Fibroblast Growth Factor 20 Gene Polymorphism in Parkinson’s Disease in Asian Population: A Meta-Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. Genetic Analysis
2.3. Statistical Analysis
2.4. Meta-Analysis
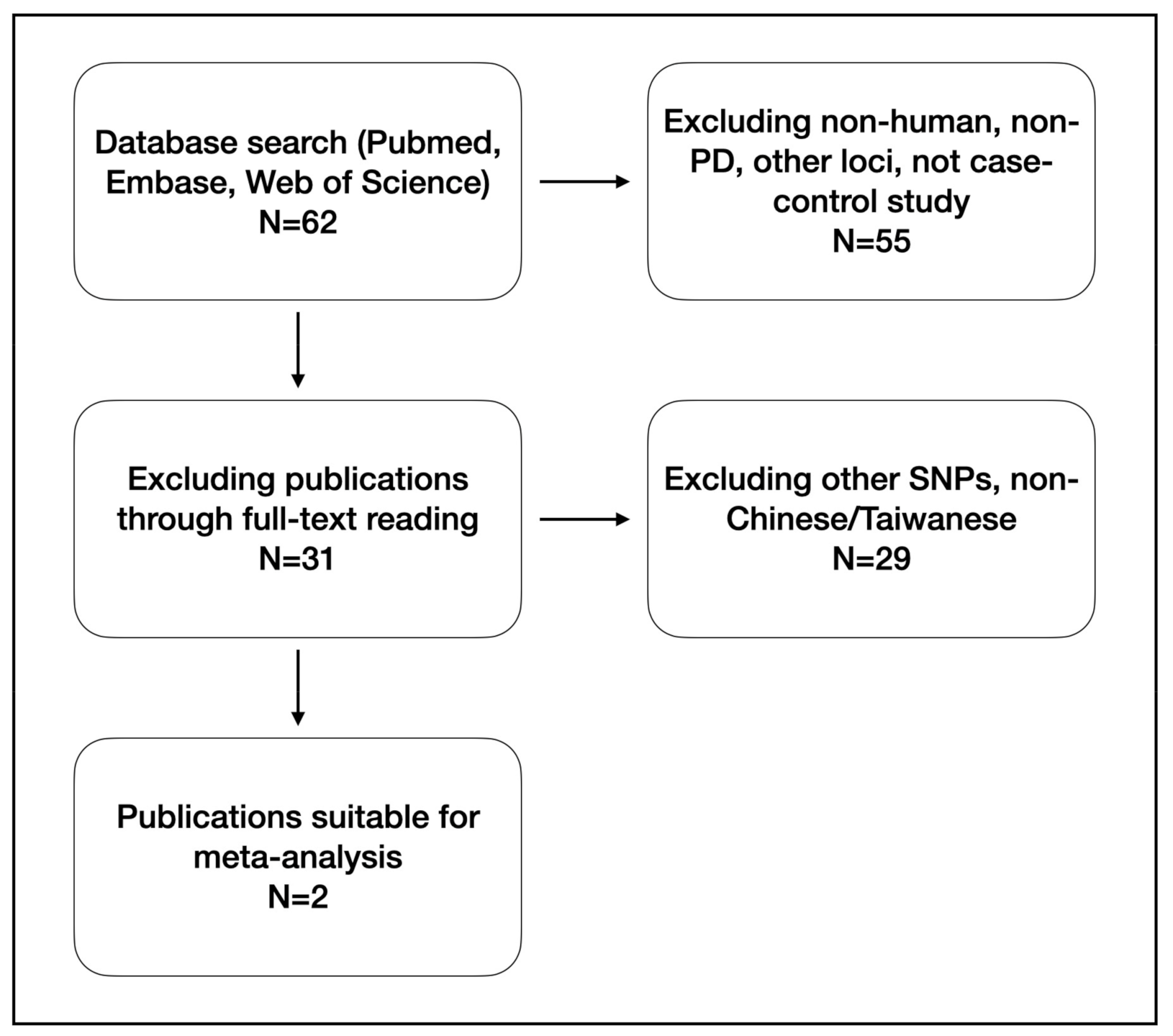
2.4.1. Literature Search and Inclusion
2.4.2. Statistical Analysis
3. Results
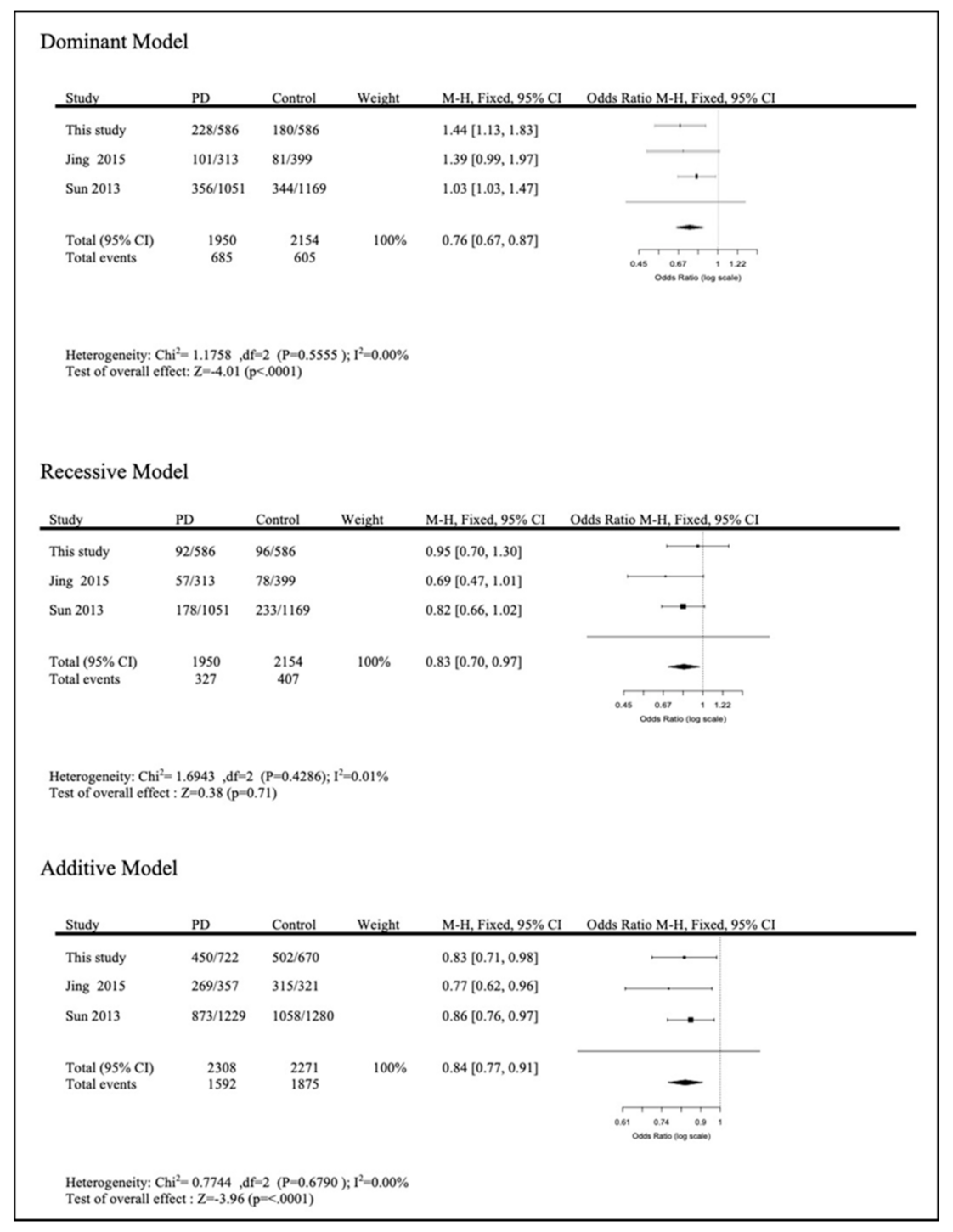
Meta-Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kalia, L.V.; Lang, A.E. Parkinson’s disease. Lancet 2015, 386, 896–912. [Google Scholar] [CrossRef]
- Emamzadeh, F.N.; Surguchov, A. Parkinson’s Disease: Biomarkers, Treatment, and Risk Factors. Front. Neurosci. 2018, 12, 612. [Google Scholar] [CrossRef] [PubMed]
- Benskey, M.J.; Perez, R.G.; Manfredsson, F.P. The contribution of α synuclein to neuronal survival and function—Implications for Parkinson’s disease. J. Neurochem. 2016, 137, 331–359. [Google Scholar] [CrossRef] [PubMed]
- Surguchev, A.A.; Surguchov, A. Synucleins and Gene Expression: Ramblers in a Crowd or Cops Regulating Traffic? Front. Mol. Neurosci. 2017, 10, 224. [Google Scholar] [CrossRef]
- Chang, D.; Nalls, M.A.; Hallgrimsdottir, I.B.; Hunkapiller, J.; van der Brug, M.; Cai, F.; Kerchner, G.A.; Ayalon, G.; International Parkinson’s Disease Genomics Consortium; 23andMe Research Team; et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat. Genet. 2017, 49, 1511–1516. [Google Scholar] [CrossRef]
- International Parkinson’s Disease Genomics Consortium (IPDGC); Wellcome Trust Case Control Consortium 2 (WTCCC2). A two-stage meta-analysis identifies several new loci for Parkinson’s disease. PLoS Genet. 2011, 7, e1002142. [Google Scholar] [CrossRef]
- Nalls, M.A.; Pankratz, N.; Lill, C.M.; Do, C.B.; Hernandez, D.G.; Saad, M.; DeStefano, A.L.; Kara, E.; Bras, J.; Sharma, M.; et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 2014, 46, 989–993. [Google Scholar] [CrossRef]
