miR-21-5p Regulates the Proliferation and Differentiation of Skeletal Muscle Satellite Cells by Targeting KLF3 in Chicken
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Standards
2.2. Animals and Samples
2.3. Cell Culture
2.4. RNA Oligonucleotides, Vectors, and Transfection
2.5. RNA Isolation, Complementary DNA (cDNA) Synthesis, and Real-Time Quantitative PCR (qRT-PCR)
2.6. Protein Extraction and Western Blot Analysis
2.7. Cell Counting Kit 8 (CCK-8) Assay
2.8. Immunofluorescence Assay
2.9. Luciferase Reporter Assay
2.10. Bioinformatic Analysis
2.11. Statistical Analysis
3. Results
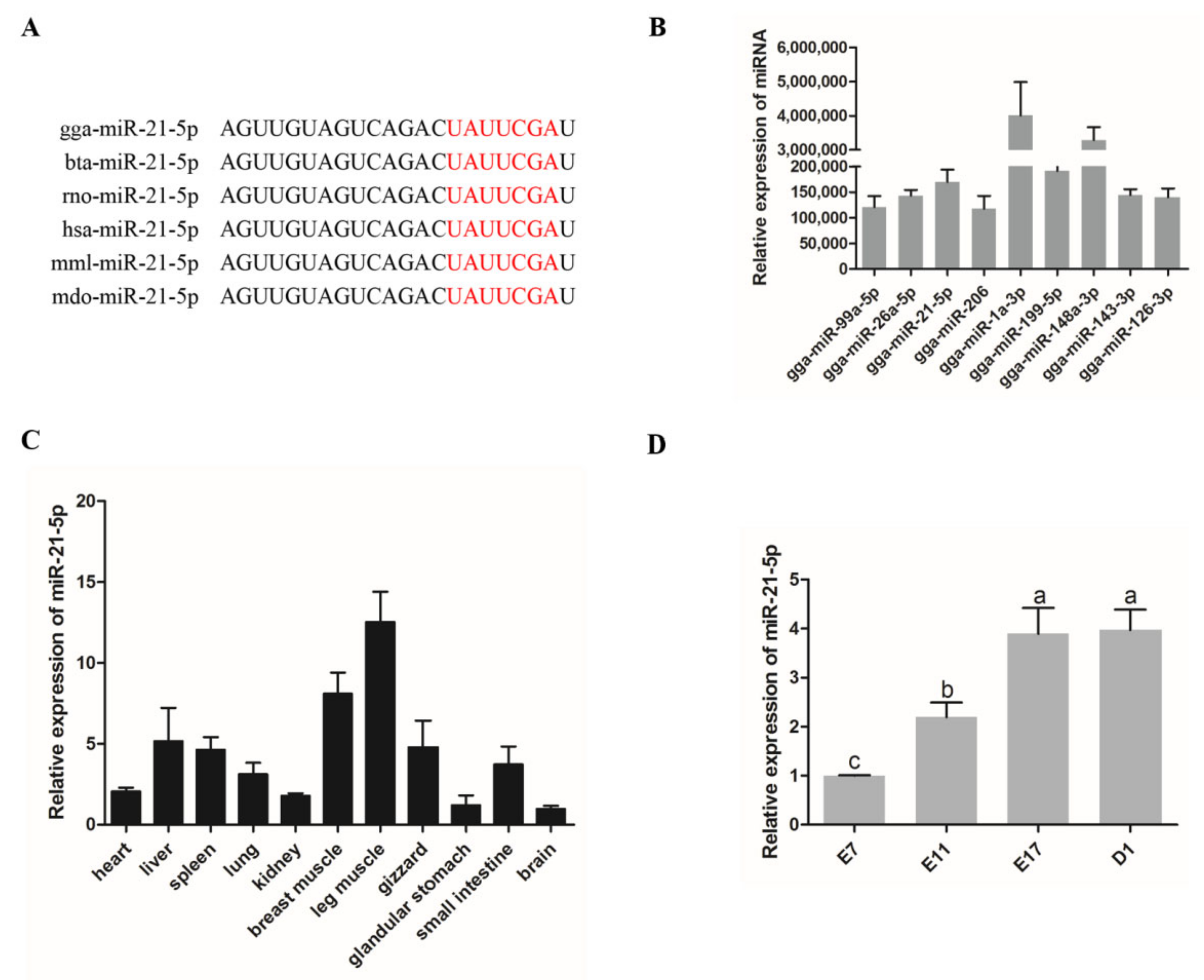
3.1. Expression of miR-21-5p in Chickens
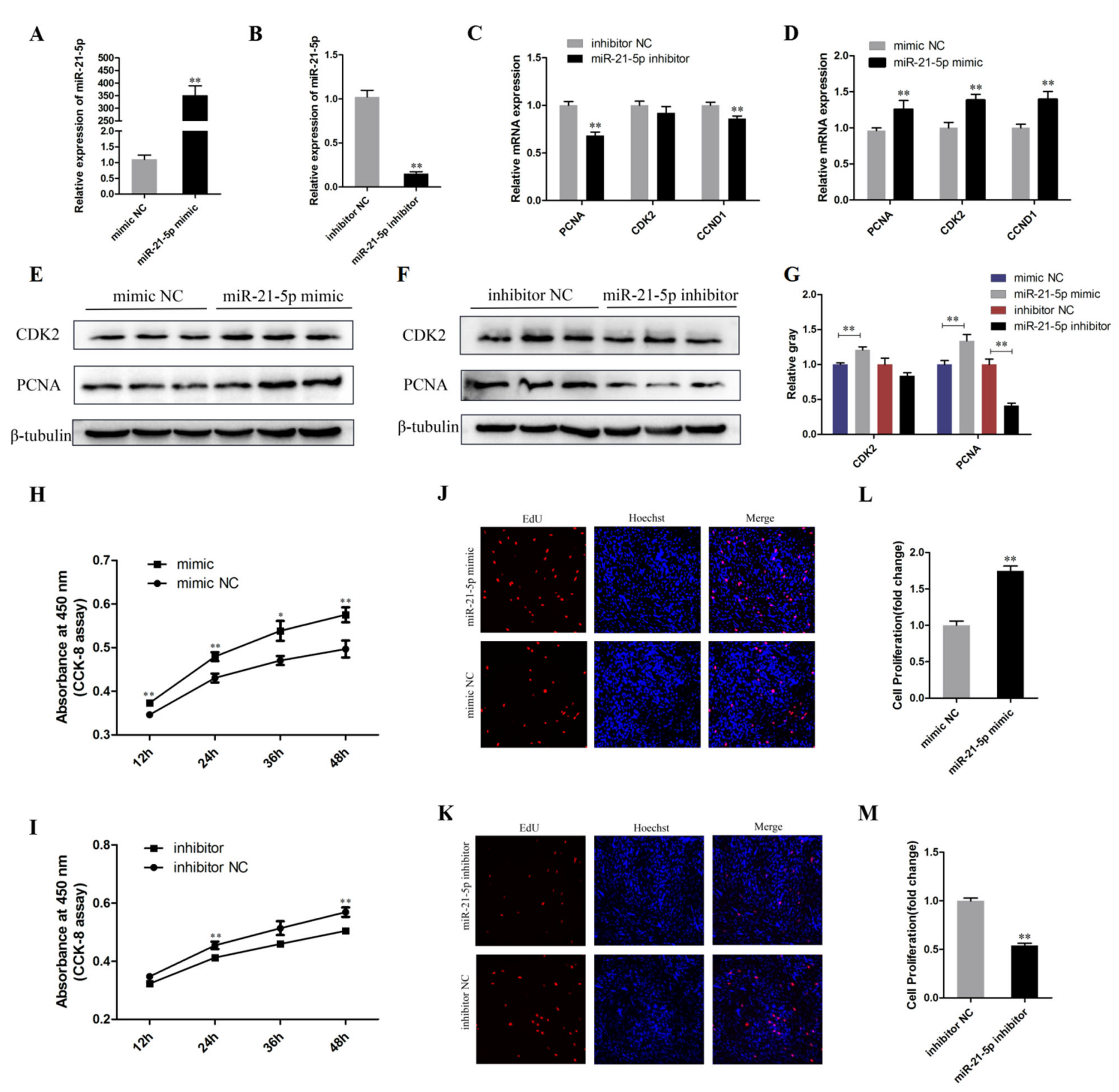
3.2. miR-21-5p Promotes the Proliferation of Chicken SMSCs
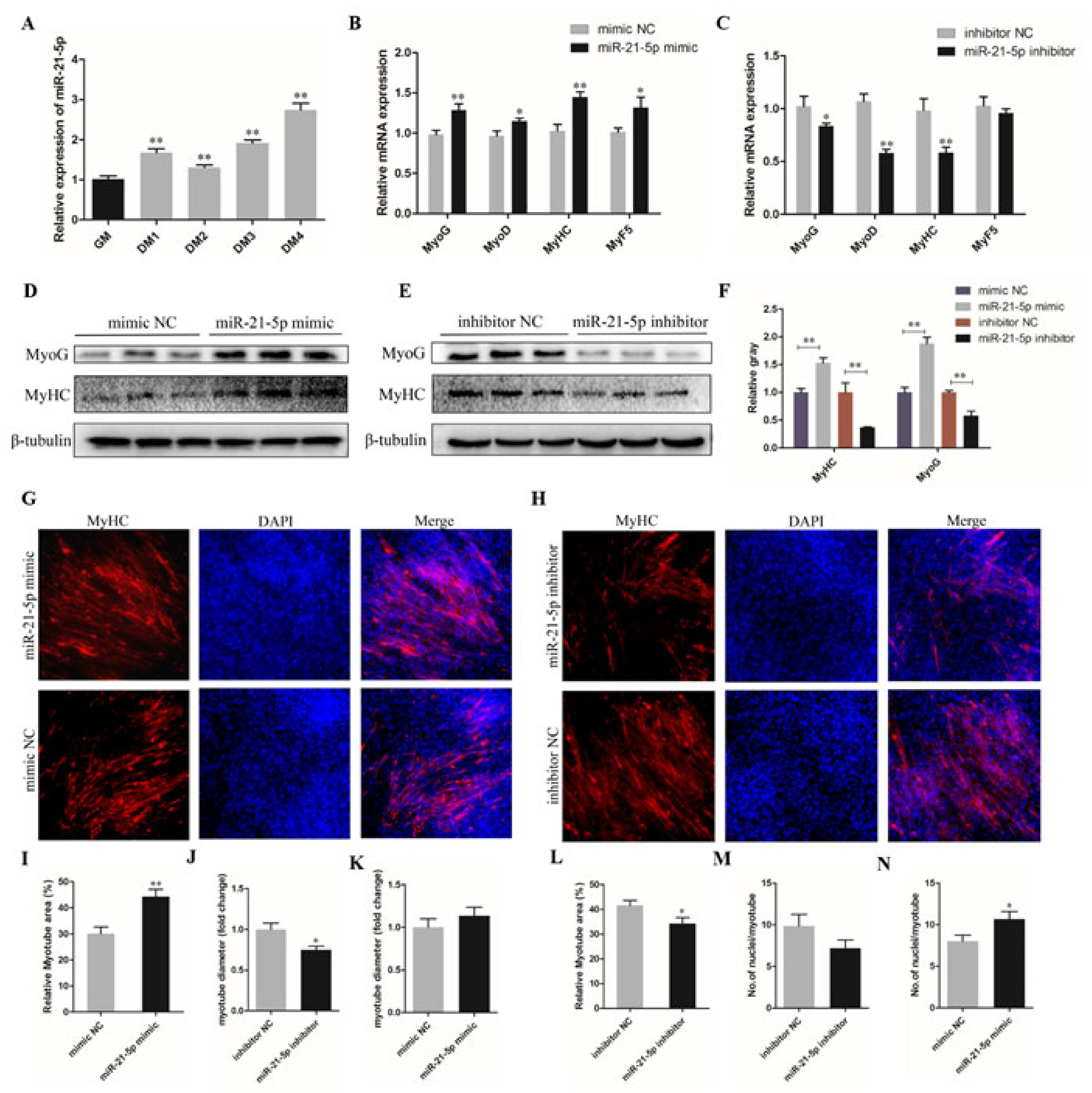
3.3. miR-21-5p Promotes the Differentiation of Chicken SMSCs
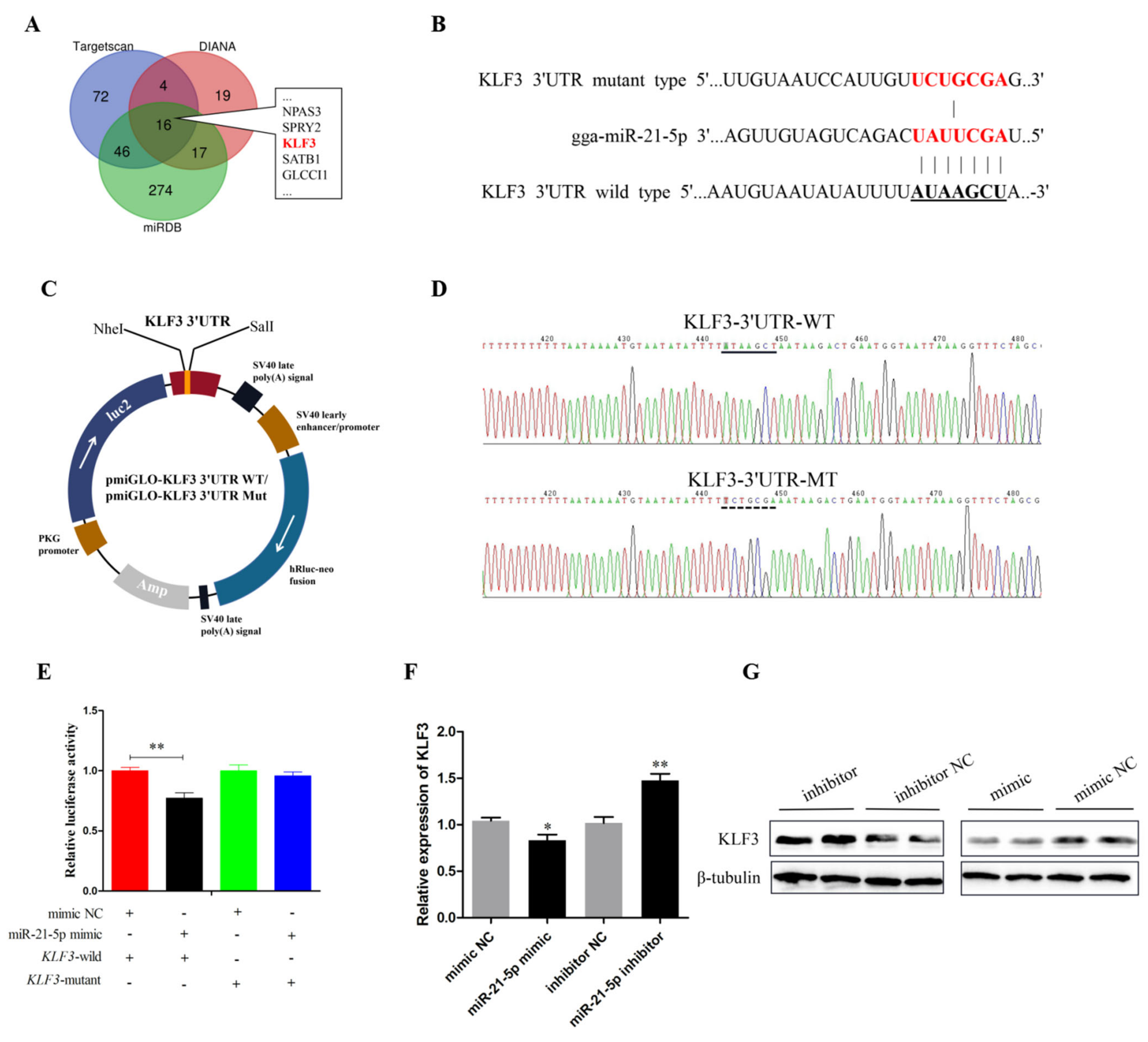
3.4. miR-21-5p Targets Directly KLF3 Gene
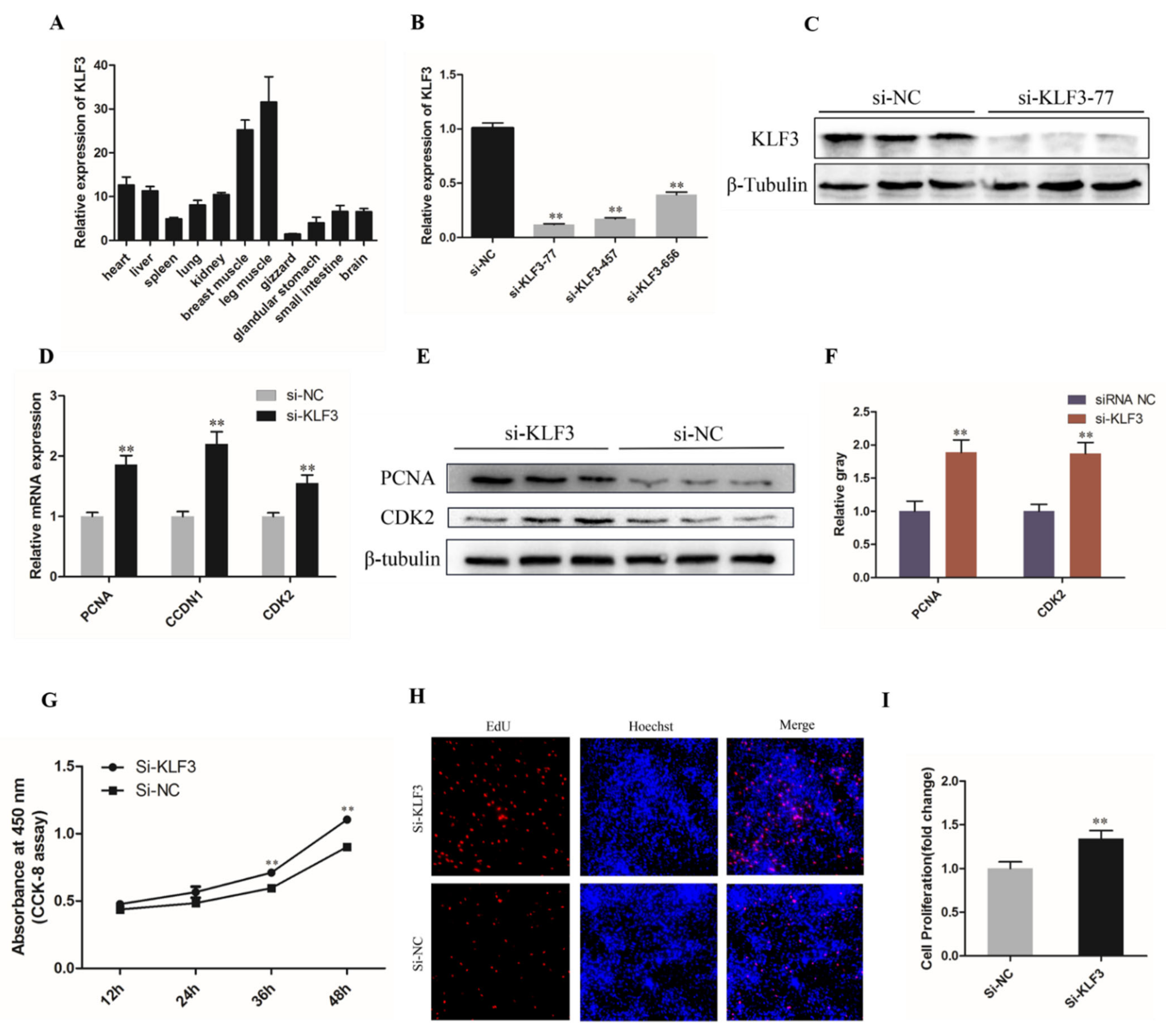
3.5. Knockdown of KLF3 Facilitates Chicken SMSCs Proliferation
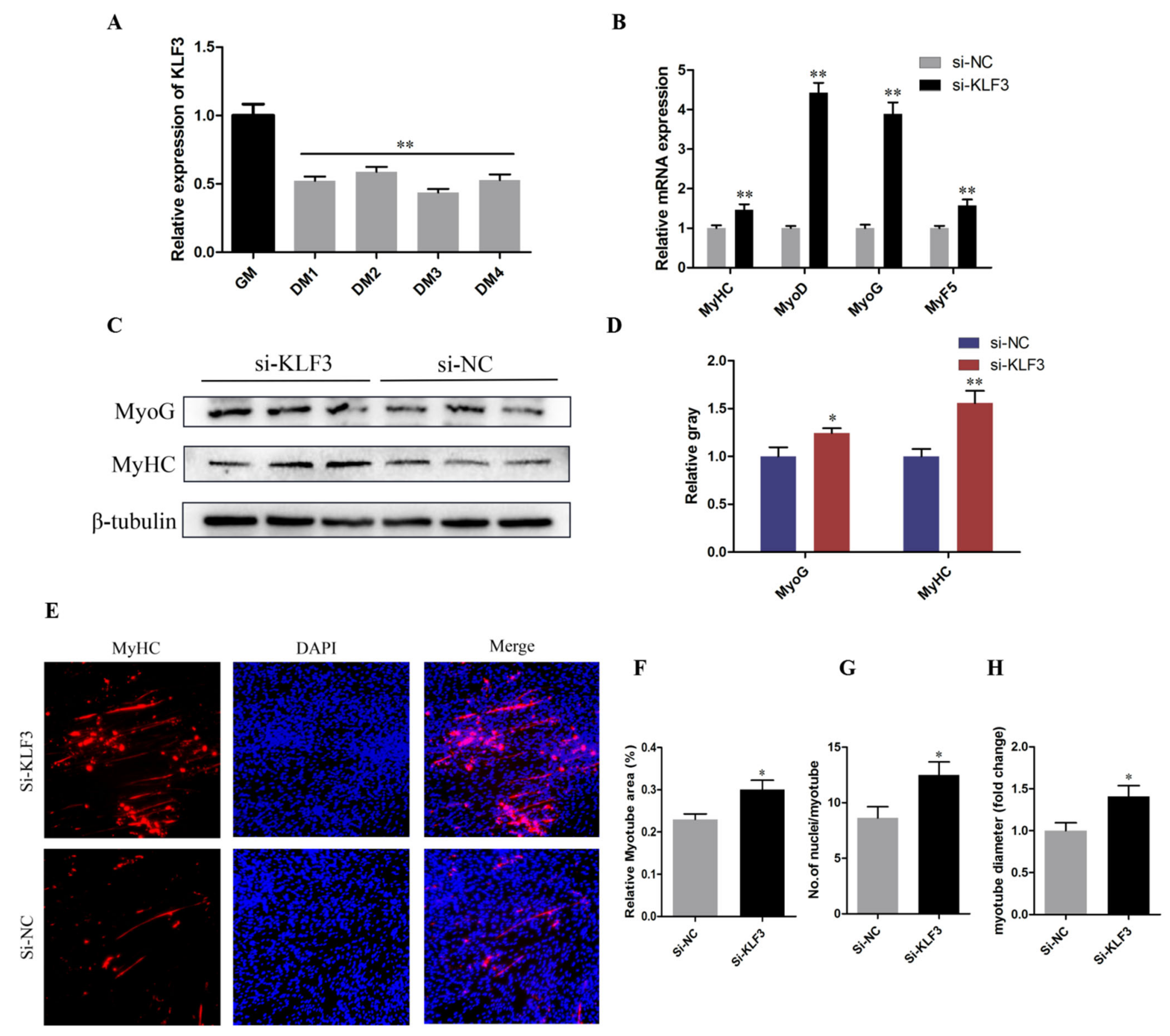
3.6. Knockdown of KLF3 Facilitates Chicken SMSCs Differentiation
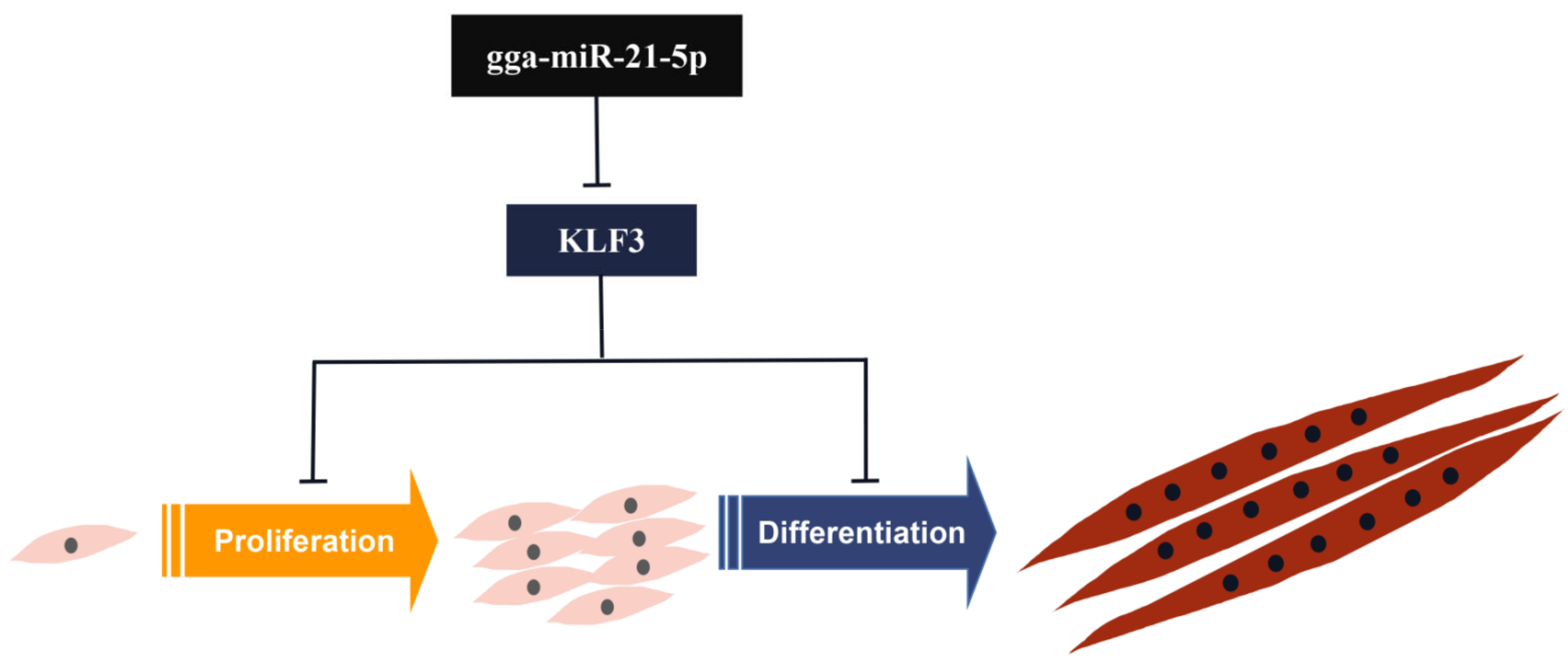
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Y.X.; Rudnicki, M. Satellite cells, the engines of muscle repair. Nat. Rev. Mol. Cell Biol. 2011, 13, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Chang, N.C.; Rudnicki, M.A. Satellite cells: The architects of skeletal muscle. Curr. Top. Dev. Biol. 2014, 107, 161–181. [Google Scholar] [PubMed]
- Relaix, F.; Montarras, D.; Zaffran, S.; Gayraud-Morel, B.; Rocancourt, D.; Tajbakhsh, S.; Mansouri, A.; Cumano, A.; Buckingham, M. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J. Cell Biol. 2006, 172, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Carrio, E.; Suelves, M. DNA methylation dynamics in muscle development and disease. Front. Aging Neurosci. 2015, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.F.; Tao, Y.; Li, J.; Deng, Z.; Yan, Z.; Xiao, X.; Wang, D.Z. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J. Cell Biol. 2010, 190, 867–879. [Google Scholar] [CrossRef] [PubMed]
