The Ribosomal Gene Loci—The Power behind the Throne
Abstract
1. Nucleoli and the rDNA Genes
2. Canonical and Non-Canonical rDNA Repeats
3. Pol I Transcription Machinery
4. Regulation of Ribosomal Gene Transcription
5. rDNA Chromatin Dynamics
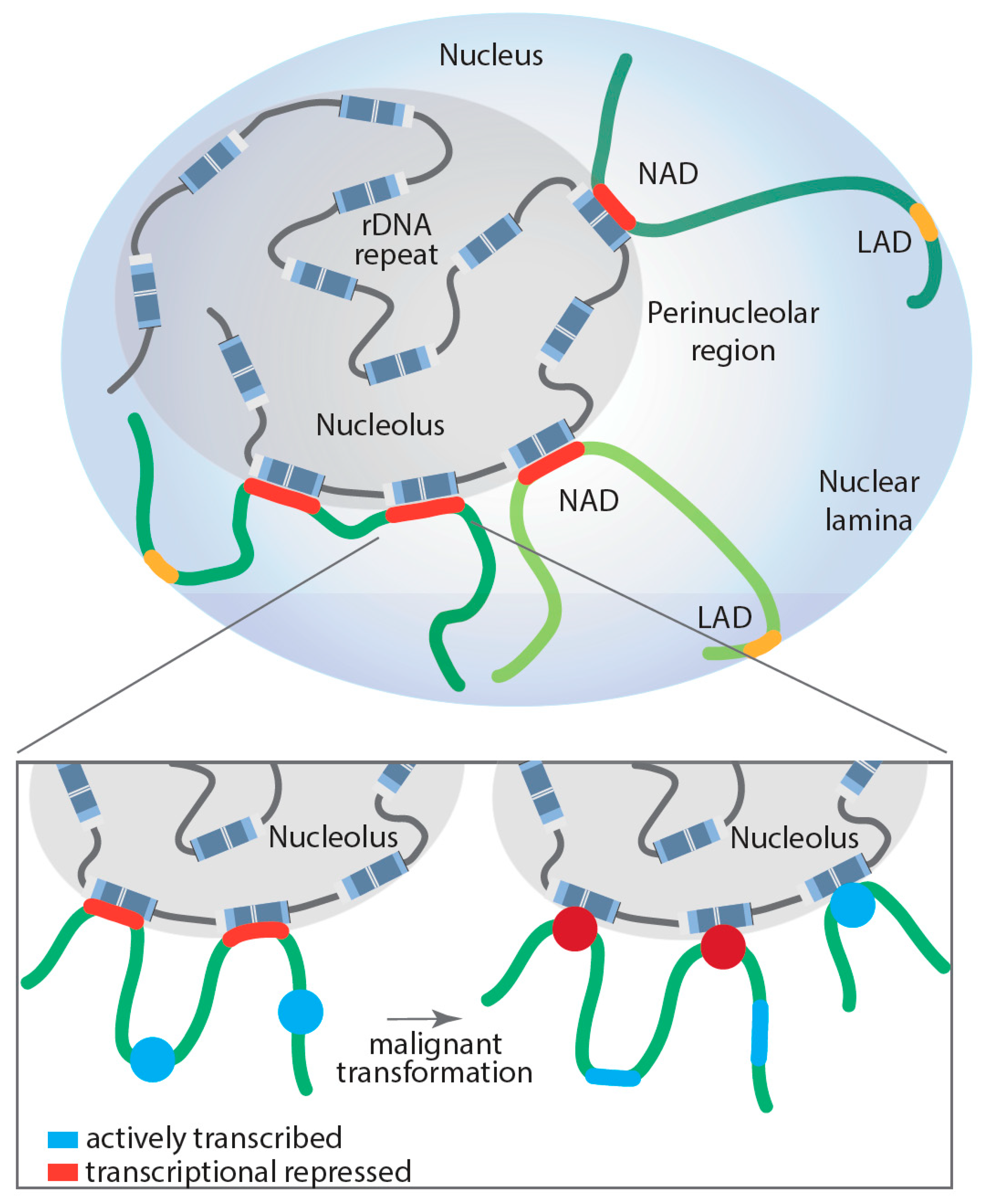
6. Role of the Nucleolus in Spatial Genome Organization and Pol II Transcription
7. Pol I Transcription in Differentiation and Development
8. How the Nucleolus and Ribosomal Genes Maintain Genome Stability
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Miller, O.L., Jr.; Beatty, B.R. Visualization of nucleolar genes. Science 1969, 164, 955–957. [Google Scholar] [CrossRef] [PubMed]
- Birch, J.L.; Zomerdijk, J.C. Structure and function of ribosomal RNA gene chromatin. Biochem. Soc. Trans. 2008, 36, 619–624. [Google Scholar] [CrossRef]
- Merz, K.; Hondele, M.; Goetze, H.; Gmelch, K.; Stoeckl, U.; Griesenbeck, J. Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev. 2008, 22, 1190–1204. [Google Scholar] [CrossRef] [PubMed]
- Marechal, V.; Elenbaas, B.; Piette, J.; Nicolas, J.C.; Levine, A.J. The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol. Cell Biol. 1994, 14, 7414–7420. [Google Scholar] [CrossRef] [PubMed]
- Lohrum, M.A.; Ludwig, R.L.; Kubbutat, M.H.; Hanlon, M.; Vousden, K.H. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 2003, 3, 577–587. [Google Scholar] [CrossRef]
- Zhang, Y.; Wolf, G.W.; Bhat, K.; Jin, A.; Allio, T.; Burkhart, W.A.; Xiong, Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell Biol. 2003, 23, 8902–8912. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.S.; Lu, H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 2004, 279, 44475–44482. [Google Scholar] [CrossRef]
- Pederson, T. The plurifunctional nucleolus. Nucleic Acids Res. 1998, 26, 3871–3876. [Google Scholar] [CrossRef]
- Nemeth, A.; Conesa, A.; Santoyo-Lopez, J.; Medina, I.; Montaner, D.; Peterfia, B.; Solovei, I.; Cremer, T.; Dopazo, J.; Langst, G. Initial genomics of the human nucleolus. PLoS Genet. 2010, 6, e1000889. [Google Scholar] [CrossRef]
- Diesch, J.; Bywater, M.J.; Sanij, E.; Cameron, D.P.; Schierding, W.; Brajanovski, N.; Son, J.; Sornkom, J.; Hein, N.; Evers, M.; et al. Changes in long-range rDNA-genomic interactions associate with altered RNA polymerase II gene programs during malignant transformation. Commun. Biol. 2019, 2, 39. [Google Scholar] [CrossRef]
- van Koningsbruggen, S.; Gierlinski, M.; Schofield, P.; Martin, D.; Barton, G.J.; Ariyurek, Y.; den Dunnen, J.T.; Lamond, A.I. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol. Biol. Cell 2010, 21, 3735–3748. [Google Scholar] [CrossRef] [PubMed]
- Frottin, F.; Schueder, F.; Tiwary, S.; Gupta, R.; Korner, R.; Schlichthaerle, T.; Cox, J.; Jungmann, R.; Hartl, F.U.; Hipp, M.S. The nucleolus functions as a phase-separated protein quality control compartment. Science 2019, 365, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Lemos, B. Ribosomal DNA copy number amplification and loss in human cancers is linked to tumor genetic context, nucleolus activity, and proliferation. PLoS Genet. 2017, 13, e1006994. [Google Scholar] [CrossRef]
- Parks, M.M.; Kurylo, C.M.; Dass, R.A.; Bojmar, L.; Lyden, D.; Vincent, C.T.; Blanchard, S.C. Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Sci. Adv. 2018, 4, eaao0665. [Google Scholar] [CrossRef]
- Gonzalez, I.L.; Sylvester, J.E. Complete sequence of the 43-kb human ribosomal DNA repeat: Analysis of the intergenic spacer. Genomics 1995, 27, 320–328. [Google Scholar] [CrossRef]
- Learned, R.M.; Learned, T.K.; Haltiner, M.M.; Tjian, R.T. Human rRNA transcription is modulated by the coordinate binding of two factors to an upstream control element. Cell 1986, 45, 847–857. [Google Scholar] [CrossRef]
- Haltiner, M.M.; Smale, S.T.; Tjian, R. Two distinct promoter elements in the human rRNA gene identified by linker scanning mutagenesis. Mol. Cell Biol. 1986, 6, 227–235. [Google Scholar] [CrossRef]
- Kuhn, A.; Grummt, I. A novel promoter in the mouse rDNA spacer is active in vivo and in vitro. EMBO J. 1987, 6, 3487–3492. [Google Scholar] [CrossRef]
- Harrington, C.A.; Chikaraishi, D.M. Transcription of spacer sequences flanking the rat 45S ribosomal DNA gene. Mol. Cell Biol. 1987, 7, 314–325. [Google Scholar] [CrossRef]
