Selection and Verification of Appropriate Reference Genes for Expression Normalization in Cryptomeria fortunei under Abiotic Stress and Hormone Treatments
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. RNA Extraction and cDNA Preparation
2.3. Selection of Candidate RGs and Primer Design
2.4. RT-PCR and qRT-PCR Analysis
2.5. Gene Expression Stability Analysis
2.6. Validation of RGs by qRT-PCR
3. Results
3.1. Assessment of Primer Specificity and PCR Efficiency
3.2. Expression Profiles of Candidate RGs
3.3. Expression Stability of Candidate RGs
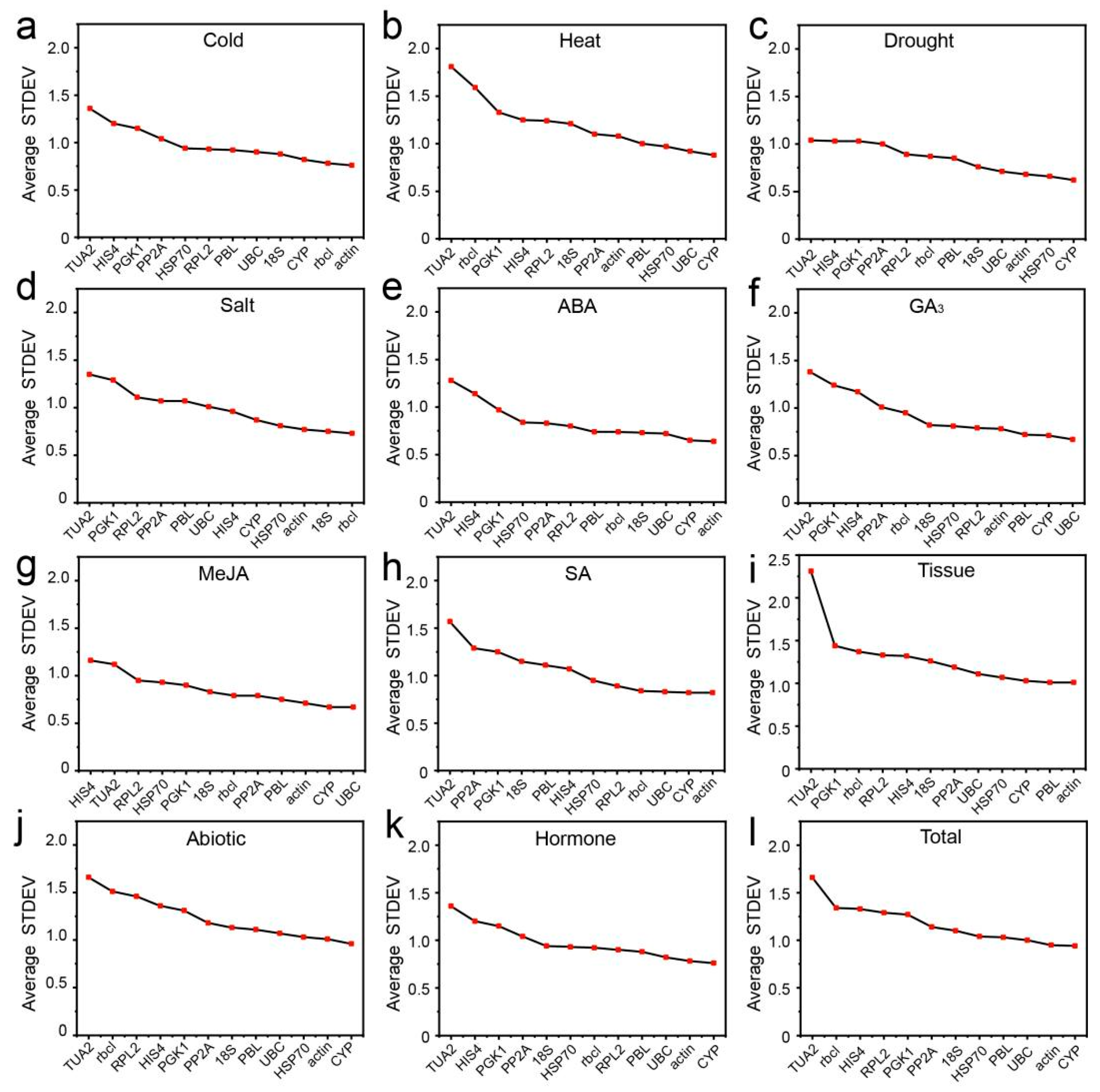
3.3.1. Delta Ct Analysis
3.3.2. GeNorm Analysis
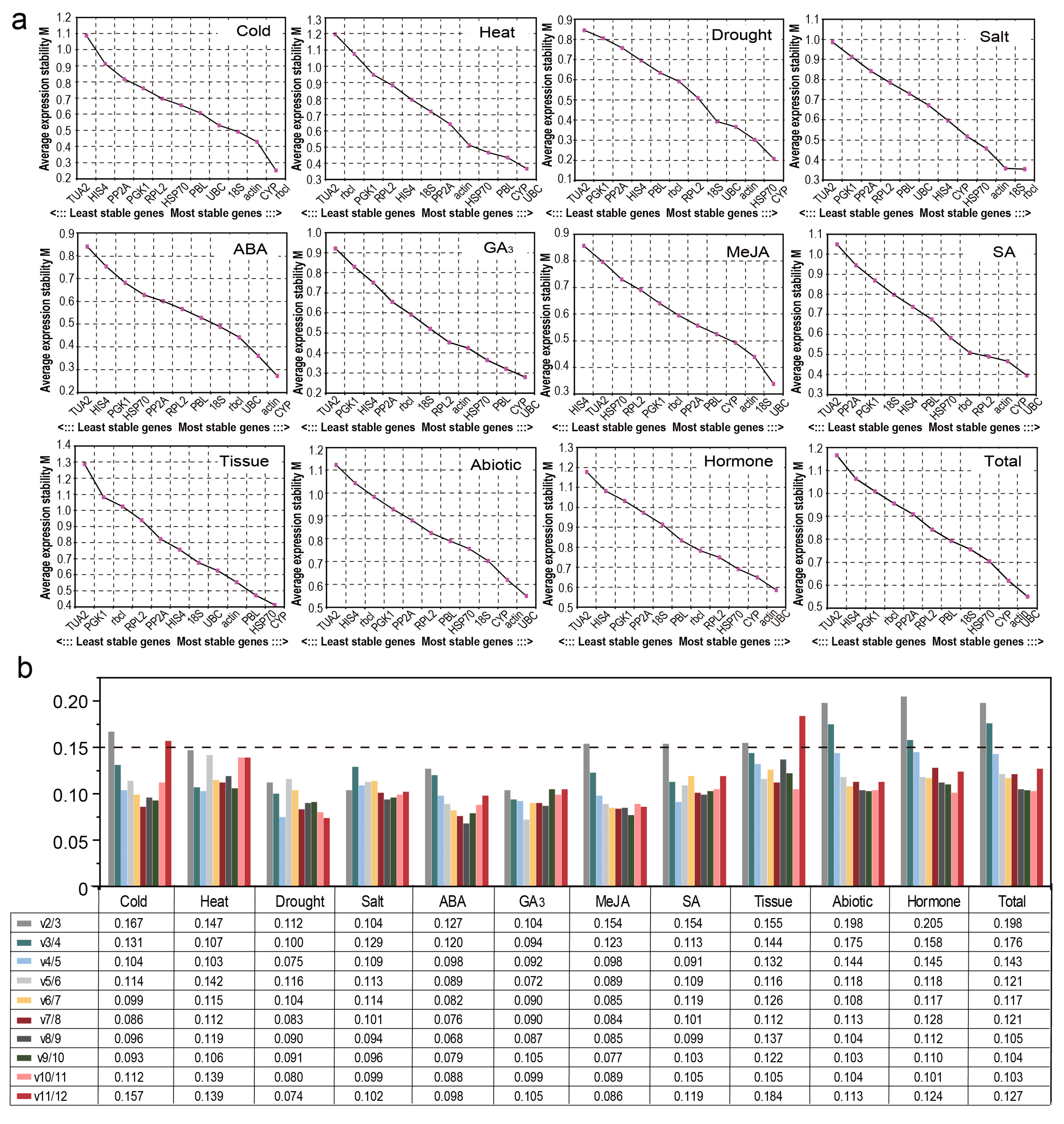
3.3.3. NormFinder Analysis
3.3.4. BestKeeper Analysis
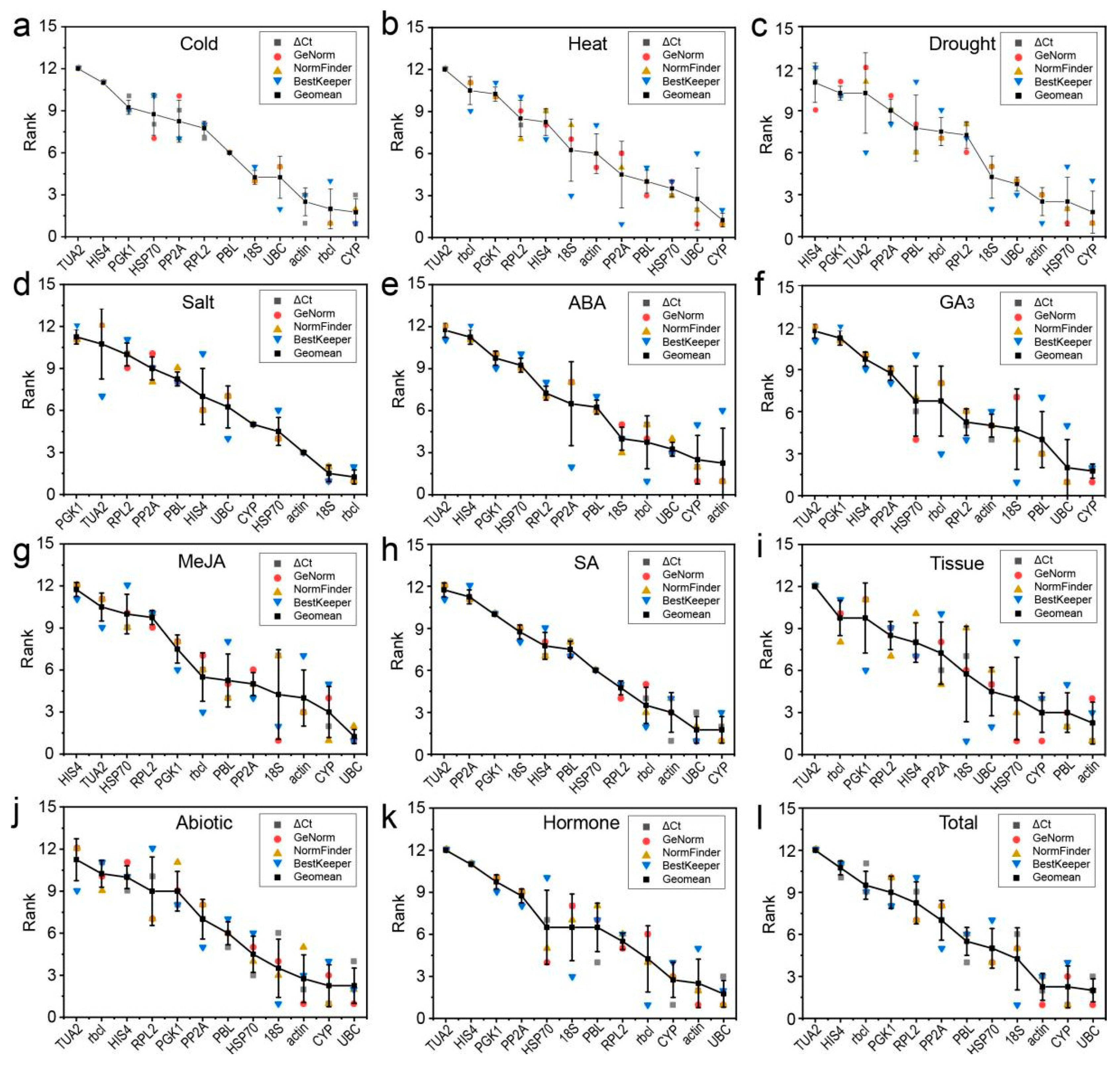
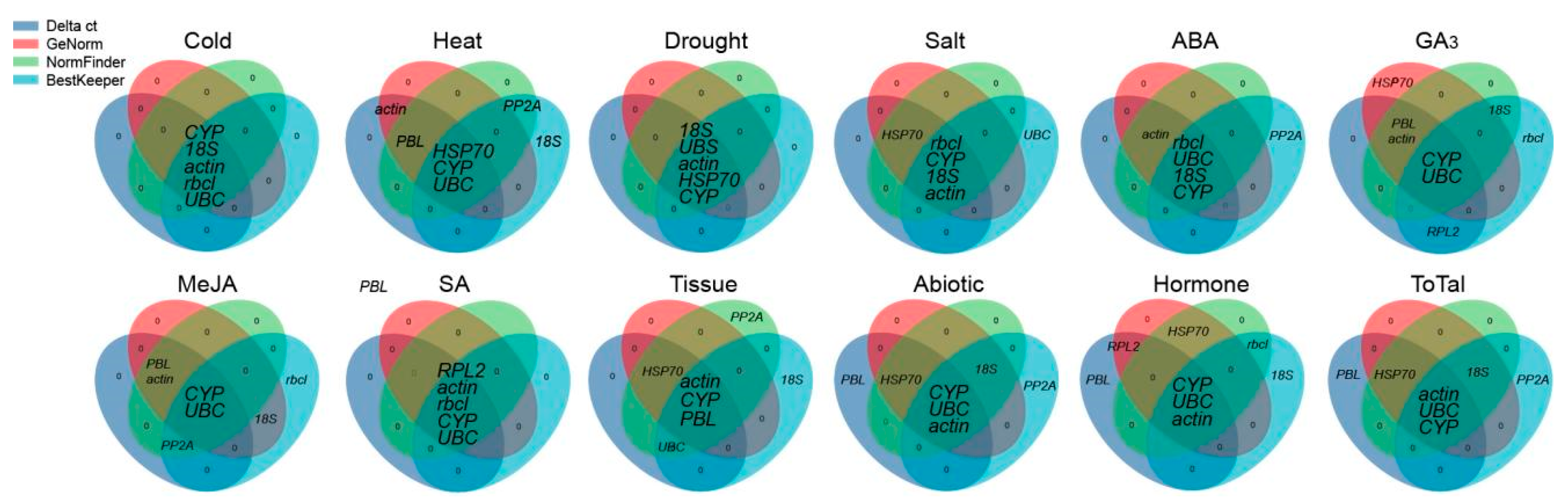
3.4. Comprehensive Stability Analysis of the RGs
3.5. Validation of the Stability of RGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ni, L.; Wang, Z.; Liu, L.; Guo, J.; Li, H.; Gu, C. Selection and verification of candidate reference genes for gene expression by quantitative RT-PCR in Hibiscus hamabo Sieb. et Zucc. Trees 2019, 33, 1591–1601. [Google Scholar] [CrossRef]
- Hu, R.; Fan, C.; Li, H.; Zhang, Q.; Fu, Y.F. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol. Biol. 2009, 10, 93. [Google Scholar] [CrossRef]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, Q.; Jiang, Y.; Li, Y.; Zhang, H.; Li, R. Reference genes identification for normalization of qPCR under multiple stresses in Hordeum brevisubulatum. Plant Methods 2018, 14, 110. [Google Scholar] [CrossRef] [PubMed]
- Saddhe, A.A.; Malvankar, M.R.; Kumar, K. Selection of reference genes for quantitative real-time PCR analysis in halophytic plant Rhizophora apiculata. PeerJ 2018, 6, e5226. [Google Scholar] [CrossRef] [PubMed]
- Menon, T.G.; Soniya, E.V. Isolation and characterization of salt-induced genes from Rhizophora apiculata Blume, a true mangrove by suppression subtractive hybridization. Curr. Sci. India 2014, 107, 650–655. [Google Scholar]
- Xu, X.; Yang, Z.; Sun, X.; Zhang, L.; Fang, Z. Selection of reference genes for quantitative real-time PCR during flower bud development in CMS7311 of heading Chinese cabbage (Brassica rapa L. ssp. pekinensis). Acta Physiol. Plant 2014, 36, 809–814. [Google Scholar] [CrossRef]