- Bassel-Duby, R.; Olson, E.N. Signaling Pathways in Skeletal Muscle Remodeling. Annu. Rev. Biochem. 2006, 75, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Saunders, M.A.; Liang, H.; Li, W.-H. Human polymorphism at microRNAs and microRNA target sites. Proc. Natl. Acad. Sci. USA 2007, 104, 3300–3305. [Google Scholar] [CrossRef]
- Lai, E.C. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002, 30, 363–364. [Google Scholar] [CrossRef]
- Giral, H.; Kratzer, A.; Landmesser, U. MicroRNAs in lipid metabolism and atherosclerosis. Best Pract. Res. Clin. Endocrinol. Metab. 2016, 30, 665–676. [Google Scholar] [CrossRef]
- Güller, I.; Russell, A.P. MicroRNAs in skeletal muscle: Their role and regulation in development, disease and function. J. Physiol. 2010, 588, 4075–4087. [Google Scholar] [CrossRef]
- Gross, N.; Kropp, J.; Khatib, H. MicroRNA Signaling in Embryo Development. Biology 2017, 6, 34. [Google Scholar] [CrossRef]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef]
- Chen, J.-F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.-Z. The Role of MicroRNA-1 and MicroRNA-133 in Skeletal Muscle Proliferation and Differentiation. Nat. Genet. 2005, 38, 228–233. [Google Scholar] [CrossRef]
- Niu, Z.; Li, A.; Zhang, S.X.; Schwartz, R.J. Serum response factor micromanaging cardiogenesis. Curr. Opin. Cell Biol. 2007, 19, 618–627. [Google Scholar] [CrossRef]
- Kim, H.K.; Lee, Y.S.; Sivaprasad, U.; Malhotra, A.; Dutta, A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J. Cell Biol. 2006, 174, 677–687. [Google Scholar] [CrossRef]
- Selcuklu, S.D.; Donoghue, M.T.; Spillane, C. miR-21 as a key regulator of oncogenic processes. Biochem. Soc. Trans. 2009, 37, 918–925. [Google Scholar] [CrossRef]
