Abstract
Fuzzless mutants are ideal materials to decipher the regulatory network and mechanism underlying fuzz initiation and formation. In this study, we utilized two Gossypium arboreum accessions differing in fuzz characteristics to explore expression pattern differences and discriminate genes involved in fuzz development using RNA sequencing. Gene ontology (GO) analysis was conducted and found that DEGs were mainly enriched in the regulation of transcription, metabolic processes and oxidation–reduction-related processes. Weighted gene co-expression network analysis discerned the MEmagenta module highly associated with a fuzz/fuzzless trait, which included a total of 50 hub genes differentially expressed between two materials. GaFZ, which negatively regulates trichome and fuzz formation, was found involved in MEmagenta cluster1. In addition, twenty-eight hub genes in MEmagenta cluster1 were significantly up-regulated and expressed in fuzzless mutant DPL972. It is noteworthy that Ga04G1219 and Ga04G1240, which, respectively, encode Fasciclin-like arabinogalactan protein 18(FLA18) and transport protein, showed remarkable differences of expression level and implied that they may be involved in protein glycosylation to regulate fuzz formation and development. This module and hub genes identified in this study will provide new insights on fiber and fuzz formation and be useful for the molecular design breeding of cotton genetic improvement.
1. Introduction
Trichomes are highly specialized cells originating and extending from the epidermal surface of leaves, stems, petal bases and seeds in plants [1,2]. They contribute to a variety of unessential but important functions for plant growth, including protections against pathogens, insects or even herbivores, water regulation and increasing the tolerance to extreme high temperature and UV irradiation [3,4]. Trichomes could be characterized and classified into various types by their morphology and nature, e.g., uni-/multi-cellular, non-/branched or non-/glandular [3,5,6]. Numerous molecular genetic studies have provided much insight into the molecular genetic mechanisms and regulatory networks associated with trichome formation. Plenty of functional genes were identified to regulate the mode of epidermal cell development in Arabidopsis, rice, maize, tomato, Artemisia annua, such as MYB-bHLH-WD40(MBW) complex [7,8], OsHL6 [9], OsWOX3 [10], ZmOCL4 [11], ZmSPL10/14/26 [12], SlMYC1 [13], SlWo [14], AaHD1 [15].
Cotton fibers are single-celled and unbranched trichomes that outgrow from the seed epidermal cells and successively undergo a highly polarized elongation similar to leaf trichomes [16,17]. Therefore, cell fate determination and elongation patterns unveiled from Arabidopsis trichomes may provide a useful framework involved in cotton fiber initiation and elongation. It has been reported that the orthologue members in the Arabidopsis MBW complex also evolved crucial roles in cotton fiber formation [18]. Besides, recent studies have uncovered various genes involved in multiple pathways to govern fiber initiation and development. GhPIN3a [19,20], GhARF2/18 [21] and GhHD1 [22,23] were reported as crucial members to regulate fiber initiation in cotton by responding to various phytohormones. Furthermore, GaDEL65 was predominantly associated with fuzz initiation and may interact with GhMYB2/3 and GhTTG3 to regulate cotton fiber elongation [24]. According to previous surveys, GhPEL76 [25], GhSK13 [26], GhAP2L [26], GhHDZ5 [26] and Gh_DNF_YB19 [27], have been identified to extensively regulate fiber elongation responsive to the phytohormone signaling pathway, respectively. AKR2A modulates the biosynthesis pathway of very long-chain fatty acids and participates in fiber elongation regulation [28]. More recently, a set of transcription factors, such as GhFSN1 [29], GhHOX3 [30] and the miR319-targeted GhTCP4 [31], were identified to remarkably playing positive or negative roles in the second cell wall deposition stage.
Fiber initiation stage determines the final fiber yield. Hence it is essential and necessary to highly explore the orchestrating network governing fiber cell differentiation and fiber formation [32]. According to the initiating time and final length, cotton fibers can be divided into lint and fuzz. Though most research studies focused on fiber initiation and elongation, the essential functional genes and the mechanisms underlying fuzz initiation and development remain largely unknown.
Fiber mutant and natural species exhibit diverse or significant fiber characteristics and are great resources to decipher the mechanisms underlying fiber and fuzz formation. With the release of high-quality sequencing data from multiple cotton species, bioinformatics-based analyses have also facilitated functional gene mapping and network regulation exploration. GhMML3 and GhMML4 have been proposed to be responsible for the fuzzless or fiberless phenotypes in tetraploid fiber mutant lines [32,33,34,35,36,37]. Compared with tetraploid cotton, diploid G. arboreum, a relatively simple genome, is potentially highly suitable for digging candidate genes corresponding to mutant traits. It has been reported that transcript splicing mistakes occurred because of a critical point mutation in GaHD1 in a recessive fiber mutant sma-4(ha) and further affects fiber and trichome initiation and retards elongation by cellular H2O2 and Ca2+ signals [38]. Meanwhile, through whole genome re-sequencing and genetic linkage analyses, Liu et al. proposed GaGIR1 and GaMYB25-like as crucial candidate genes controlling fuzz development and established GaHD-1 to be the candidate gene responsible for the lintless trait in seven fuzzless accessions and one lintless accession of G. arboreum [39].
In a previous study, we fine-mapped a dominant fuzzless gene to a 70-kb region and speculated GaGIR1 (GLABRA2-interacting repressor, Ga08G0121) as the likely candidate [40]. Recently, based on a genome-wide association study(GWAS), a 6.2 kb insertion, larINDELFZ, located ~18 kb upstream of GaFZ (Ga08G0121) has been identified to be an enhancer to elevate the expression level of GaFZ and negatively regulate fuzz and trichome formation [41]. Although much progress has been achieved, the mechanism and regulation network remains mysterious and elusive.
In this study, we utilized RNA-Seq for screening genes related to fuzz initiation development in two G. arboreum accessions and validated our data with qPCR. We also performed weighted co-expression gene network analysis to identify the modules and hub genes co-expressing with GaFZ and contributing to the fuzz development. Our results provide valuable gene resources and novel insights into mechanisms and regulatory networks of fuzz-associated genes.
2. Materials and Methods
2.1. Plant Materials
Plants of G. arboreum DPL971 with normal fuzz and lint fibers and its isogenic fuzzless mutant line DPL972 were grown under standard agronomic field conditions at the Institute of Cotton Research, Chinese Academy of Agricultural Sciences (CAAS), Anyang (E 114°48′, N 36°06′), China. All lines were strictly self-pollinated. Flowers were firstly tagged at 0 DPA, and cotton bolls were collected in morning (9:00–11:00) at 1, 3 and 5 DPA, respectively; the ovules were dissected and frozen in liquid nitrogen for RNA extraction.
2.2. RNA Extraction and RNA-Seq
All samples were finely ground before RNA isolation. Total RNA was extracted following the manufacturer’s instructions (TIANGEN BIOTECH Beijing, China). Two biological replicates from each sample were used for this experiment. The quantity and quality of total RNA were monitored by NanoDrop 2000 spectrophotometer and 1% agarose gel electrophoresis. RNA samples of good quality were used for subsequent deep sequencing and quantitative real-time PCR (qRT-PCR) analysis, and cDNA libraries were constructed and subjected to 101-cycle paired-end sequencing on an Illumina HiSeq4000 platform at Berry Genomics (Beijing, China). All raw data were deposited to the NCBI’s Sequence Read Archive (SRA) and available with the accession number PRJNA729876.