- Jing, C.C.; Luo, X.G.; Cui, H.G.; Li, F.R.; Li, P.; Jiang, E.Z.; Ren, Y.; Pang, H. Screening of polymorphisms located in the FGF20 and TMEM175 genes in North Chinese Parkinson’s disease patients. Genet. Mol. Res. 2015, 14, 13679–13687. [Google Scholar] [CrossRef]
- Sun, X.Y.; Wang, L.; Cheng, L.; Li, N.N.; Lu, Z.J.; Li, J.Y.; Peng, R. Genetic analysis of FGF20 in Chinese patients with Parkinson’s disease. Neurol. Sci. 2017, 38, 887–891. [Google Scholar] [CrossRef]
- Itoh, N.; Ohta, H. Roles of FGF20 in dopaminergic neurons and Parkinson’s disease. Front. Mol. Neurosci. 2013, 6, 15. [Google Scholar] [CrossRef]
- Wang, G.; van der Walt, J.M.; Mayhew, G.; Li, Y.J.; Zuchner, S.; Scott, W.K.; Martin, E.R.; Vance, J.M. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of α-synuclein. Am. J. Hum. Genet. 2008, 82, 283–289. [Google Scholar] [CrossRef]
- Hughes, A.J.; Daniel, S.E.; Kilford, L.; Lees, A.J. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: A clinico-pathological study of 100 cases. J. Neurol. Neurosurg. Psychiatry 1992, 55, 181–184. [Google Scholar] [CrossRef]
- Chang, K.H.; Wu, Y.R.; Chen, Y.C.; Fung, H.C.; Lee-Chen, G.J.; Chen, C.M. STK39, But Not BST1, HLA-DQB1, and SPPL2B Polymorphism, Is Associated With Han-Chinese Parkinson’s Disease in Taiwan. Medicine 2015, 94, e1690. [Google Scholar] [CrossRef]
- Chen, C.M.; Chen, Y.C.; Chiang, M.C.; Fung, H.C.; Chang, K.H.; Lee-Chen, G.J.; Wu, Y.R. Association of GCH1 and MIR4697, but not SIPA1L2 and VPS13C polymorphisms, with Parkinson’s disease in Taiwan. Neurobiol. Aging 2016, 39, 221.e1–221.e5. [Google Scholar] [CrossRef]
- Liu, T.W.; Wu, Y.R.; Chen, Y.C.; Fung, H.C.; Chen, C.M. Association of RIT2 and RAB7L1 with Parkinson’s disease: A case-control study in a Taiwanese cohort and a meta-analysis in Asian populations. Neurobiol. Aging 2020, 87, 140.e5–140.e11. [Google Scholar] [CrossRef]