- Sarkar, J.; Gou, D.; Turaka, P.; Viktorova, E.; Ramchandran, R.; Raj, J.U. MicroRNA-21 plays a role in hypoxia-mediated pulmonary artery smooth muscle cell proliferation and migration. Am. J. Physiol. Cell. Mol. Physiol. 2010, 299, L861–L871. [Google Scholar] [CrossRef]
- Bai, L.; Liang, R.; Yang, Y.; Hou, X.; Wang, Z.; Zhu, S.; Wang, C.; Tang, Z.; Li, K. MicroRNA-21 Regulates PI3K/Akt/mTOR Signaling by Targeting TGFβI during Skeletal Muscle Development in Pigs. PLoS ONE 2015, 10, e0119396. [Google Scholar] [CrossRef]
- Haldar, S.M.; Ibrahim, O.A.; Jain, M.K. Kruppel-like Factors (KLFs) in muscle biology. J. Mol. Cell. Cardiol. 2007, 43, 1–10. [Google Scholar] [CrossRef]
- Haldar, S.M. Emerging Role of Kruppel-Like Factors (KLFs) in Skeletal Muscle Biology; Yodosha: Tokyo, Japan, 2013. [Google Scholar]
- Parakati, R.; DiMario, J.X. Repression of Myoblast Proliferation and Fibroblast Growth Factor Receptor 1 Promoter Activity by KLF10 Protein. J. Biol. Chem. 2013, 288, 13876–13884. [Google Scholar] [CrossRef] [PubMed]
- Himeda, C.L.; Ranish, J.A.; Pearson, R.C.M.; Crossley, M.; Hauschka, S.D. KLF3 Regulates Muscle-Specific Gene Expression and Synergizes with Serum Response Factor on KLF Binding Sites. Mol. Cell. Biol. 2010, 30, 3430–3443. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Yang, X.; Zhang, C.; Zhang, X.; Shi, M.; Wang, S.; Mi, L.; Zhao, Y.; Cao, H.; Wang, Y. KLF3 promotes the 8-cell-like transcriptional state in pluripotent stem cells. Cell Prolif. 2020, 53, 12914. [Google Scholar] [CrossRef] [PubMed]
- Bai, C.; Hou, L.; Li, F.; He, X.; Zhang, M.; Guan, W. Isolation and Biological Characteristics of Beijing Fatty Chicken Skeletal Muscle Satellite Cells. Cell Commun. Adhes. 2012, 19, 69–77. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Cui, Z.; Liu, L.; Amevor, F.K.; Zhu, Q.; Wang, Y.; Li, D.; Shu, G.; Tian, Y.; Zhao, X. High Expression of miR-204 in Chicken Atrophic Ovaries Promotes Granulosa Cell Apoptosis and Inhibits Autophagy. Front. Cell Dev. Biol. 2020, 8, 580072. [Google Scholar] [CrossRef]