2.3. RNA-Seq Data Analysis
Raw sequences obtained from the 12 sample libraries were assessed and filtered by Fastp (version = 0.20.1). The GC content and Q30 were calculated by FastQC. Hisat2 (version = 2.2.1) was used to align the remaining clean data to reference genome of G. arboreum [42]. The number of fragments per kilobase of transcript per million mapped reads (FPKM) of each transcript in the fuzzless mutant DPL972 and wild-type DPL971 was calculated using StringTie (version = 2.1.1) software. Based on read counts obtained from StringTie, differentially expressed genes (DEGs) between the mutant and the wild type were identified by using the DESeq2 with false discovery rate (FDR) < 0.05 and |log2 (ratio)| ≥ 1. The KMeans-clustering method was used for clustering DEGs. KMeans method was performed by python package Sklearn (version = 0.19.2). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were conducted via Cotton Functional Genomics Database (https://cottonfgd.org/, accessed on 9 March 2021) to determine the main biological functions and pathways of the significant DEGs.
2.4. qRT-PCR
To verify the reproducibility and authenticity of RNA-Seq, qRT-PCR was also performed in this study. The extracted total RNA samples with 1 µg concentration were used as template to reverse transcription using TransScript All-in-one First-strand cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China) according to the manufacturer’s instructions. QRT-PCR amplifications were conducted using TransStart TOP Green qPCR SuperMix (TransGen Biotech, China) on an ABI QuantStudio5 Real-time PCR System (Applied Biosystems, Foster City, CA, USA). DEG expressions were normalized using GaHis3 as reference gene. A total of eleven pairs of specific primers were designed from with NCBI Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/, accessed on 9 March 2021) and qPCR Primer Database (https://biodb.swu.edu.cn/qprimerdb/, accessed on 9 March 2021). Primer sequences were provided in Supplementary Table S1. Each qRT-PCR reaction was performed in triplicate and relative expression levels were calculated using the 2−∆∆ct method.
2.5. Construction of Co-Expression Network and Network Analyses
To construct the co-expression network underlying two accessions, transcripts with FPKM count ≥ 5 count were used and all the outliers were removed. The WGCNA (weighted gene co-expression network analysis) R package was implemented to analyze co-expression networks and select hub genes which highly correlated with fuzz characteristic [43,44,45]. The dendrogram was generated using the cutreeDynamicTree algorithm, with a limits of minimum module size of 30 genes. A weighted correlation threshold of ≥0.85 was set to screen modules correlated with fuzz/fuzzless traits. The Sankey diagram was performed by Pyecharts (version = 1.9.0). The interaction networks among genes in selected modules were constructed using Cytoscape and visualized by Pyecharts. The membership (KME) values threshold of ≥0.80 was set to screen the module hub genes. Heatmap for visualization of expression was generated by python package Seaborn (version = 0.9.0).
3. Results
3.1. Data Processing and Analysis
To capture a global view and the temporal gene expression changes during fiber and fuzz initiation development, a transcriptome profiling experiment was performed from 12 cotton ovule libraries including wild type DPL971 and the fuzzless mutant DPL972 during the fuzz and fiber initiation stage (+1DPA, +3DPA and +5 DPA). Two biological replicates were adopted for each material in each time point. After removing adaptor and low-quality reads, a total of 90.27 Gb of cleaning data was generated, including 30.1 million reads, averaging 7.52 G and 25,077,291 reads per sample (Supplementary Table S2). For all samples, the GC content varied from 44% to 45%, and Q30 altered from 95.24% to 95.51%. All clean reads were aligned to the newly published reference G. arboreum genome, and 89.00% to 93.02% of the sequenced reads from each library could be perfectly mapped. These results indicated that our transcriptome sequencing data were of high quality and good enough to further identify DEGs and construct the regulatory network.
3.2. Differential Gene Expression in DPL971 and DPL972 during Fuzz Initiation
A total of 1076 DEGs were detected using the criteria of false discovery rate (FDR) ≤0.05 and |log2 Ratio| ≥ 1. The number of DEGs between the WT and mutant during fuzz initiation stages (+1DPA, +3 DPA and +5 DPA) was 428, 330 and 614, respectively (Figure 1). Besides, eighty-two genes shared overlapping and differentially co-expression across the three different time points, suggesting that the transcriptome variation between the two accessions maintains a stable state through fuzz determination and the formation stage.
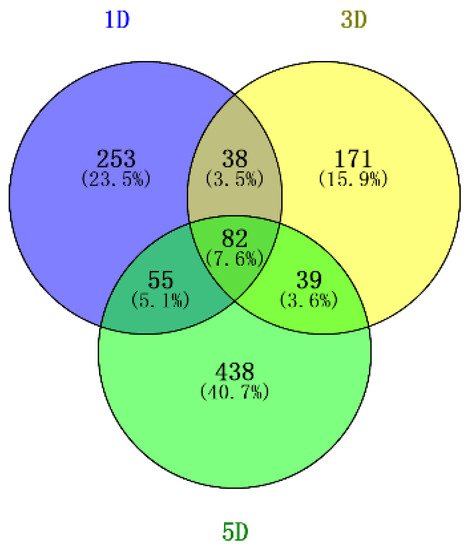
Figure 1.
Venn diagram of DEGs identified from two diploid cotton accessions; 1D, 3D, 5D represents 1 Days post-anthesis (DPA), 3 DPA, 5 DPA.
To verify the reproducibility and authenticity of the RNA-seq data, qRT-PCR was performed on selected 11 DEGs with gene-specific primers (Supplementary Table S2). As shown in Figure 2, all 11 genes showed a consistent expression pattern between the RNA-seq data and the qRT-PCR results. Genes significantly up-regulated in RNA-Seq data also exhibited an up-regulation in qPCR, and vice versa. These results also supported the reproducibility and authenticity of transcriptomic data, which were utilized in downstream co-expression network and functional enrichment analysis.
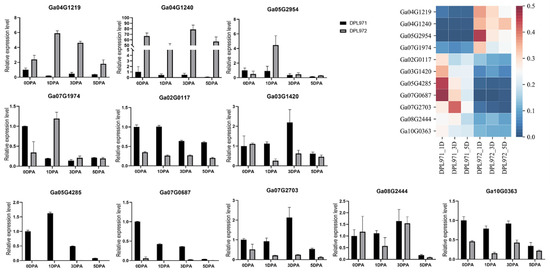
Figure 2.
Validation of DEGs identified from transcriptome analysis with qRT-PCR. The heatmap on the top right of figure represents the relative transcription abundances (based on FPKM).
