationships among Raphanus and Brassica species revealed by the analysis of trnK/matK sequence. Breed. Sci. 2008, 58, 15–22. [Google Scholar] [CrossRef]
- Kim, N.; Jeong, Y.-M.; Jeong, S.; Kim, G.-B.; Baek, S.; Kwon, Y.-E.; Cho, A.; Choi, S.-B.; Kim, J.; Lim, W.-J.; et al. Identification of candidate domestication regions in the radish genome based on high-depth resequencing analysis of 17 genotypes. Theor. Appl. Genet. 2016, 129, 1797–1814. [Google Scholar] [CrossRef] [PubMed]
- Yamane, K.; Lü, N.; Ohnishi, O. Multiple origins and high genetic diversity of cultivated radish inferred from polymorphism in chloroplast simple sequence repeats. Breed. Sci. 2009, 59, 55–65. [Google Scholar] [CrossRef]
- Armstrong, G.; Armstrong, J.K. Common hosts for Fusarium oxysporum formae speciales spinaciae and betae. Phytopathology 1976, 66, 542–545. [Google Scholar] [CrossRef]
- Bosland, P.W.; Williams, P.H. An evaluation of Fusarium oxysporum from crucifers based on pathogenicity, isozyme polymorphism, vegetative compatibility, and geographic origin. Can. J. Bot. 1987, 65, 2067–2073. [Google Scholar] [CrossRef]
- Garibaldi, A.; Gilardi, G.; Gullino, M.L. Evidence for an expanded host range of Fusarium oxysporum f.sp. raphani. Phytoparasitica 2006, 34, 115–121. [Google Scholar] [CrossRef]
- Yu, X.; Choi, S.R.; Ramchiary, N.; Miao, X.; Lee, S.H.; Sun, H.J.; Kim, S.; Ahn, C.H.; Lim, Y.P. Comparative mapping of Raphanus sativus genome using Brassica markers and quantitative trait loci analysis for the Fusarium wilt resistance trait. Theor. Appl. Genet. 2013, 126, 2553–2562. [Google Scholar] [CrossRef] [PubMed]
- Kendrick, J.; Snyder, W. Fusarium wilt of radish. Phytopathology 1942, 32, 1031–1033. [Google Scholar]
- Knepper, C.; Day, B. From Perception to Activation: The Molecular-Genetic and Biochemical Landscape of Disease Resistance Signaling in Plants. Arab. Book/Am. Soc. Plant Biol. 2010, 8, e012. [Google Scholar] [CrossRef] [PubMed]
- Peterson, J.L.; Pound, G. Studies on resistance in Radish to Fusarium oxysporum f. conglutinans. Phytopathology 1960, 50, 807–816. [Google Scholar]
- Ashizawa, M. Studies on the breeding of Fusarium resistance in radish. I. Screening of radish varieties for Fusarium resistance. Bull. Veg. Ornam. Crops Res. Stn. Jpn. A 1979, 6, 39–70. [Google Scholar]
- Shirasawa, K.; Oyama, M.; Hirakawa, H.; Sato, S.; Tabata, S.; Fujioka, T.; Kimizuka-Takagi, C.; Sasamoto, S.; Watanabe, A.; Kato, M.; et al. An EST-SSR Linkage Map of Raphanus sativus and Comparative Genomics of the Brassicaceae. DNA Res. 2011, 18, 221–232. [Google Scholar] [CrossRef]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Peng, Z.; Yang, X.; Wang, W.; Fu, J.; Wang, J.; Han, Y.; Chai, Y.; Guo, T.; Yang, N.; et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 2013, 45, 43–50. [Google Scholar] [CrossRef]