- Yuan, J.S.; Wang, D.; Stewart, C.N. Statistical methods for efficiency adjusted real-time PCR quantification. Biotechnol. J. 2008, 3, 112–123. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, J.; Qiu, W.; Zhang, J.; Feng, S.; Zhou, X.; Wang, X.; Jin, L.; Long, K.; Liu, L.; et al. Guanidinoacetic Acid Regulates Myogenic Differentiation and Muscle Growth Through miR-133a-3p and miR-1a-3p Co-mediated Akt/mTOR/S6K Signaling Pathway. Int. J. Mol. Sci. 2018, 19, 2837. [Google Scholar] [CrossRef]
- Song, C.; Yang, J.; Jiang, R.; Yang, Z.; Li, H.; Huang, Y.; Lan, X.; Lei, C.; Ma, Y.; Qi, X.; et al. miR-148a-3p regulates proliferation and apoptosis of bovine muscle cells by targeting KLF. J. Cell. Physiol. 2019, 234, 15742–15750. [Google Scholar] [CrossRef]
- Kang, T.; Xing, W.; Xi, Y.; Chen, K.; Zhan, M.; Tang, X.; Wang, Y.; Zhang, R.; Lei, M. MiR-543 regulates myoblast proliferation and differentiation of C2C12 cells by targeting KLF. J. Cell. Biochem. 2020, 121, 4827–4837. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, F.; Wu, P.; Li, T.; He, M.; Yin, X.; Shi, H.; Duan, Y.; Zhang, T.; Wang, J.; et al. MicroRNA-7 Targets the KLF4 Gene to Regulate the Proliferation and Differentiation of Chicken Primary Myoblasts. Front. Genet. 2020, 11, 842. [Google Scholar] [CrossRef]
- Berti, F.; Nogueira, J.M.; Wöhrle, S.; Sobreira, D.R.; Hawrot, K.; Dietrich, S. Time course and side-by-side analysis of mesodermal, pre-myogenic, myogenic and differentiated cell markers in the chicken model for skeletal muscle formation. J. Anat. 2015, 227, 361–382. [Google Scholar] [CrossRef]
- Hirst, C.E.; Marcelle, C. The avian embryo as a model system for skeletal myogenesis. Vertebr. Myogenesis 2015, 56, 99–122. [Google Scholar]