3.3. Functional Enrichment of DEGs
To estimate the main biological functions and regulations of identified DEGs, Gene ontology enrichment analysis and KEGG were performed on gene IDs that involved in each developmental points. As shown in Figure 3, DEGs generated from 1D were enriched in the following Molecular Function terms “transcription factor activity, sequence-specific DNA-binding” and “DNA-binding”. They were also enriched in terms of “photosystem II stabilization” and “regulation of transcription, DNA-templated” from the biological process category. The enrichment of cellular component annotations included the terms of photosystem II reaction and extracellular region. While DEGs identified from 3D were mainly enriched in ATP synthesis and metabolic process and proton transporting associated terms, and DEGs identified from 5D were clustered into terms of “oxidoreductase activity” and “oxidation-reduction process”. Further, the pathway enrichment analysis revealed that cell cycle, oxidative phosphorylation, biosynthesis of secondary metabolites, and glycolysis/gluconeogenesis were enriched among the DEGs and might be involved in fuzz initiation in cotton epidermal cell differentiation and development (Supplementary Table S3).
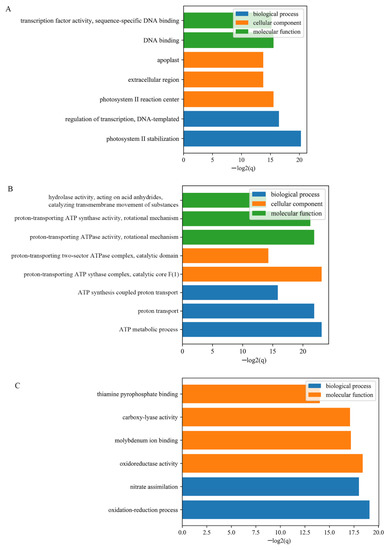
Figure 3.
GO enrichment analysis of DEGs at three fiber developmental stages; (A), DEGs detected from 1DPA; (B), DEGs detected from 1DPA; (C) DEGs detected from 1DPA.
3.4. Co-Expression Network Construction and Module Mining
To uncover important genes and regulation pathways involved in fuzz and fiber initiation stage, we performed the R WGCNA package on all 12 samples. A soft-threshold power of 25 was introduced into the network topology to reveal the scale independence and mean connectivity of the network (Figure 4A,B). Based on the fragments per kilobase per million mapped (FPKM) expression matrix and phenotypic traits including fuzz or fuzzless characteristics and three developmental points, a total of 20,601 genes in 12 samples were clustered and divided into 10 modules which decorated with diacritical colors (Figure 4C). The outliers and genes whose FPKM values <5 in all samples were excluded in this analysis.
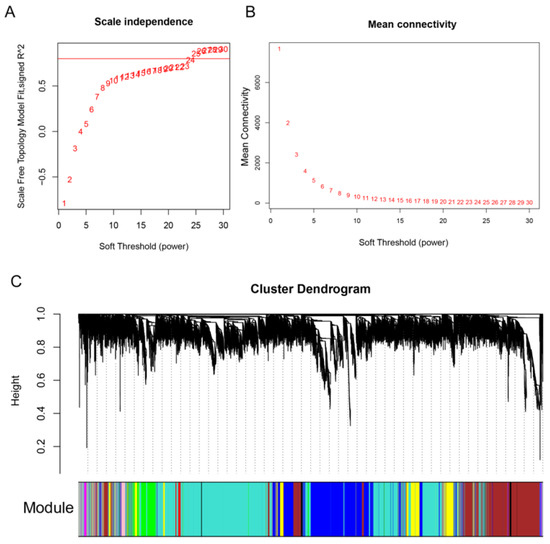
Figure 4.
Module identification by weighted gene co-expression network analysis (WGCNA). (A,B) represent the soft threshold with scale independence and mean connectivity. (C), Hierarchical dendrogram reveals co-expression modules identified by WGCNA. Each leaf represents one gene. Ten modules were identified based on calculation of eigengenes; each module was decorated with a different color.
Heatmap was graphed to unveil the correlation between modules and traits, p-values were presented in Supplementary Table S4. After calculating the correlation coefficient, one module was strongly associated with fuzz and fuzzless characteristics, indicating it might be highly contributed to fuzz initiation and formation (Figure 5). The MEmagenta module exhibited extremely exclusive correlations with fuzz/fuzzless traits (r = 0.996, p-value = 6.27 × 10−12). This module was analyzed further. In addition, we also identified red, brown and green modules significantly and substantially correlated with fiber development of 1, 3 and 5 DPA, respectively, suggesting genes involved in these modules most probably participated in fiber initiation and elongation during fiber developmental stages. Besides, the Sankey diagram was generated to detect the distribution of DEGs in each module, revealing that DEGs from MEmagenta distributed evenly at 1, 3, 5 DPA (Figure 6).
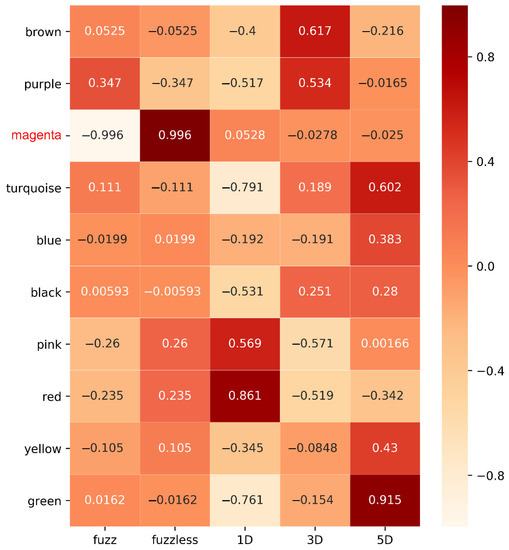
Figure 5.
The WGCNA showed the MEmagenta module is significantly associated with fuzz formation. Each row means a module, and the correlation coefficient are shown in each square and the p-value was list in Table S4. The names in red in left represent the module highly associated with fuzz development.
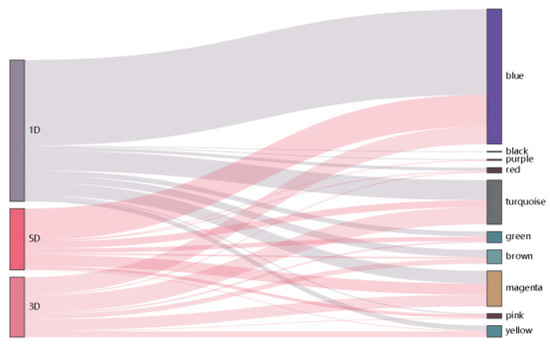
Figure 6.
The Sankey diagram represents the distributions of DEGs in each module.
3.5. Functional Enrichment and Expression Analysis of Fuzz-Associated Hub Genes
To obtain more key genes involving the process of fuzz initiation, we did further analysis on the MEmagenta module which comprises 98 genes. To better understand gene functions and regulatory networks, GO and KEGG enrichment analyses were also used to analyze the fuzz-correlated module. The top terms were, respectively, presented in Supplementary Table S5 and Supplementary Figure S1. The GO results demonstrate genes in the MEmagenta module were enriched in transferase activity and transport protein, and the KEGG revealed genes in this module were best matched with the NADH dehydrogenase (ubiquinone) 1 α sub-complex item.