- Zhao, K.; Tung, C.-W.; Eizenga, G.C.; Wright, M.; Ali, M.L.; Price, A.H.; Norton, G.J.; Islam, M.R.; Reynolds, A.R.; Mezey, J.G.; et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2011, 2, 467. [Google Scholar] [CrossRef]
- Wu, J.; Feng, F.; Lian, X.; Teng, X.; Wei, H.; Yu, H.; Xie, W.; Yan, M.; Fan, P.; Li, Y.; et al. Genome-wide Association Study (GWAS) of mesocotyl elongation based on re-sequencing approach in rice. BMC Plant Biol. 2015, 15, 218. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; Ebbels, D.; Garber, R.; Kappelman, A. Fusarium Wilt of Cotton. Fusarium: Diseases, Biology, and Taxonomy; Pennsylvania State University Press: Philadelphia, PA, USA, 1981; pp. 29–38. [Google Scholar]
- De Donato, M.; Peters, S.O.; Mitchell, S.E.; Hussain, T.; Imumorin, I.G. Genotyping-by-Sequencing (GBS): A Novel, Efficient and Cost-Effective Genotyping Method for Cattle Using Next-Generation Sequencing. PLoS ONE 2013, 8, e62137. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [PubMed]
- Herten, K.; Hestand, M.S.; Vermeesch, J.R.; Van Houdt, J.K.J. GBSX: A toolkit for experimental design and demultiplexing genotyping by sequencing experiments. BMC Bioinform. 2015, 16, 73. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.-M.; Kim, N.; Ahn, B.O.; Oh, M.; Chung, W.-H.; Chung, H.; Jeong, S.; Lim, K.-B.; Hwang, Y.-J.; Kim, G.-B.; et al. Elucidating the triplicated ancestral genome structure of radish based on chromosome-level comparison with the Brassica genomes. Theor. Appl. Genet. 2016, 129, 1357–1372. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Browning, B.L.; Browning, S.R. Genotype Imputation with Millions of Reference Samples. Am. J. Hum. Genet. 2016, 98, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Zheng, X.; Zheng, M.X. Package ‘SNPRelate’. Available online: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.368.8598&rep=rep1&type=pdf (accessed on 29 August 2016).
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Ren, J.; Sun, D.; Chen, L.; You, F.M.; Wang, J.; Peng, Y.; Nevo, E.; Sun, D.; Luo, M.-C.; Peng, J. Genetic Diversity Revealed by Single Nucleotide Polymorphism Markers in a Worldwide Germplasm Collection of Durum Wheat. Int. J. Mol. Sci. 2013, 14, 7061–7088. [Google Scholar] [CrossRef] [PubMed]
- Juliana, P.; Rutkoski, J.; Poland, J.; Singh, R.P.; Murugasamy, S.; Natesan, S.; Barbier, H.; Sorrells, M.E. Genome-Wide Association Mapping for Leaf Tip Necrosis and Pseudo-black Chaff in Relation to Durable Rust Resistance in Wheat. Plant Genome 2015, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bastien, M.; Sonah, H.; Belzile, F. Genome wide association mapping of resistance in soybean with a genotyping-by-sequencing approach. Plant Genome 2014, 7, 195. [Google Scholar] [CrossRef]