- Biolo, G.; Cederholm, T.; Muscaritoli, M. Muscle contractile and metabolic dysfunction is a common feature of sarcopenia of aging and chronic diseases: From sarcopenic obesity to cachexia. Clin. Nutr. 2014, 33, 737–748. [Google Scholar] [CrossRef]
- Zhu, L.; Hou, L.; Ou, J.; Xu, G.; Jiang, F.; Hu, C.; Wang, C. MiR-199b represses porcine muscle satellite cells proliferation by targeting JAG1. Gene 2019, 691, 24–33. [Google Scholar] [CrossRef]
- Huang, W.; Guo, L.; Zhao, M.; Zhang, D.; Xu, H.; Nie, Q. The Inhibition on MDFIC and PI3K/AKT Pathway Caused by miR-146b-3p Triggers Suppression of Myoblast Proliferation and Differentiation and Promotion of Apoptosis. Cells 2019, 8, 656. [Google Scholar] [CrossRef]
- Naguibneva, I.; Ameyar-Zazoua, M.; Polesskaya, A.; Ait-Si-Ali, S.; Groisman, S.; Souidi, M.; Cuvellier, S.; Harel-Bellan, A. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat. Cell Biol. 2006, 8, 278–284. [Google Scholar] [CrossRef]
- Chen, P.Y.; Manninga, H.; Slanchev, K.; Chien, M.; Russo, J.J.; Ju, J.; Sheridan, R.; John, B.; Marks, D.S.; Gaidatzis, D.; et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev. 2005, 19, 1288–1293. [Google Scholar] [CrossRef]
- Jiao, G.; Pan, B.; Zhou, Z.; Zhou, L.; Li, Z.; Zhang, Z. MicroRNA-21 regulates cell proliferation and apoptosis in H2O2-stimulated rat spinal cord neurons. Mol. Med. Rep. 2015, 12, 7011–7016. [Google Scholar] [CrossRef][Green Version]
- Meng, Y.-B.; Li, X.; Li, Z.-Y.; Zhao, J.; Yuan, X.-B.; Ren, Y.; Cui, Z.-D.; Liu, Y.-D.; Yang, X.-J. microRNA-21 promotes osteogenic differentiation of mesenchymal stem cells by the PI3K/β-catenin pathway. J. Orthop. Res. 2015, 33, 957–964. [Google Scholar] [CrossRef]