According to different expression profiles, but similar functions, genes in the MEmagenta module could be divided into two clusters using the K-means clustering method (Figure 7). Based on the kME value, the intramodular connectivity ranks and genes with kME value >0.8 were selected as hub genes that would determine the network centrality and potential crucial roles in fuzz/fuzzless characteristics. These hub genes were listed in Supplementary Table S6 and complex interaction networks in the co-expression module were respectively shown in Supplementary Figure S2 to highlight the potential networks underlying fuzz development.
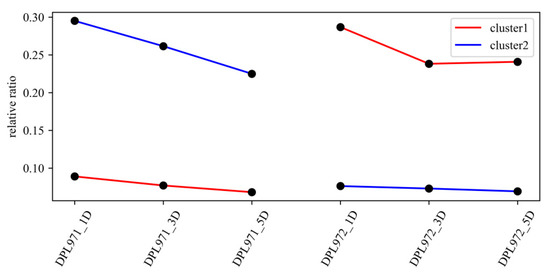
Figure 7.
K-means clustering of DEGs in the MEmagenta module.
To uncover the expression patterns of hub genes, we compared DEGs and genes in the selected module. Interestingly, a total of 50 out of 56 identified hub genes exhibited highly differential expression levels between DPL971 and DPL972 (Figure 8), indicating these hub genes play important roles in regulating fuzz initiation and development. In addition, previous studies reported GaFZ (Ga08G0121) differentially expressed between DPL971 and DPL972 due to a large insertion and thus negatively regulates fuzz formation [40,41]. The gene was also identified and involved in MEmagenta module cluster1, which we selected as main cluster negatively correlating with fuzz trait (positively correlating with fuzzless trait), suggesting the reliability of the interested module and network identified from WGCNA. What is more, similar to GaFZ, Ga04G1219 encoding Fasciclin-like arabinogalactan protein 18(FLA18) and Ga04G1240 encoding transport protein exhibited dramatic up-regulated expression in fuzzless DPL972 compared to DPL971 (Figure 2). It suggested that they and their interaction network may be involved in fuzz formation and development via controlling protein-glycosylation and responding to phytohormones.
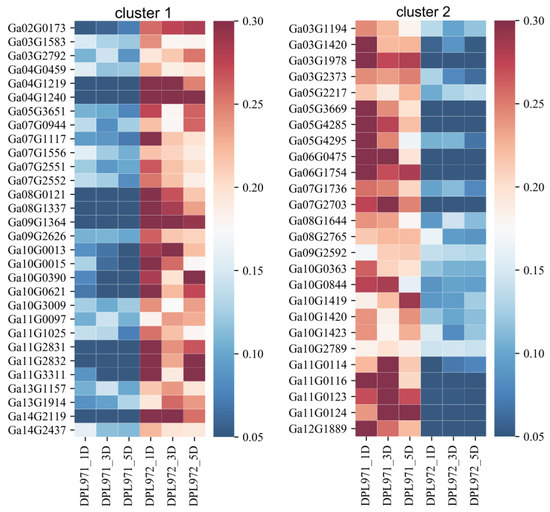
Figure 8.
The expression heatmaps of hub genes in the MEmagenta module. The right side of figure represents the relative transcription abundances (based on FPKM). The gene IDs were listed on the left and sample names were displayed on the bottom.
4. Discussion
In the present study, we investigated gene expressions and regulatory networks in two G. arboreum accessions with a significant difference in fuzz characteristic. By performing transcriptomic analysis of fiber-attached ovules collected from 1, 3 and 5 DPA, DEGs were identified, respectively. GO enrichment analysis presented that DEGs in 1 DPA were mainly enriched in transcription factor activity, and DEGs in 3 and 5 DPA were clustered into items of ATP synthesis and proton transporting, oxidoreductase activity, respectively. Further, we established a weighted gene co-expression network analysis underlying fuzz initiation and formation. WGCNA revealed that only one co-expression module, MEmagenta, had a dominantly high correlation with fuzz trait, and the genes in these modules were significantly enriched in items of transferase and transport protein based on the GO enrichment analysis.
4.1. Hub Genes in MEgrey60 Cluster Might Be Involved in Regulatory Network Responsible to Fuzz Initiation
Fiber and fuzz initiation undergo rather complex and obscure processes. Until now, multiple loci and factors have been identified to control or regulate fiber and fuzz initiation. In previous studies, larINDELFZ was screened out to be responsible for the fuzzless phenotype in various diploid accessions through genome wide association study (GWAS) and enhanced the expression level of GaFZ [41]. Overexpression of GaFZ in tetraploid cotton presented a reduction of trichomes and fuzz, suggesting GaFZ might be contributed to fuzz initiation and formation. In our study, GaFZ significantly differentially expressed in two accessions and was involved in MEmagenta module cluster1with a correlation of 0.816. By conducting the prediction and comparison of protein structure, we found this protein shared a high similarity with DNA-directed RNA polymerase II subunit RPB12, which functions as part of activated transcription complex (Supplementary Figure S2). It implies GaFZ might act in this way to regulate other genes to affect fuzz initiation. Besides, a comparison of the results of WGCNA and DEGs analysis indicated that 93 out of 98 genes in the MEmagenta module exhibited differential expression levels. In addition, 50 out of 56 hub genes, which were identified as critical genes to be significantly correlated with fuzz initiation and formation, presented notably differential expression patterns. In previous studies, glycosyltransferase31 could influence fiber initiation and elongation via controlling the glycosylation of FLAs and regulating pectin biosynthesis [46,47,48,49,50,51,52,53]. Interestingly, genes encoding FLA, UGT, RLK, transport protein were also identified in hub genes in MEmagenta cluster1. Together with GaFZ, these differentially expressed hub genes may be the key regulators underlying fuzz initiation and development in G. arboreum by protein glycosylation and phytohormone response. These results also indicated that the combination of transcriptomic analysis and WGCNA is highly efficient and superior in detecting and discerning functionally associated genes as well as gene regulatory networks.
4.2. Multiple Protein Kinase Genes may Regulate Fuzz and Plant Development in G. arboreum
Cotton is a great model for detecting the mechanism of cell differentiation and elongation. Understanding the molecular pathway regulating fiber and fuzz initiation can provide essential and valuable information for improving the fiber production and prompting the process of the molecular design breeding. Various protein kinases are important and essential in response to biotic and abiotic stresses as well as regulating plant and fiber development [54,55,56,57,58,59]. Mitogen-activated protein kinase (MAPK) cascades have been reported to alter pathogen resistance through gibberellins (GA) signaling pathway and participate in cotton fiber elongation by phosphorylating its downstream proteins [60,61]. Ectopic expression of receptor-like kinases (RLKs) improved cotton tolerance and resistance to fungal pathogen infections [62,63,64,65,66]. Calcineurin B-like protein and CBL-interacting protein kinases (CIPKs) were proved to be associated with cotton oil and sugar content and stress responses [59,67,68,69,70]. Glycosyltransferases (UGT) were identified to participate and function in biosynthesis process of hemicellulose glucuronoxylan xylan during fiber and plant development whereas xyloglucan endo-transglycosylases /hydrolases (XTH) would limit the elongation of cotton fiber [56,71,72]. Glutathione S-transferase genes had an important role in resistance of Verticillium wilt, and sulfotransferases (SOTs) genes might be involved in cotton fiber development [73,74]. In the present study, CBL-interacting protein kinase 9 (CIPK9), UDP-glycosyltransferase 83A1 (UGT83A1), MDIS1-interacting receptor-like kinase 2 (MIK2), probable inactive histone-lysine N-methyltransferase (SUVR1) and Inositol-tetrakisphosphate 1-kinase 2 (ITPK2) were identified as hub genes in the MEmagenta module, indicating that these protein kinase-related genes might be crucial and potential targets for the regulation of fiber/fuzz initiation and development in G. arboreum, and they are well worth further and deeper understanding and research.