- Cassidy, B.G.; Yang-Yen, H.F.; Rothblum, L.I. Transcriptional role for the nontranscribed spacer of rat ribosomal DNA. Mol. Cell Biol. 1986, 6, 2766–2773. [Google Scholar] [CrossRef]
- Gorski, J.J.; Pathak, S.; Panov, K.; Kasciukovic, T.; Panova, T.; Russell, J.; Zomerdijk, J.C. A novel TBP-associated factor of SL1 functions in RNA polymerase I transcription. EMBO J. 2007, 26, 1560–1568. [Google Scholar] [CrossRef]
- Grummt, I.; Maier, U.; Ohrlein, A.; Hassouna, N.; Bachellerie, J.P. Transcription of mouse rDNA terminates downstream of the 3′ end of 28S RNA and involves interaction of factors with repeated sequences in the 3′ spacer. Cell 1985, 43, 801–810. [Google Scholar] [CrossRef]
- Smid, A.; Finsterer, M.; Grummt, I. Limited proteolysis unmasks specific DNA-binding of the murine RNA polymerase I-specific transcription termination factor TTFI. J. Mol. Biol. 1992, 227, 635–647. [Google Scholar] [CrossRef]
- Herdman, C.; Mars, J.C.; Stefanovsky, V.Y.; Tremblay, M.G.; Sabourin-Felix, M.; Lindsay, H.; Robinson, M.D.; Moss, T. A unique enhancer boundary complex on the mouse ribosomal RNA genes persists after loss of Rrn3 or UBF and the inactivation of RNA polymerase I transcription. PLoS Genet. 2017, 13, e1006899. [Google Scholar] [CrossRef]
- Kuhn, A.; Deppert, U.; Grummt, I. A 140-base-pair repetitive sequence element in the mouse rRNA gene spacer enhances transcription by RNA polymerase I in a cell-free system. Proc. Natl. Acad. Sci. USA 1990, 87, 7527–7531. [Google Scholar] [CrossRef]
- Tseng, H.; Chou, W.; Wang, J.; Zhang, X.; Zhang, S.; Schultz, R.M. Mouse ribosomal RNA genes contain multiple differentially regulated variants. PLoS ONE 2008, 3, e1843. [Google Scholar] [CrossRef]
- Santoro, R.; Schmitz, K.M.; Sandoval, J.; Grummt, I. Intergenic transcripts originating from a subclass of ribosomal DNA repeats silence ribosomal RNA genes in trans. EMBO Rep. 2010, 11, 52–58. [Google Scholar] [CrossRef]
- Arnheim, N.; Southern, E.M. Heterogeneity of the ribosomal genes in mice and men. Cell 1977, 11, 363–370. [Google Scholar] [CrossRef]
- Caburet, S.; Conti, C.; Schurra, C.; Lebofsky, R.; Edelstein, S.J.; Bensimon, A. Human ribosomal RNA gene arrays display a broad range of palindromic structures. Genome Res. 2005, 15, 1079–1085. [Google Scholar] [CrossRef]
- Ide, S.; Miyazaki, T.; Maki, H.; Kobayashi, T. Abundance of ribosomal RNA gene copies maintains genome integrity. Science 2010, 327, 693–696. [Google Scholar] [CrossRef]
- Udugama, M.; Sanij, E.; Voon, H.P.J.; Son, J.; Hii, L.; Henson, J.D.; Chan, F.L.; Chang, F.T.M.; Liu, Y.; Pearson, R.B.; et al. Ribosomal DNA copy loss and repeat instability in ATRX-mutated cancers. Proc. Natl. Acad. Sci. USA 2018, 115, 4737–4742. [Google Scholar] [CrossRef]
- Comai, L.; Tanese, N.; Tjian, R. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell 1992, 68, 965–976. [Google Scholar] [CrossRef]
- Zomerdijk, J.C.; Beckmann, H.; Comai, L.; Tjian, R. Assembly of transcriptionally active RNA polymerase I initiation factor SL1 from recombinant subunits. Science 1994, 266, 2015–2018. [Google Scholar] [CrossRef]
- Hannan, K.M.; Hannan, R.D.; Rothblum, L.I. Transcription by RNA polymerase I. Front. Biosci. 1998, 3, d376–d398. [Google Scholar] [CrossRef]
- Stefanovsky, V.Y.; Pelletier, G.; Bazett-Jones, D.P.; Crane-Robinson, C.; Moss, T. DNA looping in the RNA polymerase I enhancesome is the result of non-cooperative in-phase bending by two UBF molecules. Nucleic Acids Res. 2001, 29, 3241–3247. [Google Scholar] [CrossRef]
- Panov, K.I.; Friedrich, J.K.; Russell, J.; Zomerdijk, J.C. UBF activates RNA polymerase I transcription by stimulating promoter escape. EMBO J. 2006, 25, 3310–3322. [Google Scholar] [CrossRef]
- Friedrich, J.K.; Panov, K.I.; Cabart, P.; Russell, J.; Zomerdijk, J.C. TBP-TAF complex SL1 directs RNA polymerase I pre-initiation complex formation and stabilizes upstream binding factor at the rDNA promoter. J. Biol. Chem. 2005, 280, 29551–29558. [Google Scholar] [CrossRef]
- van de Nobelen, S.; Rosa-Garrido, M.; Leers, J.; Heath, H.; Soochit, W.; Joosen, L.; Jonkers, I.; Demmers, J.; van der Reijden, M.; Torrano, V.; et al. CTCF regulates the local epigenetic state of ribosomal DNA repeats. Epigenetics Chromatin 2010, 3, 19. [Google Scholar] [CrossRef]