- Chen, X.; Song, M.; Chen, W.; Dimitrova-Shumkovska, J.; Zhao, Y.; Cao, Y.; Song, Y.; Yang, W.; Wang, F.; Xiang, Y.; et al. MicroRNA-21 Contributes to Liver Regeneration by Targeting PTEN. Med. Sci. Monit. 2016, 22, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chan, W.; Leung, W.; Wang, Y.; Xu, C. MicroRNA-21 promotes proliferation of rat hepatocyte BRL-3A by targeting FASLG. Genet. Mol. Res. 2015, 14, 4150–4160. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Wang, G.; Hu, C.; Shi, Y.; Liao, L.; Shi, S.; Cai, Y.; Cheng, S.; Wang, X.; Liu, Y.; et al. Tumor necrosis factor α suppresses the mesenchymal stem cell osteogenesis promoter miR-21 in estrogen deficiency-induced osteoporosis. J. Bone Miner. Res. 2012, 28, 559–573. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liang, A.; Yang, F.; Wang, Z.; Fu, Q. MicroRNA-21 promotes osteogenic differentiation by targeting small mothers against decapentaplegic. Mol. Med. Rep. 2012, 12, 1561–1567. [Google Scholar] [CrossRef]
- Hayashi, S.; Manabe, I.; Suzuki, Y.; Relaix, F.; Oishi, Y. Klf5 regulates muscle differentiation by directly targeting muscle-specific genes in cooperation with MyoD in mice. eLife 2016, 5, e17462. [Google Scholar] [CrossRef]
- Fabian, M.R.; Sonenberg, N. The mechanics of miRNA-mediated gene silencing: A look under the hood of miRISC. Nat. Struct. Mol. Biol. 2012, 19, 586–593. [Google Scholar] [CrossRef]
- Della Gaspera, B.; Chesneau, A.; Weill, L.; Charbonnier, F.; Chanoine, C. Xenopus SOX5 enhances myogenic transcription indirectly through transrepression. Dev. Biol. 2018, 442, 262–275. [Google Scholar] [CrossRef]
- Chuang, H.N.; Hsiao, K.M.; Chang, H.Y.; Wu, C.C.; Pan, H. The homeobox transcription factor Irxl1 negatively regulates MyoD expression and myoblast differentiation. FEBS J. 2014, 281, 2990–3003. [Google Scholar] [CrossRef]
- Xu, G.; Lu, X.; Huang, T.; Fan, J. ARHGAP24 inhibits cell cycle progression, induces apoptosis and suppresses invasion in renal cell carcinoma. Oncotarget 2016, 7, 51829–51839. [Google Scholar] [CrossRef]