5. Conclusions
In this study, we collected the fiber-attached ovules samples at three fiber developmental stages (1, 3 and 5 DPA) for RNA-Seq from a pair of G. arboreum accessions with different fuzz traits, and through transcriptome comparisons, 428 DEGs at 1DPA, 330 DEGs at 3 DPA and 614 DEGs at 5 DPA were identified, respectively. WGCNA identified a MEmagenta module highly associated with fuzz/fuzzless development. Within this module, a total of 93 genes exhibited significantly differential expression patterns between two accessions. GaFZ, which has been reported to negatively regulate trichome and fuzz formation, was found involved in MEmagenta cluster1. Besides, the expression levels of twenty-eight hub genes in MEmagenta cluster1 were notably up-regulated and twenty-two hub genes in cluster2 were dominantly down-regulated in fuzzless mutant DPL972. Furthermore, the coding genes of FLA18, UGT and transport protein showed remarkable differences of expression profiles and implied that they may be involved in protein glycosylation to regulate fuzz development. This study thus unveils the regulatory mechanisms underlying fuzz initiation and the development stage in G. arboreum, and may facilitate the molecular design breeding of cotton with improved fiber characteristics.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes12050753/s1, Figure S1: KEGG enrichment analysis of genes in the MEmagenta module; Figure S2: Diagram of interaction networks of genes in the MEmagenta module; Figure S3: Comparison of protein structures between GaFZ and RPB12, Table S1: Primers used in qRT-PCR, Table S2: Statistics and alignments of RNA-seq reads mapped to reference genome, Table S3: KEGG enrichment analysis of DEGs obtained from two accessions, Table S4: p-values of relationships between module and sample, Table S5: GO enrichment analysis of hub genes in the MEmagenta module, Table S6: Hub genes in the MEmagenta module.
Author Contributions
Conceptualization, X.F. and G.S.; data curation, X.F.; formal analysis, X.F., S.L., H.C., D.Z., Y.Z. and L.L.; funding acquisition, G.S.; investigation, G.S.; methodology, H.C.; project administration, G.S.; resources, H.C., D.Z. and Q.W.; software, S.L.; supervision, G.S.; visualization, S.L. and H.C.; writing—original draft, X.F., S.L.; writing—review and editing, S.L. and H.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by grants from the National Natural Science Foundation of China (No. 31621005), the National Natural Science Foundation of China (No. 31901581) and the National Key R & D Plan of China (No. 2018YFD0100402).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the article and supplementary material.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dai, X.; Zhou, L.; Zhang, W.; Cai, L.; Guo, H.; Tian, H.; Schiefelbein, J.; Wang, S. A single amino acid substitution in the R3 domain of GLABRA1 leads to inhibition of trichome formation in Arabidopsis without affecting its interaction with GLABRA3. Plant Cell Environ. 2015, 39, 897–907. [Google Scholar] [CrossRef]
- Tan, J.; Walford, S.A.; Dennis, E.S.; Llewellyn, D.J. Trichomes at the Base of the Petal Are Regulated by the Same Transcription Factors as Cotton Seed Fibers. Plant Cell Physiol. 2020, 61, 1590–1599. [Google Scholar] [CrossRef]
- Matias-Hernandez, L.; Aguilar-Jaramillo, A.E.; Cigliano, R.A.; Sanseverino, W.; Pelaz, S. Flowering and trichome development share hormonal and transcription factor regulation. J. Exp. Bot. 2016, 67, 1209–1219. [Google Scholar] [CrossRef]
- Yang, C.; Ye, Z. Trichomes as models for studying plant cell differentiation. Cell Mol. Life Sci. 2013, 70, 1937–1948. [Google Scholar] [CrossRef]
- Hulskamp, M. Plant trichomes: A model for cell differentiation. Nat. Rev. Mol. Cell Biol. 2004, 5, 471–480. [Google Scholar] [CrossRef]
- Balkunde, R.; Pesch, M.; Hülskamp, M. Chapter Ten-Trichome Patterning in Arabidopsis thaliana: From Genetic to Molecular Models. In Current Topics in Developmental Biology; Marja, C.P.T., Ed.; Academic Press: Cambridge, MA, USA, 2010; Volume 91, pp. 299–321. [Google Scholar]
- Pattanaik, S.; Patra, B.; Singh, S.K.; Yuan, L. An overview of the gene regulatory network controlling trichome development in the model plant, Arabidopsis. Front. Plant Sci. 2014, 5, 259. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Sano, R.; Wada, T.; Takabayashi, J.; Okada, K. Jasmonic acid control of GLABRA3 links inducible defense and trichome patterning in Arabidopsis. Development 2009, 136, 1039–1048. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Gao, D.; Xiong, Y.; Tang, X.; Xiao, X.; Wang, C.; Yu, S. Hairy Leaf 6, an AP2/ERF transcription factor, interacts with OsWOX3B and regulates trichome formation in rice. Mol. Plant 2017, 10, 1417–1433. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, K.; Wang, Y.; Peng, Y.; Hu, F.; Wen, L.; Han, B.; Qian, Q.; Teng, S. A WUSCHEL-like homeobox gene, OsWOX3B responses to NUDA/GL-1 locus in rice. Rice J. 2012, 5, 30. [Google Scholar] [CrossRef]
- Vernoud, V.; Laigle, G.; Rozier, F.; Meeley, R.B.; Perez, P.; Rogowsky, P.M. The HD-ZIP IV transcription factor OCL4 is necessary for trichome patterning and anther development in maize. Plant J. Cell Mol. Biol. 2009, 59, 883–894. [Google Scholar] [CrossRef]