- Visioni, A.; Tondelli, A.; Francia, E.; Pswarayi, A.; Malosetti, M.; Russell, J.; Thomas, W.; Waugh, R.; Pecchioni, N.; Romagosa, I.; et al. Genome-wide association mapping of frost tolerance in barley (Hordeum vulgare L.). BMC Genom. 2013, 14, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Broman, K.W.; Wu, H.; Sen, Ś.; Churchill, G.A. R/qtl: QTL mapping in experimental crosses. Bioinformatics 2003, 19, 889–890. [Google Scholar] [CrossRef] [PubMed]
- Shehzad, T.; Okuno, K. Genetic analysis of QTLs controlling allelopathic characteristics in sorghum. PLoS ONE 2020, 15, e0235896. [Google Scholar] [CrossRef]
- Tomita, A.; Fukuta, Y. QTL analysis for soil-surface roots originating from a New Plant Type rice (Oryza sativa L.). Plant Breed. 2019, 138, 154–162. [Google Scholar] [CrossRef]
- Sakiroglu, M.; Brummer, E.C. Identification of loci controlling forage yield and nutritive value in diploid alfalfa using GBS-GWAS. Theor. Appl. Genet. 2016, 130, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Iwata, H.; Ebana, K.; Uga, Y.; Hayashi, T.; Jannink, J.-L. Genome-wide association study of grain shape variation among Oryza sativa L. germplasms based on elliptic Fourier analysis. Mol. Breed. 2010, 25, 203–215. [Google Scholar] [CrossRef]
- Pasam, R.K.; Sharma, R.; Malosetti, M.; Van Eeuwijk, F.A.; Haseneyer, G.; Kilian, B.; Graner, A. Genome-wide association studies for agronomical traits in a world wide spring barley collection. BMC Plant Biol. 2012, 12, 16. [Google Scholar] [CrossRef] [PubMed]
- Byrne, S.; Czaban, A.; Studer, B.; Panitz, F.; Bendixen, C.; Asp, T. Genome Wide Allele Frequency Fingerprints (GWAFFs) of Populations via Genotyping by Sequencing. PLoS ONE 2013, 8, e57438. [Google Scholar] [CrossRef] [PubMed]
- Baik, S.-Y.; Kim, J.-C.; Jang, K.-S.; Choi, Y.-H.; Choi, G.-J. Development of effective screening method and evaluation of radish cultivars for resistance to Fusarium oxysporum f.sp. raphani. Res. Plant Dis. 2010, 16, 148–152. [Google Scholar] [CrossRef]
- Gracia-Garza, J.A.; Fravel, D.R. Effect of Relative Humidity on Sporulation of Fusarium oxysporum in Various Formulations and Effect of Water on Spore Movement Through Soil. Phytopathology 1998, 88, 544–549. [Google Scholar] [CrossRef] [PubMed]
- Cruz, D.R.; Leandro, L.F.S.; Munkvold, G.P. Effects of Temperature and pH on Fusarium oxysporum and Soybean Seedling Disease. Plant Dis. 2019, 103, 3234–3243. [Google Scholar] [CrossRef]