- Mou, S.; Zhang, X.; Miao, J.; Zheng, Z.; Xu, D.; Ye, N. Reference genes for gene expression normalization in Chlamydomonas sp. ICE-L by quantitative real-time RT-PCR. J. Plant Biochem. Biot. 2014, 24, 276–282. [Google Scholar] [CrossRef]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 10, 71. [Google Scholar] [CrossRef]
- Remans, T.; Smeets, K.; Opdenakker, K.; Mathijsen, D.; Vangronsveld, J.; Cuypers, A. Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 2008, 227, 1343–1349. [Google Scholar] [CrossRef]
- Li, X.; Zhang, D.; Li, H.; Gao, B.; Yang, H.; Zhang, Y.; Wood, A.J. Characterization of reference genes for RT-qPCR in the desert moss Syntrichia caninervis in response to abiotic stress and desiccation/rehydration. Front. Plant Sci. 2015, 6, 38. [Google Scholar] [CrossRef]
- Yue, H.; Deng, P.; Liu, S.; Wang, M.; Song, W.; Nie, X. Selection and evaluation of reference genes for quantitative gene expression analysis in broomcorn millet (Panicum miliaceum L.). J. Plant Biol. 2016, 59, 435–443. [Google Scholar] [CrossRef]
- Borowski, J.M.; Galli, V.; Messias, R.D.S.; Perin, E.C.; Rombaldi, C.V. Selection of candidate reference genes for real-time PCR studies in lettuce under abiotic stresses. Planta 2014, 239, 1187. [Google Scholar] [CrossRef]
- Tu, Z.; Hao, Z.; Zhong, W.; Li, H. Identification of suitable reference genes for RT-qPCR assays in Liriodendron chinense (Hemsl.) Sarg. Forests 2019, 10, 441. [Google Scholar] [CrossRef]
- Zhu, P.; Ma, Y.; Zhu, L.; Chen, Y.; Ji, K. Selection of suitable reference genes in Pinus massoniana Lamb. under different abiotic stresses for qPCR normalization. Forests 2019, 10, 632. [Google Scholar] [CrossRef]
- Takenaka, C.; Yamada, T.; Iwata, S. Ethylene emission from seedlings of hinoki (Chamaecyparis obtusa) and sugi (Cryptomeria japonica) under acidic stress. J. Forest Res. 1996, 1, 183–185. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, Q.; Guo, Z.; Yang, J.; Mo, J.; Cui, J.; Hu, H.; Xu, J. Individual Cryptomeria fortunei Hooibrenk clones show varying degrees of chilling stress resistance. Forests 2020, 11, 189. [Google Scholar] [CrossRef]