- Zhai, B.; Zhang, L.; Wang, C.; Zhao, Z.; Zhang, M.; Li, X. Identification of microRNA-21 target genes associated with hair follicle development in sheep. PeerJ 2019, 7, e7167. [Google Scholar] [CrossRef]
- Pearson, R.; Fleetwood, J.; Eaton, S.; Crossley, M.; Bao, S. Krüppel-like transcription factors: A functional family. Int. J. Biochem. Cell Biol. 2008, 40, 1996–2001. [Google Scholar] [CrossRef]
- Crossley, M.; Whitelaw, E.; Perkins, A.; Williams, G.; Fujiwara, Y.; Orkin, S.H. Isolation and characterization of the cDNA encoding BKLF/TEF-2, a major CACCC-box-binding protein in erythroid cells and selected other cells. Mol. Cell. Biol. 1996, 16, 1695–1705. [Google Scholar] [CrossRef]
- Pearson, R.C.; Funnell, A.P.; Crossley, M. The mammalian zinc finger transcription factor Krüppel-like factor 3 (KLF3/BKLF). IUBMB Life 2011, 63, 86–93. [Google Scholar] [CrossRef]
- Kitamura, T.; Suzuki, H.; Tamura, T.-A. Mouse Wee1 Gene Is Repressed by Krüppel-Like Factor 3 (KLF3) via Interaction with Multiple Upstream Elements. Gene 2012, 492, 361–367. [Google Scholar] [CrossRef]
- Eaton, S.A.; Funnell, A.P.W.; Sue, N.; Nicholas, H.; Pearson, R.C.M.; Crossley, M. A Network of Krüppel-like Factors (Klfs): Klf8 is repressed by klf3 and activated by klf1 in vivo. J. Biol. Chem. 2008, 283, 26937–26947. [Google Scholar] [CrossRef]
| Fragment Name | Sequences (5′-3′) |
|---|---|
| KLF3 siRNA-77 | F:GACCAAAUGAAGCCAAACATT R:UGUUUGGCUUCAUUUGGUCTT |
| KLF3 siRNA-457 | F:GCCAGUUCCUUUCAUGUAUTT R: AUACAUGAAAGGAACUGGCTT |
| KLF3 siRNA-656 | F:CUUCCAAUGACCUCAUUGUTT R:ACAAUGAGGUCAUUGGAGGTT |
| siRNA NC | F: UUCUCCGAACGUGUCACGUTT R: ACGUGACACGUUCGGAGAATT |
| miR-21-5p mimic | UAGCUUAUCAGACUGAUGUUGA |
| Mimic NC | UUGUACUACACAAAAGUACUG |
| miR-21-5p inhibitor | UCAACAUCAGUCUGAUAAGCUA |
| Inhibitor NC | CAGUACUUUUGUGUAGUACAA |
| Primer Name | Primer Sequences (5′-3′) | Length (bp) | Accession Number |
|---|---|---|---|
| MyoG | F:CGTGTGCCACAGCCAATG R:CCGCCGGAGAGAGACCTT | 63 | NM_204184.1 |