- Blattner, C.; Jennebach, S.; Herzog, F.; Mayer, A.; Cheung, A.C.; Witte, G.; Lorenzen, K.; Hopfner, K.P.; Heck, A.J.; Aebersold, R.; et al. Molecular basis of Rrn3-regulated RNA polymerase I initiation and cell growth. Genes Dev. 2011, 25, 2093–2105. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Panov, K.I.; Friedrich, J.K.; Trinkle-Mulcahy, L.; Lamond, A.I.; Zomerdijk, J.C. hRRN3 is essential in the SL1-mediated recruitment of RNA Polymerase I to rRNA gene promoters. EMBO J. 2001, 20, 1373–1382. [Google Scholar] [CrossRef]
- Milkereit, P.; Tschochner, H. A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J. 1998, 17, 3692–3703. [Google Scholar] [CrossRef] [PubMed]
- Cavanaugh, A.H.; Evans, A.; Rothblum, L.I. Mammalian Rrn3 is required for the formation of a transcription competent preinitiation complex containing RNA polymerase I. Gene Expr. 2008, 14, 131–147. [Google Scholar] [PubMed]
- Stepanchick, A.; Zhi, H.; Cavanaugh, A.H.; Rothblum, K.; Schneider, D.A.; Rothblum, L.I. DNA binding by the ribosomal DNA transcription factor rrn3 is essential for ribosomal DNA transcription. J. Biol. Chem. 2013, 288, 9135–9144. [Google Scholar] [CrossRef]
- Panov, K.I.; Panova, T.B.; Gadal, O.; Nishiyama, K.; Saito, T.; Russell, J.; Zomerdijk, J.C. RNA polymerase I-specific subunit CAST/hPAF49 has a role in the activation of transcription by upstream binding factor. Mol. Cell Biol. 2006, 26, 5436–5448. [Google Scholar] [CrossRef][Green Version]
- O’Mahony, D.J.; Xie, W.Q.; Smith, S.D.; Singer, H.A.; Rothblum, L.I. Differential phosphorylation and localization of the transcription factor UBF in vivo in response to serum deprivation. In vitro dephosphorylation of UBF reduces its transactivation properties. J. Biol. Chem. 1992, 267, 35–38. [Google Scholar] [CrossRef]
- Kihm, A.J.; Hershey, J.C.; Haystead, T.A.; Madsen, C.S.; Owens, G.K. Phosphorylation of the rRNA transcription factor upstream binding factor promotes its association with TATA binding protein. Proc. Natl. Acad. Sci. USA 1998, 95, 14816–14820. [Google Scholar] [CrossRef] [PubMed]
- Voit, R.; Grummt, I. Phosphorylation of UBF at serine 388 is required for interaction with RNA polymerase I and activation of rDNA transcription. Proc. Natl. Acad. Sci. USA 2001, 98, 13631–13636. [Google Scholar] [CrossRef]
- Voit, R.; Hoffmann, M.; Grummt, I. Phosphorylation by G1-specific cdk-cyclin complexes activates the nucleolar transcription factor UBF. EMBO J. 1999, 18, 1891–1899. [Google Scholar] [CrossRef]
- Kermekchiev, M.; Workman, J.L.; Pikaard, C.S. Nucleosome binding by the polymerase I transactivator upstream binding factor displaces linker histone H1. Mol. Cell Biol. 1997, 17, 5833–5842. [Google Scholar] [CrossRef]
- Sanij, E.; Poortinga, G.; Sharkey, K.; Hung, S.; Holloway, T.P.; Quin, J.; Robb, E.; Wong, L.H.; Thomas, W.G.; Stefanovsky, V.; et al. UBF levels determine the number of active ribosomal RNA genes in mammals. J. Cell Biol. 2008, 183, 1259–1274. [Google Scholar] [CrossRef]
- Roussel, P.; Andre, C.; Masson, C.; Geraud, G.; Hernandez-Verdun, D. Localization of the RNA polymerase I transcription factor hUBF during the cell cycle. J. Cell Sci. 1993, 104 Pt 2, 327–337. [Google Scholar] [CrossRef]
- Mais, C.; Wright, J.E.; Prieto, J.L.; Raggett, S.L.; McStay, B. UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes Dev. 2005, 19, 50–64. [Google Scholar] [CrossRef] [PubMed]
- Prieto, J.L.; McStay, B. Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev. 2007, 21, 2041–2054. [Google Scholar] [CrossRef] [PubMed]
- Birch, J.L.; Tan, B.C.; Panov, K.I.; Panova, T.B.; Andersen, J.S.; Owen-Hughes, T.A.; Russell, J.; Lee, S.C.; Zomerdijk, J.C. FACT facilitates chromatin transcription by RNA polymerases I and III. EMBO J. 2009, 28, 854–865. [Google Scholar] [CrossRef] [PubMed]
- Beckouet, F.; Labarre-Mariotte, S.; Albert, B.; Imazawa, Y.; Werner, M.; Gadal, O.; Nogi, Y.; Thuriaux, P. Two RNA polymerase I subunits control the binding and release of Rrn3 during transcription. Mol. Cell Biol. 2008, 28, 1596–1605. [Google Scholar] [CrossRef] [PubMed]
- Cavanaugh, A.H.; Hirschler-Laszkiewicz, I.; Hu, Q.; Dundr, M.; Smink, T.; Misteli, T.; Rothblum, L.I. Rrn3 phosphorylation is a regulatory checkpoint for ribosome biogenesis. J. Biol. Chem. 2002, 277, 27423–27432. [Google Scholar] [CrossRef] [PubMed]
- Peyroche, G.; Milkereit, P.; Bischler, N.; Tschochner, H.; Schultz, P.; Sentenac, A.; Carles, C.; Riva, M. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J. 2000, 19, 5473–5482. [Google Scholar] [CrossRef]