- Wang, N.; Kitamoto, N.; Ohsawa, R.; Fujimura, T. Genetic diversity of radish (Raphanus sativus) germplasms and relationships among worldwide accessions analyzed with AFLP markers. Breed. Sci. 2008, 58, 107–112. [Google Scholar] [CrossRef]
- Hyten, D.L.; Cannon, S.B.; Song, Q.; Weeks, N.; Fickus, E.W.; Shoemaker, R.C.; Specht, J.E.; Farmer, A.D.; May, G.D.; Cregan, P.B. High-throughput SNP discovery through deep resequencing of a reduced representation library to anchor and orient scaffolds in the soybean whole genome sequence. BMC Genom. 2010, 11, 38. [Google Scholar] [CrossRef] [PubMed]
- Vuong, T.D.; Sonah, H.; Meinhardt, C.G.; Deshmukh, R.K.; Kadam, S.D.; Nelson, R.L.; Shannon, J.G.; Nguyen, H.T. Genetic architecture of cyst nematode resistance revealed by genome-wide association study in soybean. BMC Genom. 2015, 16, 593. [Google Scholar] [CrossRef]
- Xing, H.T.; Guo, P.; Xia, X.L.; Yin, W.L. PdERECTA, a leucine-rich repeat receptor-like kinase of poplar, confers enhanced water use efficiency in Arabidopsis. Planta 2011, 234, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Shah, T.; Warburton, M.L.; Buckler, E.S.; McMullen, M.D.; Crouch, J. Genetic characterization and linkage disequilibrium estimation of a global maize collection using SNP markers. PLoS ONE 2009, 4, e8451. [Google Scholar] [CrossRef] [PubMed]
- Flint-Garcia, S.A.; Thornsberry, J.M.; Buckler IV, E.S. Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 2003, 54, 357–374. [Google Scholar] [CrossRef]
- Delourme, R.; Falentin, C.; Fomeju, B.F.; Boillot, M.; Lassalle, G.; André, I.; Duarte, J.; Gauthier, V.; Lucante, N.; Marty, A.; et al. High-density SNP-based genetic map development and linkage disequilibrium assessment in Brassica napus L. BMC Genom. 2013, 14, 120. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wei, X.; Sang, T.; Zhao, Q.; Feng, Q.; Zhao, Y.; Li, C.; Zhu, C.; Lu, T.; Zhang, Z.; et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010, 42, 961–967. [Google Scholar] [CrossRef]
- Zhang, Z.; Ersoz, E.; Lai, C.-Q.; Todhunter, R.J.; Tiwari, H.K.; Gore, M.; Bradbury, P.J.; Yu, J.; Arnett, D.K.; Ordovas, J.M.; et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Choi, D. Comparative transcriptome analysis of pepper (Capsicum annuum) revealed common regulons in multiple stress conditions and hormone treatments. Plant Cell Rep. 2013, 32, 1351–1359. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, M.; Fujimoto, R.; Ying, H.; Pu, Z.-j.; Ebe, Y.; Kawanabe, T.; Saeki, N.; Taylor, J.M.; Kaji, M.; Dennis, E.S. Identification of candidate genes for Fusarium yellows resistance in Chinese cabbage by differential expression analysis. Plant Mol. Biol. 2014, 85, 247–257. [Google Scholar] [CrossRef]