- Konpka, B.; Noguchi, K.; Sakata, T.; Takahashi, M.; Konôpková, Z. Effects of simulated drought stress on the fine roots of Japanese cedar (Cryptomeria japonica) in a plantation forest on the Kanto Plain, eastern Japan. J. Forest Res. 2007, 12, 143–151. [Google Scholar] [CrossRef]
- Li, Q.; Wang, Y.; Yan, L.; Liu, C.; Wu, D.; Han, B.; Li, L. Evaluation of cold resistance of different genotypes of Cryptomeria japonica under low temperature stress. Shandong Agr. Sci. 2020, 52, 50–53. (In Chinese) [Google Scholar] [CrossRef]
- Futamura, N.; Tani, N.; Tsumura, Y.; Nakajima, N.; Sakaguchi, M.; Shinohara, K. Characterization of genes for novel thaumatin-like proteins in Cryptomeria japonica. Tree Physiol. 2006, 26, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Hua, H.; Xu, J.; Mo, J.; Zhao, H.; Yang, J. Cloning and functional analysis of lignin biosynthesis genes Cf4CL and CfCCoAOMT in Cryptomeria fortunei. Genes 2019, 10, 619. [Google Scholar] [CrossRef] [PubMed]
- Mishima, K.; Fujiwara, T.; Iki, T.; Kuroda, K.; Yamashita, K.; Tamura, M.; Fujisawa, Y.; Watanabe, A. Transcriptome sequencing and profiling of expressed genes in cambial zone and differentiating xylem of Japanese cedar (Cryptomeria japonica). BMC Genom. 2014, 15, 219. [Google Scholar] [CrossRef]
- Alzohairy, A.M. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Whistler, T.; Chiang, C.; Lonergan, W.; Hollier, M.; Unger, E.R. Implementation of exon arrays: Alternative splicing during T-cell proliferation as determined by whole genome analysis. BMC Genom. 2010, 11, 496. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, h31–h34. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2014, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, S.; Yang, Z.; Yang, Y.; Liang, L.; Hu, L. CsCAT3, a catalase gene from Cucumis sativus, confers resistance to a variety of stresses to Escherichia coli. Biotechnol. Biotec. Eq. 2017, 31, 886–896. [Google Scholar] [CrossRef]
- Smékalová, V.; Doskočilová, A.; Komis, G.; Šamaj, J. Crosstalk between secondary messengers, hormones and MAPK modules during abiotic stress signalling in plants. Biotechnol. Adv. 2014, 32, 2–11. [Google Scholar] [CrossRef]