- Ray, S.; Panova, T.; Miller, G.; Volkov, A.; Porter, A.C.; Russell, J.; Panov, K.I.; Zomerdijk, J.C. Topoisomerase IIalpha promotes activation of RNA polymerase I transcription by facilitating pre-initiation complex formation. Nat. Commun. 2013, 4, 1598. [Google Scholar] [CrossRef] [PubMed]
- Panova, T.B.; Panov, K.I.; Russell, J.; Zomerdijk, J.C. Casein kinase 2 associates with initiation-competent RNA polymerase I and has multiple roles in ribosomal DNA transcription. Mol. Cell Biol. 2006, 26, 5957–5968. [Google Scholar] [CrossRef][Green Version]
- Henras, A.K.; Plisson-Chastang, C.; O’Donohue, M.F.; Chakraborty, A.; Gleizes, P.E. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev. RNA 2015, 6, 225–242. [Google Scholar] [CrossRef]
- Rickards, B.; Flint, S.J.; Cole, M.D.; LeRoy, G. Nucleolin is required for RNA polymerase I transcription in vivo. Mol. Cell Biol. 2007, 27, 937–948. [Google Scholar] [CrossRef] [PubMed]
- Murano, K.; Okuwaki, M.; Hisaoka, M.; Nagata, K. Transcription regulation of the rRNA gene by a multifunctional nucleolar protein, B23/nucleophosmin, through its histone chaperone activity. Mol. Cell Biol. 2008, 28, 3114–3126. [Google Scholar] [CrossRef]
- Evers, R.; Grummt, I. Molecular coevolution of mammalian ribosomal gene terminator sequences and the transcription termination factor TTF-I. Proc. Natl. Acad. Sci. USA 1995, 92, 5827–5831. [Google Scholar] [CrossRef] [PubMed]
- Bywater, M.J.; Pearson, R.B.; McArthur, G.A.; Hannan, R.D. Dysregulation of the basal RNA polymerase transcription apparatus in cancer. Nat. Rev. Cancer 2013, 13, 299–314. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.; Zhao, J.; Yuan, X.; Grummt, I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004, 18, 423–434. [Google Scholar] [CrossRef]
- Hannan, K.M.; Brandenburger, Y.; Jenkins, A.; Sharkey, K.; Cavanaugh, A.; Rothblum, L.; Moss, T.; Poortinga, G.; McArthur, G.A.; Pearson, R.B.; et al. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol. Cell Biol. 2003, 23, 8862–8877. [Google Scholar] [CrossRef]
- Jastrzebski, K.; Hannan, K.M.; Tchoubrieva, E.B.; Hannan, R.D.; Pearson, R.B. Coordinate regulation of ribosome biogenesis and function by the ribosomal protein S6 kinase, a key mediator of mTOR function. Growth Factors 2007, 25, 209–226. [Google Scholar] [CrossRef]
- Zhao, J.; Yuan, X.; Frodin, M.; Grummt, I. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol. Cell 2003, 11, 405–413. [Google Scholar] [CrossRef]
- Claypool, J.A.; French, S.L.; Johzuka, K.; Eliason, K.; Vu, L.; Dodd, J.A.; Beyer, A.L.; Nomura, M. Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol. Biol. Cell 2004, 15, 946–956. [Google Scholar] [CrossRef]
- Hannan, K.M.; Sanij, E.; Hein, N.; Hannan, R.D.; Pearson, R.B. Signaling to the ribosome in cancer--It is more than just mTORC1. IUBMB Life 2011, 63, 79–85. [Google Scholar] [CrossRef]
- Tsang, C.K.; Bertram, P.G.; Ai, W.; Drenan, R.; Zheng, X.F. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 2003, 22, 6045–6056. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.; Bierhoff, H.; Grummt, I. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev. 2005, 19, 933–941. [Google Scholar] [CrossRef] [PubMed]
- Hoppe, S.; Bierhoff, H.; Cado, I.; Weber, A.; Tiebe, M.; Grummt, I.; Voit, R. AMP-activated protein kinase adapts rRNA synthesis to cellular energy supply. Proc. Natl. Acad. Sci. USA 2009, 106, 17781–17786. [Google Scholar] [CrossRef] [PubMed]
- Salifou, K.; Ray, S.; Verrier, L.; Aguirrebengoa, M.; Trouche, D.; Panov, K.I.; Vandromme, M. The histone demethylase JMJD2A/KDM4A links ribosomal RNA transcription to nutrients and growth factors availability. Nat. Commun. 2016, 7, 10174. [Google Scholar] [CrossRef]
- Murayama, A.; Ohmori, K.; Fujimura, A.; Minami, H.; Yasuzawa-Tanaka, K.; Kuroda, T.; Oie, S.; Daitoku, H.; Okuwaki, M.; Nagata, K.; et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell 2008, 133, 627–639. [Google Scholar] [CrossRef]
- Santoro, R.; Li, J.; Grummt, I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 2002, 32, 393–396. [Google Scholar] [CrossRef]
- Hannan, R.D.; Luyken, J.; Rothblum, L.I. Regulation of rDNA transcription factors during cardiomyocyte hypertrophy induced by adrenergic agents. J. Biol. Chem. 1995, 270, 8290–8297. [Google Scholar] [CrossRef]