- Yu, X.; Lu, L.; Ma, Y.; Chhapekar, S.S.; Yi, S.Y.; Lim, Y.P.; Choi, S.R. Fine-mapping of a major QTL (Fwr1) for Fusarium wilt resistance in radish. Theor. Appl. Genet. 2020, 133, 329–340. [Google Scholar] [CrossRef]
- Biswas, K.; Tarafdar, A.; Kumar, R.; Singhvi, N.; Ghosh, P.; Sharma, M.; Pabbi, S.; Shukla, P. Molecular Analysis of Disease-Responsive Genes Revealing the Resistance Potential Against Fusarium Wilt (Fusarium udum Butler) Dependent on Genotype Variability in the Leguminous Crop Pigeonpea. Front. Genet. 2020, 11, 862. [Google Scholar] [CrossRef] [PubMed]
- Lorenc-Kukuła, K.; Zuk, M.; Kulma, A.; Czemplik, M.; Kostyn, K.; Skala, J.; Starzycki, M.; Szopa, J. Engineering Flax with the GT Family 1 Solanum sogarandinum Glycosyltransferase SsGT1 Confers Increased Resistance to Fusarium Infection. J. Agric. Food Chem. 2009, 57, 6698–6705. [Google Scholar] [CrossRef]
- Bent, A.F.; Mackey, D. Elicitors, effectors, and R genes: The new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 2007, 45, 399–436. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Rai, A.K.; Kanwar, S.S.; Sharma, T.R. Comparative Analysis of Zinc Finger Proteins Involved in Plant Disease Resistance. PLoS ONE 2012, 7, e42578. [Google Scholar] [CrossRef]
- Diener, A.C.; Ausubel, F.M. Resistance to Fusarium oxysporum 1, a Dominant Arabidopsis Disease-Resistance Gene, Is Not Race Specific. Genetics 2005, 171, 305–321. [Google Scholar] [CrossRef] [PubMed]
- Diener, A. Visualizing and Quantifying Fusarium oxysporum in the Plant Host. Mol. Plant-Microbe Interact. 2012, 25, 1531–1541. [Google Scholar] [CrossRef] [PubMed]
- Pu, Z.-J.; Shimizu, M.; Zhang, Y.-J.; Nagaoka, T.; Hayashi, T.; Hori, H.; Matsumoto, S.; Fujimoto, R.; Okazaki, K. Genetic mapping of a Fusarium wilt resistance gene in Brassica oleracea. Mol. Breed. 2011, 30, 809–818. [Google Scholar] [CrossRef]

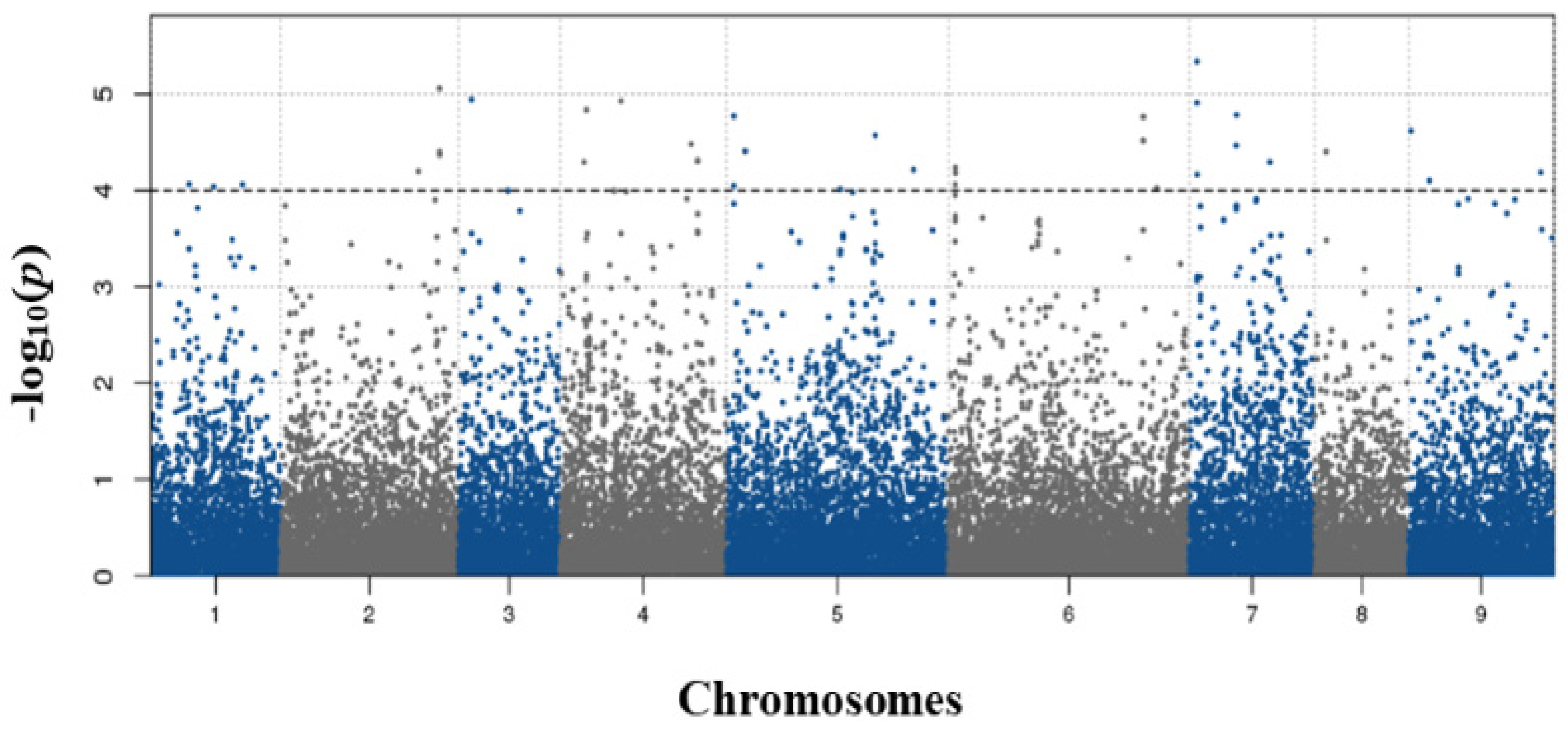
| Gene | SNP | Chromosome | Encoding | p-Value |
|---|---|---|---|---|
| Rs014470 | GBS-FW1 | 1 | Brassinosteroid signaling positive regulator (BZR1) family protein | 9.13 × 10−5 |
| Rs045120 | GBS-FW2 GBS-FW3 GBS-FW4 GBS-FW5 GBS-FW6 | 2 | 1-phosphatidylinositol-3-phosphate 5-kinases | 8.80 × 10−6, 3.98 × 10−5, 4.25 × 10−5 |
| Rs118240 | GBS-FW7 | 3 | β-galactosidase 8 | 1.13 × 10−5 |
| Rs173640 | GBS-FW8 | 4 | myb domain protein 121 | 1.17 × 10−5 |
| Rs204510 | GBS-FW9 | 4 | acyl-n-acyltransferase with ring five phd-type zinc finger domain-containing protein | 3.30 × 10−5 |
| Rs207880 | GBS-FW10 GBS-FW11 GBS-FW12 GBS-FW13 | 4 | transcription factor bhlh51 | 4.92 × 10−5 |
| Rs160720 | GBS-FW14 | 4 | pentatricopeptide repeat-containing protein | 1.46 × 10−5 |