- Raja, V.; Majeed, U.; Kang, H.; Andrabicet, K.I.; Johna, R. Abiotic stress: Interplay between ROS, hormones and MAPKs. Environ. Exp. Bot. 2017, 137, 142–157. [Google Scholar] [CrossRef]
- Liu, X.M.; Nguyen, X.C.; Kim, K.E.; Han, H.J.; Yoo, J.; Lee, K.; Kim, M.C.; Yun, D.J.; Chung, W.S. Phosphorylation of the zinc finger transcriptional regulator ZAT6 by MPK6 regulates Arabidopsis seed germination under salt and osmotic stress. Biochem. Bioph. Res. Co. 2013, 430, 1054–1059. [Google Scholar] [CrossRef]
- Ye, J.; Jin, C.; Li, N.; Liu, M.; Fei, Z.; Dong, L.; Li, L.; Li, Z. Selection of suitable reference genes for qRT-PCR normalisation under different experimental conditions in Eucommia ulmoides Oliv. Sci. Rep. 2018, 8, 15043. [Google Scholar] [CrossRef]
- Mafra, V.; Kubo, K.S.; Alves-Ferreira, M.; Ribeiro-Alves, M.; Stuart, M.R.; Boava, L.P.; Rodrigues, C.M.; Machado, M.A. Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. PLoS ONE 2012, 7, e31263. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Chen, M.; Huang, X.; Chen, H.; Tao, A.; Xu, J.; Qi, J. Reference gene selection for qRT-PCR normalization analysis in kenaf (Hibiscus cannabinus L.) under abiotic stress and Hormonal stimuli. Front. Plant Sci. 2017, 8, 771. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Yin, J.; Li, G.; Qi, L.; Yang, F.; Wang, R.; Li, G. Reference gene selection for qRT-PCR in Caragana korshinskii Kom. under different stress conditions. Mol. Biol. Rep. 2014, 41, 2325–2334. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Hu, B.; Zhao, L.; Shi, D.; Qin, Y. Differential expression analysis of reference genes in pineapple (Ananas comosus L.) during reproductive development and response to abiotic stress, hormonal stimuli. Trop. Plant Biol. 2019, 12, 67–77. [Google Scholar] [CrossRef]
- Wang, B.; Duan, H.; Chong, P.; Su, S.; Shan, L.; Yi, D.; Wang, L.; Li, Y. Systematic selection and validation of suitable reference genes for quantitative real-time PCR normalization studies of gene expression in Nitraria tangutorum. Sci. Rep. 2020, 10, 15891. [Google Scholar] [CrossRef] [PubMed]

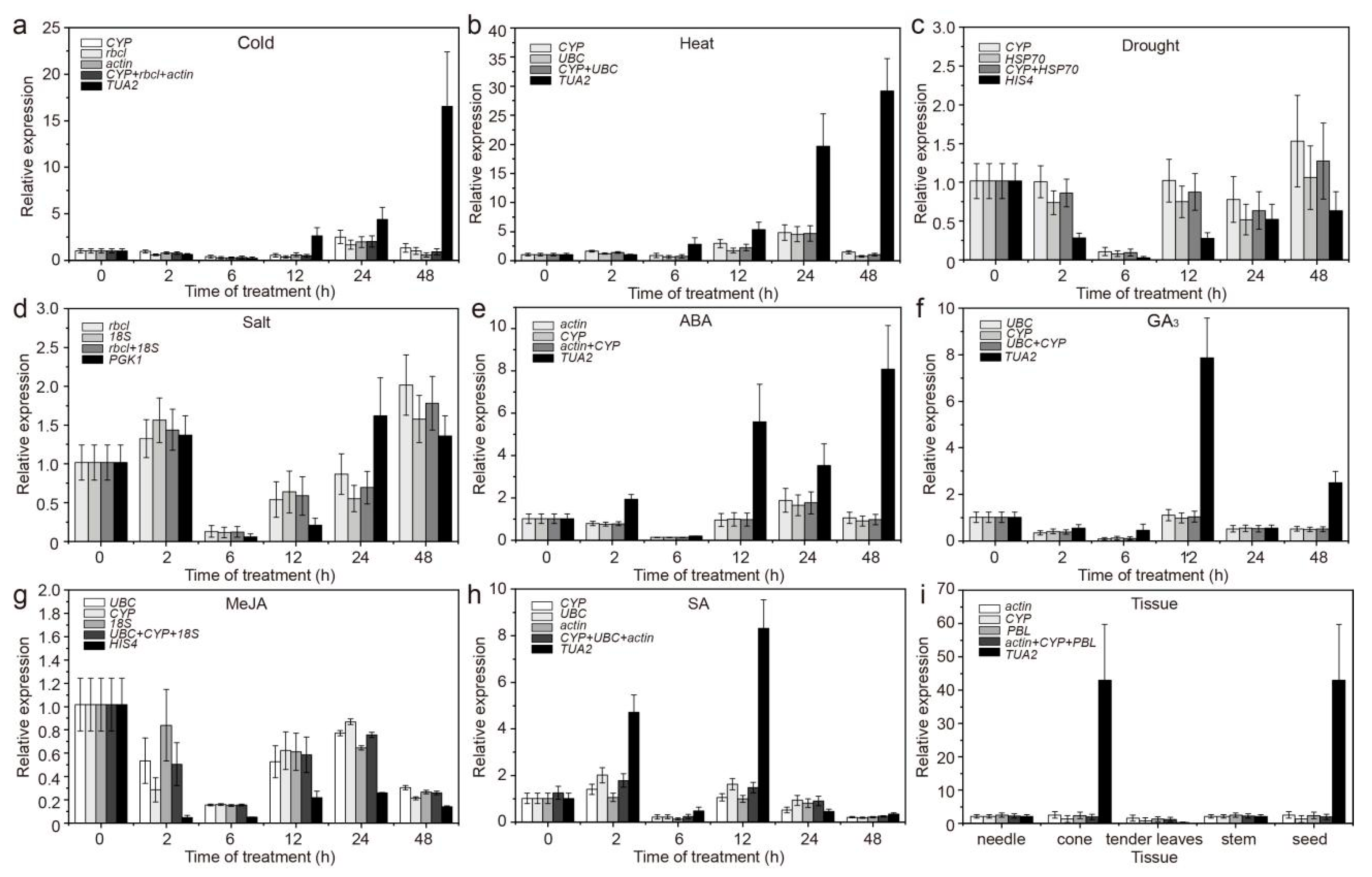
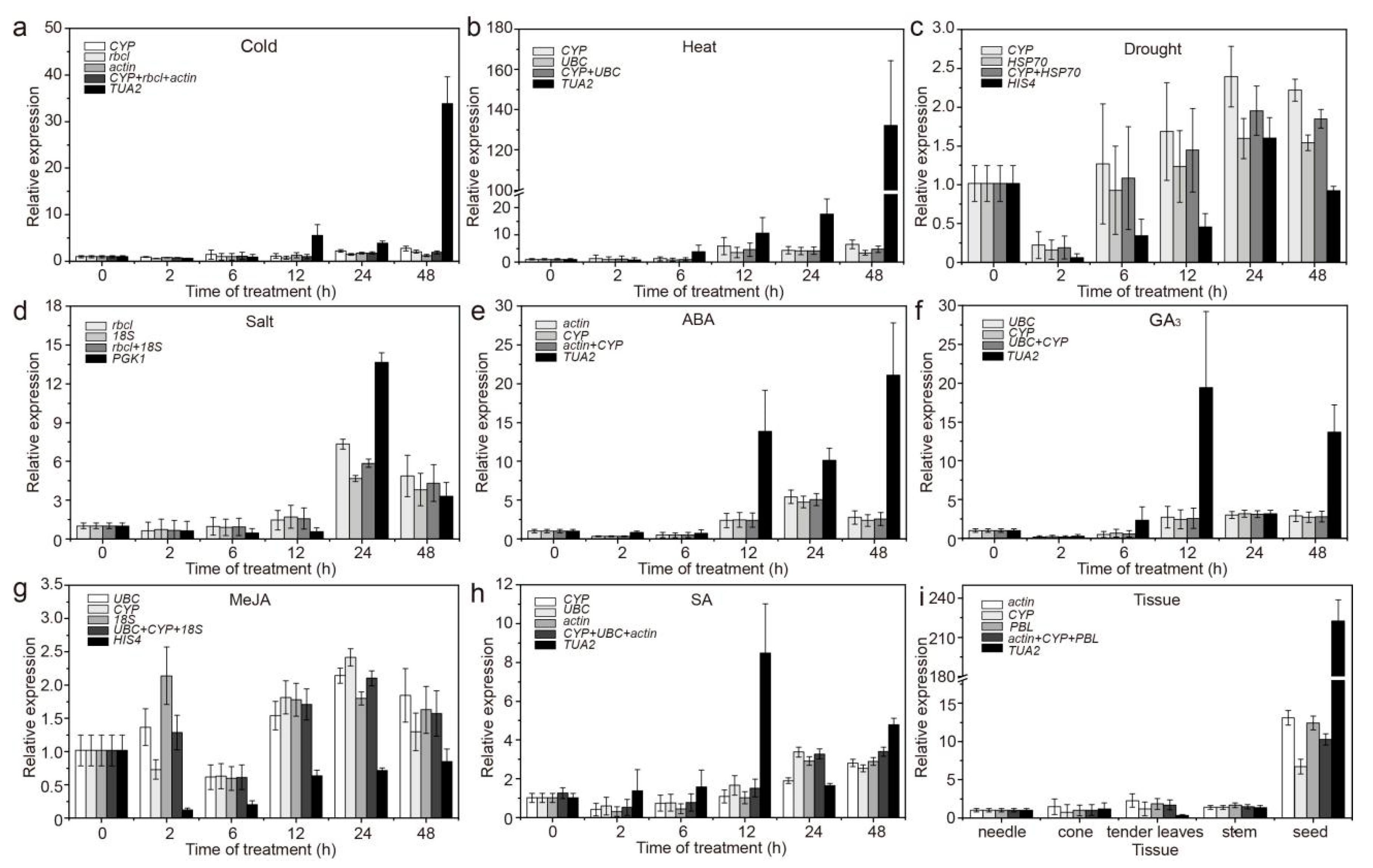