- Bakshi, R.; Zaidi, S.K.; Pande, S.; Hassan, M.Q.; Young, D.W.; Montecino, M.; Lian, J.B.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S. The leukemogenic t(8;21) fusion protein AML1-ETO controls rRNA genes and associates with nucleolar-organizing regions at mitotic chromosomes. J. Cell Sci. 2008, 121, 3981–3990. [Google Scholar] [CrossRef]
- French, S.L.; Sikes, M.L.; Hontz, R.D.; Osheim, Y.N.; Lambert, T.E.; El Hage, A.; Smith, M.M.; Tollervey, D.; Smith, J.S.; Beyer, A.L. Distinguishing the roles of Topoisomerases I and II in relief of transcription-induced torsional stress in yeast rRNA genes. Mol. Cell Biol. 2011, 31, 482–494. [Google Scholar] [CrossRef] [PubMed]
- Denissov, S.; Lessard, F.; Mayer, C.; Stefanovsky, V.; van Driel, M.; Grummt, I.; Moss, T.; Stunnenberg, H.G. A model for the topology of active ribosomal RNA genes. EMBO Rep. 2011, 12, 231–237. [Google Scholar] [CrossRef]
- Maiser, A.; Dillinger, S.; Langst, G.; Schermelleh, L.; Leonhardt, H.; Nemeth, A. Super-resolution in situ analysis of active ribosomal DNA chromatin organization in the nucleolus. Sci. Rep. 2020, 10, 7462. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.; Schmitz, K.M.; Li, J.; Grummt, I.; Santoro, R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell 2006, 22, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, K.M.; Mayer, C.; Postepska, A.; Grummt, I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010, 24, 2264–2269. [Google Scholar] [CrossRef] [PubMed]
- Guetg, C.; Lienemann, P.; Sirri, V.; Grummt, I.; Hernandez-Verdun, D.; Hottiger, M.O.; Fussenegger, M.; Santoro, R. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 2010, 29, 2135–2146. [Google Scholar] [CrossRef]
- Savic, N.; Bar, D.; Leone, S.; Frommel, S.C.; Weber, F.A.; Vollenweider, E.; Ferrari, E.; Ziegler, U.; Kaech, A.; Shakhova, O.; et al. lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell 2014, 15, 720–734. [Google Scholar] [CrossRef]
- Grummt, I.; Langst, G. Epigenetic control of RNA polymerase I transcription in mammalian cells. Biochim. Biophys. Acta 2013, 1829, 393–404. [Google Scholar] [CrossRef]
- Moss, T.; Mars, J.C.; Tremblay, M.G.; Sabourin-Felix, M. The chromatin landscape of the ribosomal RNA genes in mouse and human. Chromosome Res. 2019, 27, 31–40. [Google Scholar] [CrossRef]
- Abraham, K.J.; Khosraviani, N.; Chan, J.N.Y.; Gorthi, A.; Samman, A.; Zhao, D.Y.; Wang, M.; Bokros, M.; Vidya, E.; Ostrowski, L.A.; et al. Nucleolar RNA polymerase II drives ribosome biogenesis. Nature 2020, 585, 298–302. [Google Scholar] [CrossRef]
- Dammann, R.; Lucchini, R.; Koller, T.; Sogo, J.M. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993, 21, 2331–2338. [Google Scholar] [CrossRef]
- Osheim, Y.N.; Beyer, A.L. Electron microscopy of ribonucleoprotein complexes on nascent RNA using Miller chromatin spreading method. Methods Enzymol. 1989, 180, 481–509. [Google Scholar] [CrossRef]
- Jones, H.S.; Kawauchi, J.; Braglia, P.; Alen, C.M.; Kent, N.A.; Proudfoot, N.J. RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat. Struct Mol. Biol. 2007, 14, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Foltankova, V.; Legartova, S.; Kozubek, S.; Hofer, M.; Bartova, E. DNA-damage response in chromatin of ribosomal genes and the surrounding genome. Gene 2013, 522, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Zentner, G.E.; Saiakhova, A.; Manaenkov, P.; Adams, M.D.; Scacheri, P.C. Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Res. 2011, 39, 4949–4960. [Google Scholar] [CrossRef] [PubMed]
- Zentner, G.E.; Balow, S.A.; Scacheri, P.C. Genomic characterization of the mouse ribosomal DNA locus. G3 2014, 4, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Lemos, B. Ribosomal DNA harbors an evolutionarily conserved clock of biological aging. Genome Res. 2019, 29, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Ling, T.; Zhou, Y.; Feng, W.; Zhu, Q.; Stunnenberg, H.G.; Grummt, I.; Tao, W. The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc. Natl. Acad. Sci. USA 2012, 109, 8161–8166. [Google Scholar] [CrossRef]
- Patel, A.; Dharmarajan, V.; Vought, V.E.; Cosgrove, M.S. On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. J. Biol. Chem. 2009, 284, 24242–24256. [Google Scholar] [CrossRef]
- Feng, W.; Yonezawa, M.; Ye, J.; Jenuwein, T.; Grummt, I. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat. Struct Mol. Biol. 2010, 17, 445–450. [Google Scholar] [CrossRef]