| MyHC | F:GAAGGAGACCTCAACGAGATGG R: ATTCAGGTGTCCCAAGTCATCC | 138 | NM_001319304.1 |
| MyoD | F:GCTACTACACGGAATCACCAAAT R:RCTGGGCTCCACTGTCACTCA | 66 | NM_204214.2 |
| PCNA | F: AACACTCAGAGCAGAAGAC R: GCACAGGAGATGACAACA | 225 | NM_204170.2 |
| CDK2 | F:GCTCTTCCGTATCTTCCGCA R: ATGCGCTTGTTGGGATCGTA | 192 | NM_001199857.1 |
| CCND1 | F:CTCCTATCAATGCCTCACA R:TCTGCTTCGTCCTCTACA | 152 | NM_205381.1 |
| MyF5 | F:TTCCCTGAGGATTTCGAGCC R:CTCATAGTGGCTGCCTTCCG | 197 | NM_001030363.1 |
| KLF3 | F:CCCCGTTTCAGTGTCATACCC R:TGAGTTTCGCTTGTTCACCG | 175 | XM_015285673.3 |
| GAPDH | F:GGTGGCCATCAATGATCCCT R:CCGTTCTCAGCCTTGACAGT | 105 | NM_204305.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, D.; Ran, J.; Li, J.; Yu, C.; Cui, Z.; Amevor, F.K.; Wang, Y.; Jiang, X.; Qiu, M.; Du, H.; et al. miR-21-5p Regulates the Proliferation and Differentiation of Skeletal Muscle Satellite Cells by Targeting KLF3 in Chicken. Genes 2021, 12, 814. https://doi.org/10.3390/genes12060814
Zhang D, Ran J, Li J, Yu C, Cui Z, Amevor FK, Wang Y, Jiang X, Qiu M, Du H, et al. miR-21-5p Regulates the Proliferation and Differentiation of Skeletal Muscle Satellite Cells by Targeting KLF3 in Chicken. Genes. 2021; 12(6):814. https://doi.org/10.3390/genes12060814
Chicago/Turabian StyleZhang, Donghao, Jinshan Ran, Jingjing Li, Chunlin Yu, Zhifu Cui, Felix Kwame Amevor, Yan Wang, Xiaosong Jiang, Mohan Qiu, Huarui Du, and et al. 2021. "miR-21-5p Regulates the Proliferation and Differentiation of Skeletal Muscle Satellite Cells by Targeting KLF3 in Chicken" Genes 12, no. 6: 814. https://doi.org/10.3390/genes12060814
APA StyleZhang, D., Ran, J., Li, J., Yu, C., Cui, Z., Amevor, F. K., Wang, Y., Jiang, X., Qiu, M., Du, H., Zhu, Q., Yang, C., & Liu, Y. (2021). miR-21-5p Regulates the Proliferation and Differentiation of Skeletal Muscle Satellite Cells by Targeting KLF3 in Chicken. Genes, 12(6), 814. https://doi.org/10.3390/genes12060814