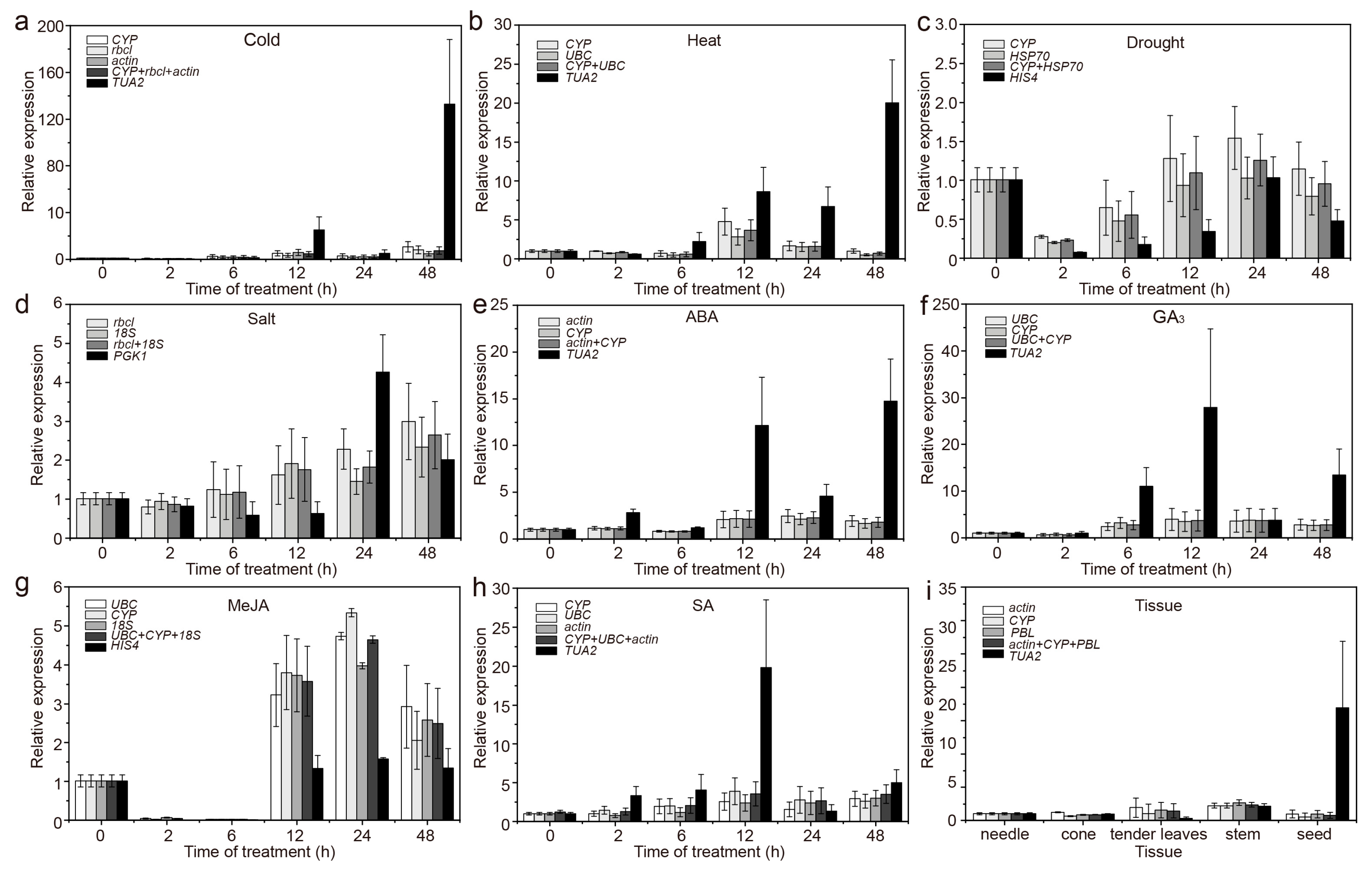
| Gene Symbol | Gene Name | Primer Sequence (5′ to 3′) | Amplicon Size (bp) | PCR Efficiency (E%) | R2 |
|---|---|---|---|---|---|
| 18S | 18S ribosomal RNA | TCTGGTCCTGTTCCGTTGG | 124 | 100.21 | 0.983 |
| GCTTTCGCAGTGGTTCGTC | |||||
| ACT | actin | GTTGCCATTCAAGCCGTTCT | 228 | 98.54 | 0.984 |
| AACAATTTCACGCTCAGCAGTAG | |||||
| CYP | cyclophilin | TCTCGGGCAGCATTTCACGC | 79 | 105.98 | 0.990 |
| AGCCGAAACTGGCGCCAACA | |||||
| HIS4 | histone H4 | TTCCAGTTGGAAGGAAAAATGTCTG | 253 | 107.83 | 0.981 |
| GGCGAGCGTGCTCAGTGTAT | |||||
| HSP70 | heat shock protein 70 | AACGCAAGGGCTTTGAGAA | 139 | 103.40 | 0.980 |
| ACCTGGCACGGGTTATGGT | |||||
| PBL | serine/threonine-protein kinase | AGTTCTGCCATGGCCCGTGA | 223 | 109.08 | 0.994 |
| TGCAGTGCCAACAACCGCTG | |||||
| PGK1 | phosphoglycerate kinase 1 | GCGGGCGAGTAAAGTGGTA | 181 | 95.14 | 0.990 |
| GGAGATCAAATACTTAATGGTGGGT | |||||
| PP2A | protein phosphatase 2A | TGAAGGAGGGAGATTTGATTGA | 128 | 107.83 | 0.988 |
| CAGTTCCGATGCACTTGGGT | |||||
| rbcl | large subunit of the ribulose-1,5-bisphosphate carboxylase/oxygenase | CGTATTACAGTTCGGTGGAGGG | 185 | 101.39 | 0.997 |
| CACAAGCGGCAGCTAGTTCA | |||||
| RPL2 | 60S ribosomal protein L2 | CCAGCATCGTTGTGGGAAAG | 70 | 102.28 | 0.980 |
| GTGACCTCCTCCTCTATGTCGTAT | |||||
| TUA2 | tubulin α-2 | CTTTCCTCGCACTCGCTGTT | 181 | 109.94 | 0.992 |
| GGTGTAGGTAGGGCGGTCAA | |||||
| UBC | ubiquitin-conjugating enzyme | CTCGCAGAATCATAAAGGAAACAC | 180 | 107.09 | 0.981 |
| CCATTGGATACTCTTCAGGCAAA |
| Sample | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cold | gene | rbcl | CYP | actin | 18S | UBC | PBL | PP2A | RPL2 | PGK1 | HSP70 | HIS4 | TUA2 |
| stability | 0.062 | 0.125 | 0.164 | 0.326 | 0.394 | 0.516 | 0.516 | 0.561 | 0.598 | 0.625 | 0.750 | 1.298 | |
| Heat | gene | CYP | UBC | HSP70 | PBL | PP2A | actin | RPL2 | 18S | HIS4 | PGK1 | rbcl | TUA2 |
| stability | 0.104 | 0.316 | 0.340 | 0.441 | 0.444 | 0.534 | 0.608 | 0.609 | 0.637 | 0.721 | 0.951 | 1.142 | |
| Drought | gene | CYP | HSP70 | actin | UBC | 18S | PBL | rbcl | RPL2 | PP2A | PGK1 | TUA2 | HIS4 |
| stability | 0.051 | 0.175 | 0.245 | 0.263 | 0.337 | 0.430 | 0.439 | 0.467 | 0.571 | 0.580 | 0.604 | 0.606 | |