- Kong, D.; Pan, X.; Jing, Y.; Zhao, Y.; Duan, Y.; Yang, J.; Wang, B.; Liu, Y.; Shen, R.; Cao, Y.; et al. ZmSPL10/14/26 are required for epidermal hair cell fate specification on maize leaf. New Phytol. 2021, 230, 1533–1549. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; van Herwijnen, Z.O.; Drager, D.B.; Sui, C.; Haring, M.A.; Schuurink, R.C. SlMYC1 Regulates Type VI Glandular Trichome Formation and Terpene Biosynthesis in Tomato Glandular Cells. Plant Cell 2018, 30, 2988–3005. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, H.; Zhang, J.; Luo, Z.; Gong, P.; Zhang, C.; Li, J.; Wang, T.; Zhang, Y.; Lu, Y.; et al. A regulatory gene induces trichome formation and embryo lethality in tomato. Proc. Natl. Acad. Sci. USA 2011, 108, 11836–11841. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Chen, M.; Shen, Q.; Li, L.; Fu, X.; Pan, Q.; Tang, Y.; Shi, P.; Lv, Z.; Jiang, W.; et al. HOMEODOMAIN PROTEIN 1 is required for jasmonate-mediated glandular trichome initiation in Artemisia annua. New Phytol. 2017, 213, 1145–1155. [Google Scholar] [CrossRef]
- Wang, L.K.D.; Ruan, Y. Looking into ‘hair tonics’ for cotton fiber initiation. New Phytol. 2020, 229, 1844–1851. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wu, S.; Nowak, J.; Wang, G.; Han, L.; Feng, Z.; Mendrinna, A.; Ma, Y.; Wang, H.; Zhang, X.; et al. Live-cell imaging of the cytoskeleton in elongating cotton fibres. Nat. Plants 2019, 5, 498–504. [Google Scholar] [CrossRef]
- Wang, L.; Wang, G.; Long, L.; Altunok, S.; Feng, Z.; Wang, D.; Khawar, K.M.; Mujtaba, M. Understanding the role of phytohormones in cotton fiber development through omic approaches; recent advances and future directions. Int. J. Biol. Macromol. 2020, 163, 1301–1313. [Google Scholar] [CrossRef]
- Mei, G.; Zhang, Z. Optimization of polar distribution of GhPIN3a in the ovule epidermis improves cotton fiber development. J. Exp. Bot. 2019, 70, 3021–3023. [Google Scholar] [CrossRef]
- Zeng, J.; Zhang, M.; Hou, L.; Bai, W.; Yan, X.; Hou, N.; Wang, H.; Huang, J.; Zhao, J.; Pei, Y. Cytokinin inhibits cotton fiber initiation by disrupting PIN3a-mediated asymmetric accumulation of auxin in the ovule epidermis. J. Exp. Bot. 2019, 70, 3139–3151. [Google Scholar] [CrossRef]
- Xiao, G.; He, P.; Zhao, P.; Liu, H.; Zhang, L.; Pang, C.; Yu, J. Genome-wide identification of the GhARF gene family reveals that GhARF2 and GhARF18 are involved in cotton fibre cell initiation. J. Exp. Bot. 2018, 69, 4323–4337. [Google Scholar] [CrossRef]
- Zhang, M.; Zeng, J.Y.; Long, H.; Xiao, Y.H.; Yan, X.Y.; Pei, Y. Auxin Regulates Cotton Fiber Initiation via GhPIN-Mediated Auxin Transport. Plant Cell Physiol. 2017, 58, 385–397. [Google Scholar] [CrossRef]
- Walford, S.A.; Wu, Y.; Llewellyn, D.J.; Dennis, E.S. Epidermal cell differentiation in cotton mediated by the homeodomain leucine zipper gene, GhHD-1. Plant J. Cell Mol. Biol. 2012, 71, 464–478. [Google Scholar] [CrossRef]
- Shangguan, X.X.; Yang, C.Q.; Zhang, X.F.; Wang, L.J. Functional characterization of a basic helix-loop-helix (bHLH) transcription factor GhDEL65 from cotton (Gossypium hirsutum). Physiol. Plant 2016, 158, 200–212. [Google Scholar] [CrossRef]
- Sun, H.; Hao, P.; Gu, L.; Cheng, S.; Wang, H.; Wu, A.; Ma, L.; Wei, H.; Yu, S. Pectate lyase-like Gene GhPEL76 regulates organ elongation in Arabidopsis and fiber elongation in cotton. Plant Sci. Int. J. Exp. Plant Biol. 2020, 293, 110395. [Google Scholar] [CrossRef]
- Wang, L.; Cheng, H.; Xiong, F.; Ma, S.; Zheng, L.; Song, Y.; Deng, K.; Wu, H.; Li, F.; Yang, Z. Comparative phosphoproteomic analysis of BR-defective mutant reveals a key role of GhSK13 in regulating cotton fiber development. Sci. China Life Sci. 2020, 63, 1905–1917. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, Z.; Xiao, Y.; Wang, P.; Wang, Y.; Ge, X.; Zhang, C.; Zhang, X.; Li, F. Genome-Wide Analysis of the NF-YB Gene Family in Gossypium hirsutum L. and Characterization of the Role of GhDNF-YB22 in Embryogenesis. Int. J. Mol. Sci. 2018, 19, 483. [Google Scholar] [CrossRef]
- Hu, W.; Chen, L.; Qiu, X.; Wei, J.; Lu, H.; Sun, G.; Ma, X.; Yang, Z.; Zhu, C.; Hou, Y.; et al. AKR2A participates in the regulation of cotton fibre development by modulating biosynthesis of very-long-chain fatty acids. Plant Biotechnol. J. 2020, 18, 526–539. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, G.-Q.; Zou, D.; Yan, J.-Q.; Li, Y.; Hu, S.; Li, X.-B. The cotton (Gossypium hirsutum) NAC transcription factor (FSN1) as a positive regulator participates in controlling secondary cell wall biosynthesis and modification of fibers. New Phytol. 2017, 217, 625–640. [Google Scholar] [CrossRef]
- Shan, C.-M.; Shangguan, X.-X.; Zhao, B.; Zhang, X.-F.; Chao, L.-m.; Yang, C.-Q.; Wang, L.-J.; Zhu, H.-Y.; Zeng, Y.-D.; Guo, W.-Z.; et al. Control of cotton fibre elongation by a homeodomain transcription factor GhHOX3. Nat. Commun. 2014, 5, 5519. [Google Scholar] [CrossRef]
- Cao, J.F.; Zhao, B.; Huang, C.C.; Chen, Z.W.; Zhao, T.; Liu, H.R.; Hu, G.J.; Shangguan, X.X.; Shan, C.M.; Wang, L.J.; et al. The miR319-Targeted GhTCP4 Promotes the Transition from Cell Elongation to Wall Thickening in Cotton Fiber. Mol. Plant 2020, 13, 1063–1077. [Google Scholar] [CrossRef]
- Tian, Y.; Zhang, T. MIXTAs and phytohormones orchestrate cotton fiber development. Curr. Opin. Plant Biol. 2021, 59, 101975. [Google Scholar] [CrossRef]
- Wan, Q.; Guan, X.; Yang, N.; Wu, H.; Pan, M.; Liu, B.; Fang, L.; Yang, S.; Hu, Y.; Ye, W.; et al. Small interfering RNAs from bidirectional transcripts of GhMML3_A12 regulate cotton fiber development. New Phytol. 2016, 210, 1298–1310. [Google Scholar] [CrossRef]