- Wu, H.C.; Chen, C.M.; Chen, Y.C.; Fung, H.C.; Chang, K.H.; Wu, Y.R. DLG2, but not TMEM229B, GPNMB, and ITGA8 polymorphism, is associated with Parkinson’s disease in a Taiwanese population. Neurobiol. Aging 2018, 64, 158.e1–158.e6. [Google Scholar] [CrossRef]
- Pihlstrom, L.; Axelsson, G.; Bjornara, K.A.; Dizdar, N.; Fardell, C.; Forsgren, L.; Holmberg, B.; Larsen, J.P.; Linder, J.; Nissbrandt, H.; et al. Supportive evidence for 11 loci from genome-wide association studies in Parkinson’s disease. Neurobiol. Aging 2013, 34, 1708.e7–1708.e13. [Google Scholar] [CrossRef]
- Liu, Z.H.; Guo, J.F.; Li, K.; Wang, Y.Q.; Kang, J.F.; Wei, Y.; Sun, Q.Y.; Xu, Q.; Wang, D.L.; Xia, K.; et al. Analysis of several loci from genome-wide association studies in Parkinson’s disease in mainland China. Neurosci. Lett. 2015, 587, 68–71. [Google Scholar] [CrossRef]
- Ohmachi, S.; Watanabe, Y.; Mikami, T.; Kusu, N.; Ibi, T.; Akaike, A.; Itoh, N. FGF-20, a novel neurotrophic factor, preferentially expressed in the substantia nigra pars compacta of rat brain. Biochem. Biophys. Res. Commun. 2000, 277, 355–360. [Google Scholar] [CrossRef]
- van der Walt, J.M.; Noureddine, M.A.; Kittappa, R.; Hauser, M.A.; Scott, W.K.; McKay, R.; Zhang, F.; Stajich, J.M.; Fujiwara, K.; Scott, B.L.; et al. Fibroblast growth factor 20 polymorphisms and haplotypes strongly influence risk of Parkinson disease. Am. J. Hum. Genet. 2004, 74, 1121–1127. [Google Scholar] [CrossRef]
- Satake, W.; Mizuta, I.; Suzuki, S.; Nakabayashi, Y.; Ito, C.; Watanabe, M.; Takeda, A.; Hasegawa, K.; Sakoda, S.; Yamamoto, M.; et al. Fibroblast growth factor 20 gene and Parkinson’s disease in the Japanese population. Neuroreport 2007, 18, 937–940. [Google Scholar] [CrossRef]
- Pan, J.; Li, H.; Wang, Y.; Ma, J.F.; Zhang, J.; Wang, G.; Liu, J.; Wang, X.J.; Xiao, Q.; Chen, S.D. Fibroblast growth factor 20 (FGF20) polymorphism is a risk factor for Parkinson’s disease in Chinese population. Parkinsonism Relat. Disord. 2012, 18, 629–631. [Google Scholar] [CrossRef]