| Salt | gene | rbcl | 18S | actin | HSP70 | CYP | HIS4 | UBC | PP2A | PBL | RPL2 | PGK1 | TUA2 |
| stability | 0.097 | 0.159 | 0.198 | 0.259 | 0.347 | 0.453 | 0.530 | 0.579 | 0.596 | 0.629 | 0.779 | 0.840 | |
| ABA | gene | actin | CYP | 18S | UBC | rbcl | PBL | RPL2 | PP2A | HSP70 | PGK1 | HIS4 | TUA2 |
| stability | 0.160 | 0.176 | 0.256 | 0.294 | 0.295 | 0.308 | 0.361 | 0.406 | 0.421 | 0.539 | 0.692 | 0.811 | |
| GA3 | gene | UBC | CYP | PBL | 18S | actin | RPL2 | HSP70 | rbcl | PP2A | HIS4 | PGK1 | TUA2 |
| stability | 0.031 | 0.165 | 0.224 | 0.311 | 0.325 | 0.327 | 0.371 | 0.487 | 0.531 | 0.680 | 0.761 | 0.863 | |
| MeJA | gene | CYP | UBC | actin | PBL | PP2A | rbcl | 18S | PGK1 | HSP70 | RPL2 | TUA2 | HIS4 |
| stability | 0.140 | 0.191 | 0.252 | 0.300 | 0.345 | 0.356 | 0.420 | 0.471 | 0.496 | 0.516 | 0.672 | 0.708 | |
| SA | gene | CYP | UBC | rbcl | actin | RPL2 | HSP70 | HIS4 | PBL | 18S | PGK1 | PP2A | TUA2 |
| stability | 0.214 | 0.233 | 0.241 | 0.255 | 0.367 | 0.408 | 0.539 | 0.585 | 0.616 | 0.716 | 0.740 | 0.982 | |
| Tissue | gene | actin | PBL | HSP70 | CYP | PP2A | UBC | RPL2 | rbcl | 18S | HIS4 | PGK1 | TUA2 |
| stability | 0.316 | 0.326 | 0.394 | 0.419 | 0.474 | 0.505 | 0.632 | 0.676 | 0.688 | 0.699 | 0.775 | 1.520 | |
| Abiotic | gene | CYP | UBC | 18S | HSP70 | actin | PBL | RPL2 | PP2A | rbcl | HIS4 | PGK1 | TUA2 |
| stability | 0.203 | 0.395 | 0.395 | 0.448 | 0.486 | 0.517 | 0.558 | 0.598 | 0.700 | 0.718 | 0.722 | 1.016 | |
| Hormone | gene | UBC | actin | CYP | rbcl | HSP70 | RPL2 | 18S | PBL | PP2A | PGK1 | HIS4 | TUA2 |
| stability | 0.207 | 0.279 | 0.289 | 0.396 | 0.415 | 0.450 | 0.467 | 0.473 | 0.538 | 0.737 | 0.788 | 0.825 | |
| Total | gene | CYP | UBC | actin | HSP70 | 18S | PBL | RPL2 | PP2A | rbcl | PGK1 | HIS4 | TUA2 |
| stability | 0.281 | 0.345 | 0.386 | 0.439 | 0.478 | 0.518 | 0.547 | 0.571 | 0.598 | 0.734 | 0.752 | 1.040 | |
| Sample | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cold | gene | CYP | UBC | actin | rbcl | 18S | PBL | PP2A | RPL2 | PGK1 | HSP70 | HIS4 | TUA2 |
| SD | 0.35 | 0.38 | 0.44 | 0.46 | 0.47 | 0.70 | 0.71 | 0.74 | 0.76 | 0.79 | 1.00 | 1.31 | |