- Chen, W.; Li, Y.; Zhu, S.; Fang, S.; Zhao, L.; Guo, Y.; Wang, J.; Yuan, L.; Lu, Y.; Liu, F.; et al. A Retrotransposon Insertion in GhMML3_D12 Is Likely Responsible for the Lintless Locus li3 of Tetraploid Cotton. Front. Plant Sci. 2020, 11, 593679. [Google Scholar] [CrossRef]
- Wu, H.; Tian, Y.; Wan, Q.; Fang, L.; Guan, X.; Chen, J.; Hu, Y.; Ye, W.; Zhang, H.; Guo, W.; et al. Genetics and evolution of MIXTA genes regulating cotton lint fiber development. New Phytol. 2017, 217, 883–895. [Google Scholar] [CrossRef]
- Zhu, Q.-H.; Yuan, Y.; Stiller, W.; Jia, Y.; Wang, P.; Pan, Z.; Du, X.; Llewellyn, D.; Wilson, I. Genetic dissection of the fuzzless seed trait in Gossypium barbadense. J. Exp. Bot. 2018, 69, 997–1009. [Google Scholar] [CrossRef] [PubMed]
- Naoumkina, M.; Thyssen, G.N.; Fang, D.D.; Li, P.; Florane, C.B. Elucidation of sequence polymorphism in fuzzless-seed cotton lines. Mol. Genet. Genom. MGG 2021, 296, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Cao, Y.; He, S.; Sun, J.; Dai, H.; Zhang, H.; Sun, C.; Jiang, Y.; Paterson, A.H.; Rong, J. GaHD1, a candidate gene for the Gossypium arboreum SMA-4 mutant, promotes trichome and fiber initiation by cellular H2O2 and Ca(2+) signals. Plant Mol. Biol. 2020, 103, 409–423. [Google Scholar] [CrossRef]
- Liu, X.; Moncuquet, P.; Zhu, Q.H.; Stiller, W.; Zhang, Z.; Wilson, I. Genetic Identification and Transcriptome Analysis of Lintless and Fuzzless Traits in Gossypium arboreum L. Int. J. Mol. Sci. 2020, 21, 1675. [Google Scholar] [CrossRef]
- Feng, X.; Cheng, H.; Zuo, D.; Zhang, Y.; Wang, Q.; Liu, K.; Ashraf, J.; Yang, Q.; Li, S.; Chen, X.; et al. Fine mapping and identification of the fuzzless gene GaFzl in DPL972 (Gossypium arboreum). TAG Theor. Appl. Genet. Theor. Angew. Genet. 2019, 132, 2169–2179. [Google Scholar] [CrossRef]
- Wang, X.; Miao, Y.; Cai, Y.; Sun, G.; Jia, Y.; Song, S.; Pan, Z.; Zhang, Y.; Wang, L.; Fu, G.; et al. Large-fragment insertion activates gene GaFZ (Ga08G0121) and is associated with the fuzz and trichome reduction in cotton (Gossypium arboreum). Plant Biotechnol. J. 2020, 1–15. [Google Scholar] [CrossRef]
- Du, X.; Huang, G.; He, S.; Yang, Z.; Sun, G.; Ma, X.; Li, N.; Zhang, X.; Sun, J.; Liu, M.; et al. Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nat. Genet. 2018, 50, 796–802. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Kuang, J.F.; Wu, C.J.; Guo, Y.F.; Walther, D.; Shan, W.; Chen, J.Y.; Chen, L.; Lu, W.J. Deciphering transcriptional regulators of banana fruit ripening by regulatory network analysis. Plant Biotechnol. J. 2021, 19, 477–489. [Google Scholar] [CrossRef]
- Zaidi, S.S.; Naqvi, R.Z.; Asif, M.; Strickler, S.; Shakir, S.; Shafiq, M.; Khan, A.M.; Amin, I.; Mishra, B.; Mukhtar, M.S.; et al. Molecular insight into cotton leaf curl geminivirus disease resistance in cultivated cotton (Gossypium hirsutum). Plant Biotechnol. J. 2020, 18, 691–706. [Google Scholar] [CrossRef]
- Huang, G.Q.; Xu, W.L.; Gong, S.Y.; Li, B.; Wang, X.L.; Xu, D.; Li, X.B. Characterization of 19 novel cotton FLA genes and their expression profiling in fiber development and in response to phytohormones and salt stress. Physiol. Plant 2008, 134, 348–359. [Google Scholar] [CrossRef]
- Bowling, A.J.; Vaughn, K.C.; Turley, R.B. Polysaccharide and glycoprotein distribution in the epidermis of cotton ovules during early fiber initiation and growth. Protoplasma 2011, 248, 579–590. [Google Scholar] [CrossRef]
- Huang, G.Q.; Gong, S.Y.; Xu, W.L.; Li, W.; Li, P.; Zhang, C.J.; Li, D.D.; Zheng, Y.; Li, F.G.; Li, X.B. A fasciclin-like arabinogalactan protein, GhFLA1, is involved in fiber initiation and elongation of cotton. Plant Physiol. 2013, 161, 1278–1290. [Google Scholar] [CrossRef]
- Kumar, S.; Kumar, K.; Pandey, P.; Rajamani, V.; Padmalatha, K.V.; Dhandapani, G.; Kanakachari, M.; Leelavathi, S.; Kumar, P.A.; Reddy, V.S. Glycoproteome of elongating cotton fiber cells. Mol. Cell Proteom. 2013, 12, 3677–3689. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Shi, R.; Wang, X.; Pan, Y.; Li, Z.; Yang, X.; Zhang, G.; Ma, Z. Characterization and expression analysis of a fiber differentially expressed Fasciclin-like arabinogalactan protein gene in Sea Island cotton fibers. PLoS ONE 2013, 8, e70185. [Google Scholar] [CrossRef]
- Qin, L.X.; Rao, Y.; Li, L.; Huang, J.F.; Xu, W.L.; Li, X.B. Cotton GalT1 encoding a putative glycosyltransferase is involved in regulation of cell wall pectin biosynthesis during plant development. PLoS ONE 2013, 8, e59115. [Google Scholar] [CrossRef] [PubMed]
- Hande, A.S.; Katageri, I.S.; Jadhav, M.P.; Adiger, S.; Gamanagatti, S.; Padmalatha, K.V.; Dhandapani, G.; Kanakachari, M.; Kumar, P.A.; Reddy, V.S. Transcript profiling of genes expressed during fibre development in diploid cotton (Gossypium arboreum L.). BMC Genom. 2017, 18, 675. [Google Scholar] [CrossRef]
- Qin, L.X.; Chen, Y.; Zeng, W.; Li, Y.; Gao, L.; Li, D.D.; Bacic, A.; Xu, W.L.; Li, X.B. The cotton β-galactosyltransferase 1 (GalT1) that galactosylates arabinogalactan proteins participates in controlling fiber development. Plant J. Cell Mol. Biol. 2017, 89, 957–971. [Google Scholar] [CrossRef]
- Yang, C.-Q.; Wu, X.-M.; Ruan, J.-X.; Hu, W.-L.; Mao, Y.-B.; Chen, X.-Y.; Wang, L.-J. Isolation and characterization of terpene synthases in cotton (Gossypium hirsutum). Phytochemistry 2013, 96, 46–56. [Google Scholar] [CrossRef]