| Rs223390 | GBS-FW15 | 5 | neutral invertase | 3.91 × 10−5 |
| Rs267700 | GBS-FW16 | 5 | basic helix-loop-helix (bHLH) DNA-binding superfamily protein | 2.68 × 10−5 |
| Rs277800 | GBS-FW17 | 5 | unknown function | 6.09 × 10−5 |
| Rs220650 | GBS-FW18 GBS-FW19 | 5 | homeobox-leucine zipper protein hdg8 | 1.69 × 10−5, 8.91 × 10−5 |
| Rs357270 | GBS-FW20 GBS-FW21 GBS-FW22 | 6 | unknown function | 1.72 × 10−5, 3.05 × 10−5 |
| Rs360300 | GBS-FW23 | 6 | P-loop containing nucleoside triphosphate hydrolases superfamily protein | 9.48 × 10−5 |
| Rs384720 | GBS-FW24 | 7 | subtilase family protein | 5.07 × 10−5 |
| Rs404770 | GBS-FW25 GBS-FW26 GBS-FW27 GBS-FW28 | 7 | glycosyltransferase family protein | 4.60 × 10−6, 1.23 × 10−5, 6.85 × 10−5 |
| Rs393590 | GBS-FW29 GBS-FW30 | 7 | Vacuolar sorting protein 9 (VPS9) domain | 1.64 × 10−5, 3.42 × 10−5 |
| Rs410280 | GBS-FW31 | 8 | ankyrin repeat family protein | 3.96 × 10−5 |
| Rs448450 | GBS-FW32 | 9 | major facilitator superfamily protein | 2.41 × 10−5 |
| Rs498780 | GBS-FW33 | 9 | tir-nbs-lrr class disease resistance protein | 6.49 × 10−5 |
| Rs454240 | GBS-FW34 | 9 | RNA polymerase II transcription elongation factor | 7.91 × 10−5 |
| Name | Chromosome | Pos (cM) | CI. Low | CI. High | LOD | PVE (%) | Additive | Dominance | p-Value (Chi2) | p-Value (F) |
| qFWR 1 | 3 | 60.1 | 57.0 | 61.9 | 2.58 | 5.75 | 0.33 | 0.02 | 1.9 × 10−3 | 2.9 × 10−3 |
| qFWR 2 | 6 | 264.0 | 254.8 | 274.1 | 5.35 | 11.93 | 0.44 | 0.10 | 4.1 × 10−6 | 9.6 × 10−6 |
| qFWR 3 | 7 | 45.7 | 43.8 | 49.5 | 6.70 | 18.84 | 0.68 | 0.11 | 8.1 × 10−9 | 2.9 × 10−8 |
| qFWR 4 | 8 | 340.5 | 336.9 | 346.3 | 3.48 | 7.90 | 0.36 | 0.09 | 2.1 × 10−4 | 3.8 × 10−4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, O.N.; Koo, H.; Yu, J.W.; Park, H.Y. Genotyping-by-Sequencing-Based Genome-Wide Association Studies of Fusarium Wilt Resistance in Radishes (Raphanus sativus L.). Genes 2021, 12, 858. https://doi.org/10.3390/genes12060858
Lee ON, Koo H, Yu JW, Park HY. Genotyping-by-Sequencing-Based Genome-Wide Association Studies of Fusarium Wilt Resistance in Radishes (Raphanus sativus L.). Genes. 2021; 12(6):858. https://doi.org/10.3390/genes12060858
Chicago/Turabian StyleLee, O New, Hyunjin Koo, Jae Woong Yu, and Han Yong Park. 2021. "Genotyping-by-Sequencing-Based Genome-Wide Association Studies of Fusarium Wilt Resistance in Radishes (Raphanus sativus L.)" Genes 12, no. 6: 858. https://doi.org/10.3390/genes12060858
APA StyleLee, O. N., Koo, H., Yu, J. W., & Park, H. Y. (2021). Genotyping-by-Sequencing-Based Genome-Wide Association Studies of Fusarium Wilt Resistance in Radishes (Raphanus sativus L.). Genes, 12(6), 858. https://doi.org/10.3390/genes12060858