| CV | 1.29 | 1.69 | 1.97 | 2.94 | 7.37 | 2.73 | 2.79 | 3.87 | 3.47 | 3.46 | 3.76 | 5.75 | |
| Heat | gene | PP2A | CYP | 18S | HSP70 | PBL | UBC | HIS4 | actin | rbcl | RPL2 | PGK1 | TUA2 |
| SD | 0.42 | 0.66 | 0.67 | 0.68 | 0.78 | 0.78 | 0.80 | 0.92 | 1.04 | 1.04 | 1.12 | 1.19 | |
| CV | 1.62 | 2.36 | 10.67 | 2.91 | 2.98 | 3.49 | 2.97 | 4.08 | 5.52 | 4.68 | 4.78 | 4.99 | |
| Drought | gene | actin | 18S | UBC | CYP | HSP70 | TUA2 | RPL2 | PP2A | rbcl | PGK1 | PBL | HIS4 |
| SD | 0.33 | 0.35 | 0.35 | 0.42 | 0.53 | 0.58 | 0.66 | 0.69 | 0.74 | 0.82 | 0.88 | 1.05 | |
| CV | 1.52 | 5.60 | 1.61 | 1.57 | 2.33 | 2.60 | 3.31 | 2.75 | 4.79 | 3.81 | 3.46 | 4.22 | |
| Salt | gene | 18S | rbcl | actin | UBC | CYP | HSP70 | TUA2 | PBL | PP2A | HIS4 | RPL2 | PGK1 |
| SD | 0.38 | 0.39 | 0.40 | 0.45 | 0.57 | 0.60 | 0.68 | 0.72 | 0.78 | 0.84 | 0.96 | 1.12 | |
| CV | 6.18 | 2.45 | 1.78 | 2.03 | 2.11 | 2.65 | 3.08 | 2.85 | 3.06 | 3.28 | 4.88 | 5.15 | |
| ABA | gene | rbcl | PP2A | UBC | 18S | CYP | actin | PBL | RPL2 | PGK1 | HSP70 | TUA2 | HIS4 |
| SD | 0.37 | 0.47 | 0.56 | 0.66 | 0.72 | 0.74 | 0.87 | 0.87 | 0.92 | 0.96 | 1.05 | 1.15 | |
| CV | 2.26 | 1.81 | 2.51 | 9.82 | 2.64 | 3.27 | 3.32 | 4.28 | 4.11 | 4.12 | 4.50 | 4.43 | |
| GA3 | gene | 18S | CYP | rbcl | RPL2 | UBC | actin | PBL | PP2A | HIS4 | HSP70 | TUA2 | PGK1 |
| SD | 0.46 | 0.47 | 0.47 | 0.53 | 0.59 | 0.65 | 0.66 | 0.69 | 0.78 | 0.78 | 0.79 | 0.96 | |
| CV | 7.20 | 1.76 | 2.88 | 2.67 | 2.71 | 2.95 | 2.56 | 2.74 | 3.00 | 3.47 | 3.51 | 4.41 | |
| MeJA | gene | UBC | 18S | rbcl | PP2A | CYP | PGK1 | actin | PBL | TUA2 | RPL2 | HIS4 | HSP70 |
| SD | 0.35 | 0.36 | 0.45 | 0.61 | 0.63 | 0.65 | 0.69 | 0.70 | 0.92 | 0.95 | 0.95 | 1.09 | |
| CV | 1.59 | 5.73 | 2.72 | 2.41 | 2.33 | 2.88 | 3.12 | 2.75 | 4.11 | 4.69 | 3.80 | 4.90 | |
| SA | gene | UBC | rbcl | CYP | actin | RPL2 | HSP70 | PBL | 18S | HIS4 | PGK1 | TUA2 | PP2A |
| SD | 0.35 | 0.35 | 0.41 | 0.44 | 0.54 | 0.64 | 0.82 | 0.85 | 0.87 | 0.91 | 0.98 | 1.05 | |
| CV | 1.54 | 2.19 | 1.52 | 1.96 | 2.74 | 2.82 | 3.24 | 14.33 | 3.43 | 4.09 | 4.33 | 4.20 | |
| Tissue | gene | 18S | UBC | actin | CYP | PBL | PGK1 | HIS4 | HSP70 | RPL2 | PP2A | rbcl | TUA2 |
| SD | 0.68 | 0.70 | 0.92 | 0.95 | 0.97 | 1.10 | 1.11 | 1.22 | 1.22 | 1.25 | 1.50 | 2.39 | |