- Yuan, X.; Feng, W.; Imhof, A.; Grummt, I.; Zhou, Y. Activation of RNA polymerase I transcription by cockayne syndrome group B protein and histone methyltransferase G9a. Mol. Cell 2007, 27, 585–595. [Google Scholar] [CrossRef]
- Shen, M.; Zhou, T.; Xie, W.; Ling, T.; Zhu, Q.; Zong, L.; Lyu, G.; Gao, Q.; Zhang, F.; Tao, W. The chromatin remodeling factor CSB recruits histone acetyltransferase PCAF to rRNA gene promoters in active state for transcription initiation. PLoS ONE 2013, 8, e62668. [Google Scholar] [CrossRef]
- Garapaty, S.; Xu, C.F.; Trojer, P.; Mahajan, M.A.; Neubert, T.A.; Samuels, H.H. Identification and characterization of a novel nuclear protein complex involved in nuclear hormone receptor-mediated gene regulation. J. Biol. Chem. 2009, 284, 7542–7552. [Google Scholar] [CrossRef] [PubMed]
- Hung, K.H.; Woo, Y.H.; Lin, I.Y.; Liu, C.H.; Wang, L.C.; Chen, H.Y.; Chiang, B.L.; Lin, K.I. The KDM4A/KDM4C/NF-kappaB and WDR5 epigenetic cascade regulates the activation of B cells. Nucleic Acids Res. 2018, 46, 5547–5560. [Google Scholar] [CrossRef] [PubMed]
- Hamdane, N.; Stefanovsky, V.Y.; Tremblay, M.G.; Nemeth, A.; Paquet, E.; Lessard, F.; Sanij, E.; Hannan, R.; Moss, T. Conditional inactivation of Upstream Binding Factor reveals its epigenetic functions and the existence of a somatic nucleolar precursor body. PLoS Genet. 2014, 10, e1004505. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, H.; Matsumori, H.; Kalendova, A.; Hozak, P.; Goldberg, I.G.; Nakao, M.; Saitoh, N.; Harata, M. The actin family protein ARP6 contributes to the structure and the function of the nucleolus. Biochem. Biophys. Res. Commun. 2015, 464, 554–560. [Google Scholar] [CrossRef]
- Zheng, Y.; John, S.; Pesavento, J.J.; Schultz-Norton, J.R.; Schiltz, R.L.; Baek, S.; Nardulli, A.M.; Hager, G.L.; Kelleher, N.L.; Mizzen, C.A. Histone H1 phosphorylation is associated with transcription by RNA polymerases I and II. J. Cell Biol. 2010, 189, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Jia, J.; Wu, C.; Yao, M.; Li, M.; Jin, J.; Jiang, C.; Cai, Y.; Pei, D.; Pan, G.; et al. Ribosomal RNA gene transcription mediated by the master genome regulator protein CCCTC-binding factor (CTCF) is negatively regulated by the condensin complex. J. Biol. Chem. 2013, 288, 26067–26077. [Google Scholar] [CrossRef] [PubMed]
- Bose, T.; Lee, K.K.; Lu, S.; Xu, B.; Harris, B.; Slaughter, B.; Unruh, J.; Garrett, A.; McDowell, W.; Box, A.; et al. Cohesin proteins promote ribosomal RNA production and protein translation in yeast and human cells. PLoS Genet. 2012, 8, e1002749. [Google Scholar] [CrossRef]
- Uuskula-Reimand, L.; Hou, H.; Samavarchi-Tehrani, P.; Rudan, M.V.; Liang, M.; Medina-Rivera, A.; Mohammed, H.; Schmidt, D.; Schwalie, P.; Young, E.J.; et al. Topoisomerase II β interacts with cohesin and CTCF at topological domain borders. Genome Biol. 2016, 17, 182. [Google Scholar] [CrossRef]
- Vertii, A.; Ou, J.; Yu, J.; Yan, A.; Pages, H.; Liu, H.; Zhu, L.J.; Kaufman, P.D. Two contrasting classes of nucleolus-associated domains in mouse fibroblast heterochromatin. Genome Res. 2019, 29, 1235–1249. [Google Scholar] [CrossRef]
- Bizhanova, A.; Kaufman, P.D. Close to the edge: Heterochromatin at the nucleolar and nuclear peripheries. Biochim. Biophys. Acta Gene Regul. Mech. 2021, 1864, 194666. [Google Scholar] [CrossRef]
- Haeusler, R.A.; Engelke, D.R. Spatial organization of transcription by RNA polymerase III. Nucleic Acids Res. 2006, 34, 4826–4836. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.F.; Huynh, K.D.; Lee, J.T. Perinucleolar targeting of the inactive X during S phase: Evidence for a role in the maintenance of silencing. Cell 2007, 129, 693–706. [Google Scholar] [CrossRef]
- Dillinger, S.; Straub, T.; Nemeth, A. Nucleolus association of chromosomal domains is largely maintained in cellular senescence despite massive nuclear reorganisation. PLoS ONE 2017, 12, e0178821. [Google Scholar] [CrossRef]
- Yu, S.; Lemos, B. The long-range interaction map of ribosomal DNA arrays. PLoS Genet. 2018, 14, e1007258. [Google Scholar] [CrossRef] [PubMed]
- Bizhanova, A.; Yan, A.; Yu, J.; Zhu, L.J.; Kaufman, P.D. Distinct features of nucleolus-associated domains in mouse embryonic stem cells. Chromosoma 2020, 129, 121–139. [Google Scholar] [CrossRef] [PubMed]