- Pak, K.; Nam, H.Y.; Shin, S.; Kim, K.; Lee, M.J.; Kim, E.J.; Lee, J.M.; Kim, S.J.; Kim, I.J. Effects of rs591323 on serotonin transporter availability in healthy male subjects. Ann. Nucl. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Joling, M.; van den Heuvel, O.A.; Berendse, H.W.; Booij, J.; Vriend, C. Serotonin transporter binding and anxiety symptoms in Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry 2018, 89, 89–94. [Google Scholar] [CrossRef] [PubMed]
| First Author | Country | PD | Controls | HWE for PD | HWE for Controls | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Allele | Genotype | Allele | ||||||||||||
| GG | GA | AA | G | A | GG | GA | AA | G | A | X2 | P | X2 | P | ||
| Chiang | Taiwan | 228 | 266 | 92 | 722 | 450 | 180 | 310 | 96 | 670 | 502 | 0.96 | 0.33 | 3.77 | 0.052 |
| Jing | China | 101 | 155 | 57 | 357 | 269 | 81 | 159 | 78 | 321 | 315 | 0.035 | 0.85 | 2.52 × 10−6 | 0.99 |
| Sun | China | 356 | 517 | 178 | 1229 | 873 | 344 | 592 | 233 | 1280 | 1058 | 0.174 | 0.67 | 0.568 | 0.45 |
| rs591323 Genotype/Allele | PD (%) | Control (%) | OR | p Value |
|---|---|---|---|---|
| This study (Taiwan) (n = 586 + 586 = 1172) | ||||
| GG | 228 (39%) | 180 (31%) | ||
| GA | 266 (45%) | 310 (53%) | ||
| AA | 92 (16%) | 96 (16%) | 0.011 | |
| G | 722 (62%) | 670 (57%) | 1 | |
| A | 450 (38%) | 502 (43%) | 0.832 (0.705, 0.981) | 0.0287 |
| Dominant model | ||||
| GA + AA | 358 (61%) | 406 (69%) | 0.696 (0.547, 0.886) | 0.0032 |
| GG | 228 (39%) | 180 (31%) | 1 | |
| Recessive model | ||||
| AA | 92 (16%) | 96 (16%) | 0.956 (0.696, 1.298) | 0.7502 |
| GA + GG | 494 (84%) | 490 (84%) | 1 | |
| Jing et al. (China) (n = 313 + 318 = 631) | ||||
| GG | 101 (32%) | 81(25%) | ||
| GA | 155 (50%) | 159 (50%) | ||
| AA | 57 (18%) | 78 (25%) | ||