| CV | 11.98 | 3.34 | 4.39 | 3.69 | 3.94 | 5.15 | 4.58 | 5.75 | 5.92 | 5.06 | 8.84 | 10.33 | |
| Abiotic | gene | 18S | UBC | actin | CYP | PP2A | HSP70 | PBL | PGK1 | TUA2 | HIS4 | rbcl | RPL2 |
| SD | 0.47 | 0.54 | 0.61 | 0.66 | 0.70 | 0.75 | 0.83 | 1.02 | 1.12 | 1.23 | 1.24 | 1.26 | |
| CV | 7.50 | 2.45 | 2.73 | 2.42 | 2.74 | 3.29 | 3.22 | 4.60 | 4.92 | 4.73 | 7.56 | 6.24 | |
| Hormone | gene | rbcl | UBC | 18S | CYP | actin | RPL2 | PBL | PP2A | PGK1 | HSP70 | HIS4 | TUA2 |
| SD | 0.50 | 0.56 | 0.60 | 0.62 | 0.67 | 0.75 | 0.78 | 0.82 | 0.88 | 0.91 | 0.96 | 1.03 | |
| CV | 3.05 | 2.51 | 9.51 | 2.29 | 3.03 | 3.73 | 3.03 | 3.21 | 3.98 | 4.01 | 3.74 | 4.52 | |
| Total | gene | 18S | UBC | actin | CYP | PP2A | PBL | HSP70 | PGK1 | rbcl | RPL2 | HIS4 | TUA2 |
| SD | 0.51 | 0.61 | 0.67 | 0.71 | 0.74 | 0.80 | 0.86 | 0.87 | 0.92 | 0.97 | 1.08 | 1.13 | |
| CV | 8.10 | 2.77 | 3.03 | 2.64 | 2.92 | 3.15 | 3.81 | 3.94 | 5.60 | 4.78 | 4.20 | 4.95 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Zhu, L.; Xue, J.; Yang, J.; Hu, H.; Cui, J.; Xu, J. Selection and Verification of Appropriate Reference Genes for Expression Normalization in Cryptomeria fortunei under Abiotic Stress and Hormone Treatments. Genes 2021, 12, 791. https://doi.org/10.3390/genes12060791
Zhang Y, Zhu L, Xue J, Yang J, Hu H, Cui J, Xu J. Selection and Verification of Appropriate Reference Genes for Expression Normalization in Cryptomeria fortunei under Abiotic Stress and Hormone Treatments. Genes. 2021; 12(6):791. https://doi.org/10.3390/genes12060791
Chicago/Turabian StyleZhang, Yingting, Lijuan Zhu, Jinyu Xue, Junjie Yang, Hailiang Hu, Jiebing Cui, and Jin Xu. 2021. "Selection and Verification of Appropriate Reference Genes for Expression Normalization in Cryptomeria fortunei under Abiotic Stress and Hormone Treatments" Genes 12, no. 6: 791. https://doi.org/10.3390/genes12060791
APA StyleZhang, Y., Zhu, L., Xue, J., Yang, J., Hu, H., Cui, J., & Xu, J. (2021). Selection and Verification of Appropriate Reference Genes for Expression Normalization in Cryptomeria fortunei under Abiotic Stress and Hormone Treatments. Genes, 12(6), 791. https://doi.org/10.3390/genes12060791