- Matheson, T.D.; Kaufman, P.D. The p150N domain of chromatin assembly factor-1 regulates Ki-67 accumulation on the mitotic perichromosomal layer. Mol. Biol. Cell 2017, 28, 21–29. [Google Scholar] [CrossRef]
- Solovei, I.; Wang, A.S.; Thanisch, K.; Schmidt, C.S.; Krebs, S.; Zwerger, M.; Cohen, T.V.; Devys, D.; Foisner, R.; Peichl, L.; et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 2013, 152, 584–598. [Google Scholar] [CrossRef]
- Shin, Y.; Chang, Y.C.; Lee, D.S.W.; Berry, J.; Sanders, D.W.; Ronceray, P.; Wingreen, N.S.; Haataja, M.; Brangwynne, C.P. Liquid Nuclear Condensates Mechanically Sense and Restructure the Genome. Cell 2018, 175, 1481–1491. [Google Scholar] [CrossRef]
- Altmann, G.G.; Leblond, C.P. Changes in the size and structure of the nucleolus of columnar cells during their migration from crypt base to villus top in rat jejunum. J. Cell Sci. 1982, 56, 83–99. [Google Scholar] [CrossRef]
- Cavanaugh, A.H.; Hempel, W.M.; Taylor, L.J.; Rogalsky, V.; Todorov, G.; Rothblum, L.I. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature 1995, 374, 177–180. [Google Scholar] [CrossRef]
- Comai, L.; Song, Y.; Tan, C.; Bui, T. Inhibition of RNA polymerase I transcription in differentiated myeloid leukemia cells by inactivation of selectivity factor 1. Cell Growth Differ. 2000, 11, 63–70. [Google Scholar]
- Alzuherri, H.M.; White, R.J. Regulation of RNA polymerase I transcription in response to F9 embryonal carcinoma stem cell differentiation. J. Biol. Chem. 1999, 274, 4328–4334. [Google Scholar] [CrossRef]
- Larson, D.E.; Xie, W.; Glibetic, M.; O’Mahony, D.; Sells, B.H.; Rothblum, L.I. Coordinated decreases in rRNA gene transcription factors and rRNA synthesis during muscle cell differentiation. Proc. Natl. Acad. Sci. USA 1993, 90, 7933–7936. [Google Scholar] [CrossRef] [PubMed]
- Neben, C.L.; Lay, F.D.; Mao, X.; Tuzon, C.T.; Merrill, A.E. Ribosome biogenesis is dynamically regulated during osteoblast differentiation. Gene 2017, 612, 29–35. [Google Scholar] [CrossRef]
- Poortinga, G.; Wall, M.; Sanij, E.; Siwicki, K.; Ellul, J.; Brown, D.; Holloway, T.P.; Hannan, R.D.; McArthur, G.A. c-MYC coordinately regulates ribosomal gene chromatin remodeling and Pol I availability during granulocyte differentiation. Nucleic Acids Res. 2011, 39, 3267–3281. [Google Scholar] [CrossRef] [PubMed]
- Tu, X.; Baffa, R.; Luke, S.; Prisco, M.; Baserga, R. Intracellular redistribution of nuclear and nucleolar proteins during differentiation of 32D murine hemopoietic cells. Exp. Cell Res. 2003, 288, 119–130. [Google Scholar] [CrossRef]
- Zhang, Q.; Shalaby, N.A.; Buszczak, M. Changes in rRNA transcription influence proliferation and cell fate within a stem cell lineage. Science 2014, 343, 298–301. [Google Scholar] [CrossRef] [PubMed]
- Vicidomini, R.; Petrizzo, A.; di Giovanni, A.; Cassese, L.; Lombardi, A.A.; Pragliola, C.; Furia, M. Drosophila dyskerin is required for somatic stem cell homeostasis. Sci. Rep. 2017, 7, 347. [Google Scholar] [CrossRef]
- You, K.T.; Park, J.; Kim, V.N. Role of the small subunit processome in the maintenance of pluripotent stem cells. Genes Dev. 2015, 29, 2004–2009. [Google Scholar] [CrossRef]
- Ali, S.A.; Zaidi, S.K.; Dacwag, C.S.; Salma, N.; Young, D.W.; Shakoori, A.R.; Montecino, M.A.; Lian, J.B.; van Wijnen, A.J.; Imbalzano, A.N.; et al. Phenotypic transcription factors epigenetically mediate cell growth control. Proc. Natl. Acad. Sci. USA 2008, 105, 6632–6637. [Google Scholar] [CrossRef]
- Hein, N.; Cameron, D.P.; Hannan, K.M.; Nguyen, N.N.; Fong, C.Y.; Sornkom, J.; Wall, M.; Pavy, M.; Cullinane, C.; Diesch, J.; et al. Inhibition of Pol I transcription treats murine and human AML by targeting the leukemia-initiating cell population. Blood 2017, 129, 2882–2895. [Google Scholar] [CrossRef]
- Hayashi, Y.; Kuroda, T.; Kishimoto, H.; Wang, C.; Iwama, A.; Kimura, K. Downregulation of rRNA transcription triggers cell differentiation. PLoS ONE 2014, 9, e98586. [Google Scholar] [CrossRef] [PubMed]
- Prakash, V.; Carson, B.B.; Feenstra, J.M.; Dass, R.A.; Sekyrova, P.; Hoshino, A.; Petersen, J.; Guo, Y.; Parks, M.M.; Kurylo, C.M.; et al. Ribosome biogenesis during cell cycle arrest fuels EMT in development and disease. Nat. Commun. 2019, 10, 2110. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, L.M.; Baserga, S.J. Crosstalk between the nucleolus and the DNA damage response. Mol. Biosyst 2017, 13, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.S.; Lyon, C.E.; Fox, A.H.; Leung, A.K.; Lam, Y.W.; Steen, H.; Mann, M.; Lamond, A.I. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002, 12, 1–11. [Google Scholar] [CrossRef]