- Zou, C.; Lu, C.; Shang, H.; Jing, X.; Cheng, H.; Zhang, Y.; Song, G. Genome-wide analysis of the Sus gene family in cotton. J. Integr. Plant Biol. 2013, 55, 643–653. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Huang, J.; Qin, L.; Huang, Y.; Zeng, W.; Rao, Y.; Li, J.; Li, X.; Xu, W. Two cotton fiber-associated glycosyltransferases, GhGT43A1 and GhGT43C1, function in hemicellulose glucuronoxylan biosynthesis during plant development. Physiol. Plant 2014, 152, 367–379. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, J.-Y. Cotton cytosolic pyruvate kinase GhPK6 participates in fast fiber elongation regulation in a ROS-mediated manner. Planta 2016, 244, 915–926. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Lu, X.K.; Yin, Z.J.; Mu, M.; Zhao, X.J.; Wang, D.L.; Wang, S.; Fan, W.L.; Guo, L.X.; Ye, W.W.; et al. Genome-wide identification and expression analysis of CIPK genes in diploid cottons. Genet. Mol. Res. 2016, 15, 10–4238. [Google Scholar] [CrossRef]
- Cui, Y.; Su, Y.; Wang, J.; Jia, B.; Wu, M.; Pei, W.; Zhang, J.; Yu, J. Genome-Wide Characterization and Analysis of CIPK Gene Family in Two Cultivated Allopolyploid Cotton Species: Sequence Variation, Association with Seed Oil Content, and the Role of GhCIPK6. Int. J. Mol. Sci. 2020, 21, 863. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; He, X.; Wang, X.; Zhang, S.; Guo, X.; Denby, K. ghr-miR5272a-mediated regulation of GhMKK6 gene transcription contributes to the immune response in cotton. J. Exp. Bot. 2017, 68, 5895–5906. [Google Scholar] [CrossRef]
- Chen, L.; Sun, H.; Wang, F.; Yue, D.; Shen, X.; Sun, W.; Zhang, X.; Yang, X. Genome-wide identification of MAPK cascade genes reveals the GhMAP3K14-GhMKK11-GhMPK31 pathway is involved in the drought response in cotton. Plant Mol. Biol. 2020, 103, 211–223. [Google Scholar] [CrossRef]
- Jun, Z.; Zhang, Z.; Gao, Y.; Zhou, L.; Fang, L.; Chen, X.; Ning, Z.; Chen, T.; Guo, W.; Zhang, T. Overexpression of GbRLK, a putative receptor-like kinase gene, improved cotton tolerance to Verticillium wilt. Sci. Rep. 2015, 5, 15048. [Google Scholar] [CrossRef]
- Yuan, N.; Rai, K.M.; Balasubramanian, V.K.; Upadhyay, S.K.; Luo, H.; Mendu, V. Genome-wide identification and characterization of LRR-RLKs reveal functional conservation of the SIF subfamily in cotton (Gossypium hirsutum). BMC Plant Biol. 2018, 18, 185. [Google Scholar] [CrossRef] [PubMed]
- Babilonia, K.; Wang, P.; Liu, Z.; Jamieson, P.; Mormile, B.; Rodrigues, O.; Zhang, L.; Lin, W.; Danmaigona Clement, C.; Menezes de Moura, S.; et al. A nonproteinaceous Fusarium cell wall extract triggers receptor-like protein-dependent immune responses in Arabidopsis and cotton. New Phytol. 2021, 230, 275–289. [Google Scholar] [CrossRef]
- Wang, P.; Zhou, L.; Jamieson, P.; Zhang, L.; Zhao, Z.; Babilonia, K.; Shao, W.; Wu, L.; Mustafa, R.; Amin, I.; et al. The Cotton Wall-Associated Kinase GhWAK7A Mediates Responses to Fungal Wilt Pathogens by Complexing with the Chitin Sensory Receptors. Plant Cell 2020, 32, 3978–4001. [Google Scholar] [CrossRef]
- Feng, H.; Li, C.; Zhou, J.; Yuan, Y.; Feng, Z.; Shi, Y.; Zhao, L.; Zhang, Y.; Wei, F.; Zhu, H. A cotton WAKL protein interacted with a DnaJ protein and was involved in defense against Verticillium dahliae. Int. J. Biol. Macromol. 2021, 167, 633–643. [Google Scholar] [CrossRef]
- Gao, P.; Zhao, P.M.; Wang, J.; Wang, H.Y.; Du, X.M.; Wang, G.L.; Xia, G.X. Co-expression and preferential interaction between two calcineurin B-like proteins and a CBL-interacting protein kinase from cotton. Plant Physiol. Biochem. PPB Soc. Fr. Physiol. Veg. 2008, 46, 935–940. [Google Scholar] [CrossRef]
- Lu, T.; Zhang, G.; Sun, L.; Wang, J.; Hao, F. Genome-wide identification of CBL family and expression analysis of CBLs in response to potassium deficiency in cotton. PeerJ 2017, 5, e3653. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Yang, X.; Sun, W.; Miao, Y.; He, L.; Zhang, X. The Calcium Sensor CBL2 and Its Interacting Kinase CIPK6 Are Involved in Plant Sugar Homeostasis via Interacting with Tonoplast Sugar Transporter TST2. Plant Physiol. 2020, 183, 236–249. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Zhang, B.; Deng, J.; Chen, L.; Ullah, A.; Yang, X. Genome-wide analysis of CBL and CIPK family genes in cotton: Conserved structures with divergent interactions and expression. Physiol. Mol. Biol. Plants 2021, 27, 359–368. [Google Scholar] [CrossRef]
- Huang, J.; Pang, C.; Fan, S.; Song, M.; Yu, J.; Wei, H.; Ma, Q.; Li, L.; Zhang, C.; Yu, S. Genome-wide analysis of the family 1 glycosyltransferases in cotton. Mol. Genet. Genom. MGG 2015, 290, 1805–1818. [Google Scholar] [CrossRef]
- Lee, J.; Burns, T.H.; Light, G.; Sun, Y.; Fokar, M.; Kasukabe, Y.; Fujisawa, K.; Maekawa, Y.; Allen, R.D. Xyloglucan endotransglycosylase/hydrolase genes in cotton and their role in fiber elongation. Planta 2010, 232, 1191–1205. [Google Scholar] [CrossRef]
- Li, Z.K.; Chen, B.; Li, X.X.; Wang, J.P.; Zhang, Y.; Wang, X.F.; Yan, Y.Y.; Ke, H.F.; Yang, J.; Wu, J.H.; et al. A newly identified cluster of glutathione S-transferase genes provides Verticillium wilt resistance in cotton. Plant J. Cell Mol. Biol. 2019, 98, 213–227. [Google Scholar] [CrossRef]
- Wang, L.; Liu, X.; Wang, X.; Pan, Z.; Geng, X.; Chen, B.; Liu, B.; Du, X.; Song, X. Identification and characterization analysis of sulfotransferases (SOTs) gene family in cotton (Gossypium) and its involvement in fiber development. BMC Plant Biol. 2019, 19, 595. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).