| G | 357 (57%) | 321 (50%) | 1 | |
| A | 269 (43%) | 315 (50%) | 0.768 (0.615, 0.959) | 0.0195 |
| Dominant model | ||||
| GA + AA | 212 (68%) | 237 (75%) | 0.717 (0.508, 1.014) | 0.0595 |
| GG | 101 (32%) | 81 (25%) | 1 | |
| Recessive model | ||||
| AA | 57 (18%) | 78 (25%) | 0.685 (0.467, 1.006) | 0.0530 |
| GA + GG | 256 (82%) | 240 (75%) | 1 | |
| Sun et al. (China) (n = 1051 + 1169 = 2220) | ||||
| GG | 356 (34%) | 344 (29%) | ||
| GA | 517 (49%) | 592 (51%) | ||
| AA | 178 (17%) | 233 (20%) | ||
| G | 1229 (58%) | 1280 (55%) | 1 | |
| A | 873 (42%) | 1058 (45%) | 0.859 (0.763, 0.968) | 0.0125 |
| Dominant model | ||||
| GA + AA | 695 (66%) | 825 (71%) | 0.814 (0.680, 0.974) | 0.0244 |
| GG | 356 (34%) | 344 (29%) | 1 | |
| Recessive model | ||||
| AA | 178 (17%) | 233 (20%) | 0.819 (0.660, 1.016) | 0.0696 |
| GA + GG | 873 (83%) | 936 (80%) | 1 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiang, H.-L.; Wu, Y.-R.; Chen, Y.-C.; Fung, H.-C.; Chen, C.-M. Fibroblast Growth Factor 20 Gene Polymorphism in Parkinson’s Disease in Asian Population: A Meta-Analysis. Genes 2021, 12, 674. https://doi.org/10.3390/genes12050674
Chiang H-L, Wu Y-R, Chen Y-C, Fung H-C, Chen C-M. Fibroblast Growth Factor 20 Gene Polymorphism in Parkinson’s Disease in Asian Population: A Meta-Analysis. Genes. 2021; 12(5):674. https://doi.org/10.3390/genes12050674
Chicago/Turabian StyleChiang, Han-Lin, Yih-Ru Wu, Yi-Chun Chen, Hon-Chung Fung, and Chiung-Mei Chen. 2021. "Fibroblast Growth Factor 20 Gene Polymorphism in Parkinson’s Disease in Asian Population: A Meta-Analysis" Genes 12, no. 5: 674. https://doi.org/10.3390/genes12050674
APA StyleChiang, H.-L., Wu, Y.-R., Chen, Y.-C., Fung, H.-C., & Chen, C.-M. (2021). Fibroblast Growth Factor 20 Gene Polymorphism in Parkinson’s Disease in Asian Population: A Meta-Analysis. Genes, 12(5), 674. https://doi.org/10.3390/genes12050674