- Andersen, J.S.; Lam, Y.W.; Leung, A.K.; Ong, S.E.; Lyon, C.E.; Lamond, A.I.; Mann, M. Nucleolar proteome dynamics. Nature 2005, 433, 77–83. [Google Scholar] [CrossRef]
- Johnston, R.; D’Costa, Z.; Ray, S.; Gorski, J.; Harkin, D.P.; Mullan, P.; Panov, K.I. The identification of a novel role for BRCA1 in regulating RNA polymerase I transcription. Oncotarget 2016, 7, 68097–68110. [Google Scholar] [CrossRef] [PubMed]
- Trainor, P.A.; Dixon, J.; Dixon, M.J. Treacher Collins syndrome: Etiology, pathogenesis and prevention. Eur J. Hum. Genet. 2009, 17, 275–283. [Google Scholar] [CrossRef]
- Larsen, D.H.; Hari, F.; Clapperton, J.A.; Gwerder, M.; Gutsche, K.; Altmeyer, M.; Jungmichel, S.; Toledo, L.I.; Fink, D.; Rask, M.B.; et al. The NBS1-Treacle complex controls ribosomal RNA transcription in response to DNA damage. Nat. Cell Biol. 2014, 16, 792–803. [Google Scholar] [CrossRef]
- Ciccia, A.; Huang, J.W.; Izhar, L.; Sowa, M.E.; Harper, J.W.; Elledge, S.J. Treacher Collins syndrome TCOF1 protein cooperates with NBS1 in the DNA damage response. Proc. Natl. Acad. Sci. USA 2014, 111, 18631–18636. [Google Scholar] [CrossRef]
- Mooser, C.; Symeonidou, I.E.; Leimbacher, P.A.; Ribeiro, A.; Shorrocks, A.K.; Jungmichel, S.; Larsen, S.C.; Knechtle, K.; Jasrotia, A.; Zurbriggen, D.; et al. Treacle controls the nucleolar response to rDNA breaks via TOPBP1 recruitment and ATR activation. Nat. Commun. 2020, 11, 123. [Google Scholar] [CrossRef] [PubMed]
- Fages, J.; Chailleux, C.; Humbert, J.; Jang, S.M.; Loehr, J.; Lambert, J.P.; Cote, J.; Trouche, D.; Canitrot, Y. JMJD6 participates in the maintenance of ribosomal DNA integrity in response to DNA damage. PLoS Genet. 2020, 16, e1008511. [Google Scholar] [CrossRef] [PubMed]
- Lindstrom, M.S.; Jurada, D.; Bursac, S.; Orsolic, I.; Bartek, J.; Volarevic, S. Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene 2018, 37, 2351–2366. [Google Scholar] [CrossRef] [PubMed]
- Paredes, S.; Angulo-Ibanez, M.; Tasselli, L.; Carlson, S.M.; Zheng, W.; Li, T.M.; Chua, K.F. The epigenetic regulator SIRT7 guards against mammalian cellular senescence induced by ribosomal DNA instability. J. Biol. Chem. 2018, 293, 11242–11250. [Google Scholar] [CrossRef]
- Sanij, E.; Diesch, J.; Lesmana, A.; Poortinga, G.; Hein, N.; Lidgerwood, G.; Cameron, D.P.; Ellul, J.; Goodall, G.J.; Wong, L.H.; et al. A novel role for the Pol I transcription factor UBTF in maintaining genome stability through the regulation of highly transcribed Pol II genes. Genome Res. 2015, 25, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Akamatsu, Y.; Kobayashi, T. The Human RNA Polymerase I Transcription Terminator Complex Acts as a Replication Fork Barrier That Coordinates the Progress of Replication with rRNA Transcription Activity. Mol. Cell Biol. 2015, 35, 1871–1881. [Google Scholar] [CrossRef]
- Santos-Pereira, J.M.; Aguilera, A. R loops: New modulators of genome dynamics and function. Nat. Rev. Genet. 2015, 16, 583–597. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.C.; Ostrowski, L.A.; Pietrobon, V.; Mekhail, K. Repetitive DNA loci and their modulation by the non-canonical nucleic acid structures R-loops and G-quadruplexes. Nucleus 2017, 8, 162–181. [Google Scholar] [CrossRef] [PubMed]
- Skourti-Stathaki, K.; Proudfoot, N.J. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014, 28, 1384–1396. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panov, K.I.; Hannan, K.; Hannan, R.D.; Hein, N. The Ribosomal Gene Loci—The Power behind the Throne. Genes 2021, 12, 763. https://doi.org/10.3390/genes12050763
Panov KI, Hannan K, Hannan RD, Hein N. The Ribosomal Gene Loci—The Power behind the Throne. Genes. 2021; 12(5):763. https://doi.org/10.3390/genes12050763
Chicago/Turabian StylePanov, Konstantin I., Katherine Hannan, Ross D. Hannan, and Nadine Hein. 2021. "The Ribosomal Gene Loci—The Power behind the Throne" Genes 12, no. 5: 763. https://doi.org/10.3390/genes12050763
APA StylePanov, K. I., Hannan, K., Hannan, R. D., & Hein, N. (2021). The Ribosomal Gene Loci—The Power behind the Throne. Genes, 12(5), 763. https://doi.org/10.3390/genes12050763





