Overexpression of OsMed16 Inhibits the Growth of Rice and Causes Spontaneous Cell Death
Abstract
1. Introduction
2. Materials and Methods
2.1. The Plant Materials and Growth Conditions
2.2. Generation of Transgenic Plants
2.3. RNA Isolation and Gene Expression Analysis
2.4. Subcellular Localization of OsMed16
2.5. Histochemical Stain
2.6. RNA-Seq Data Analysis
2.7. Phenotypic Analysis of OsMed16 Mutants and OsMed16-Overexpressing Plants
3. Results
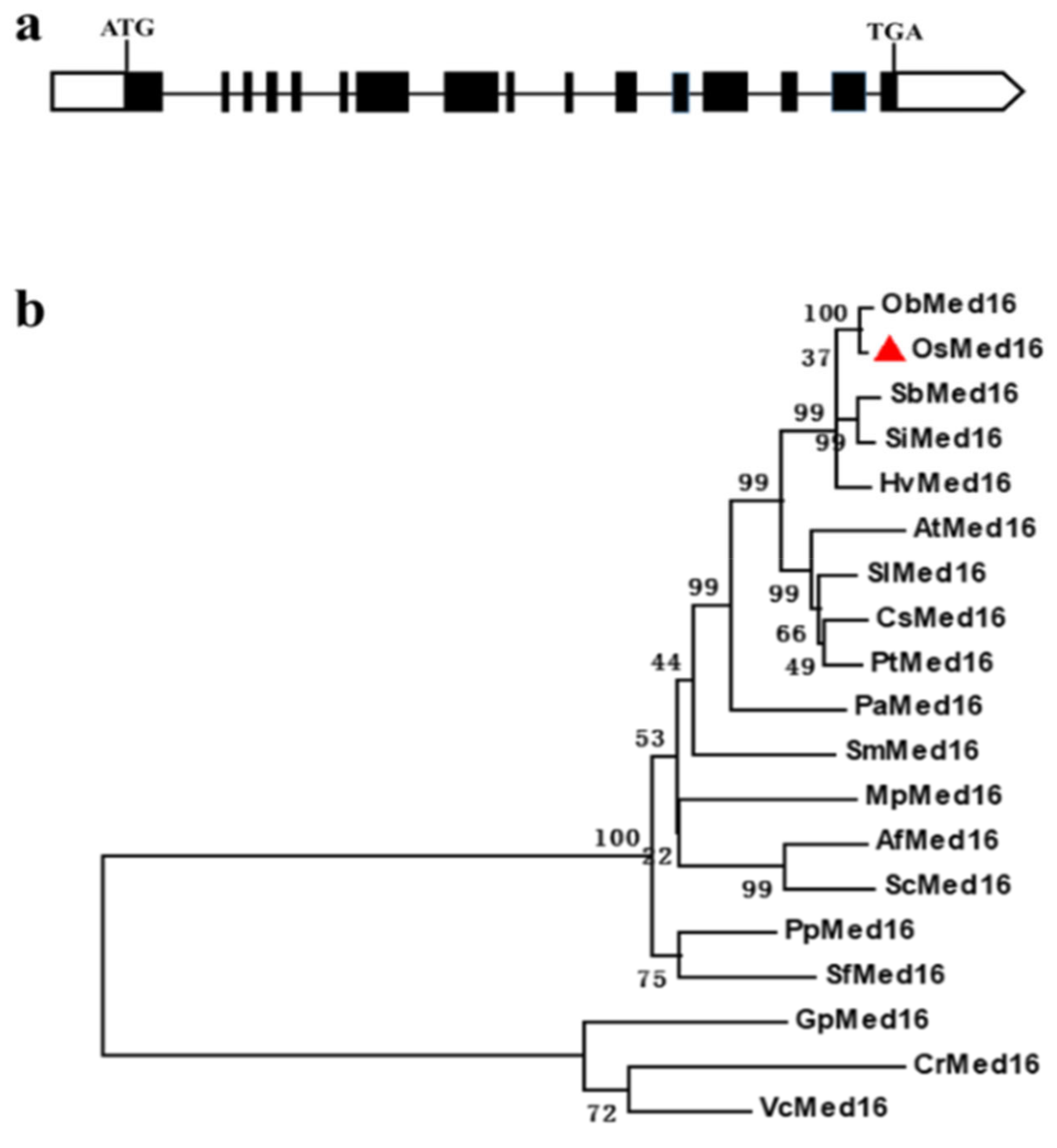
3.1. Sequence and Phylogenic Analysis of OsMed16
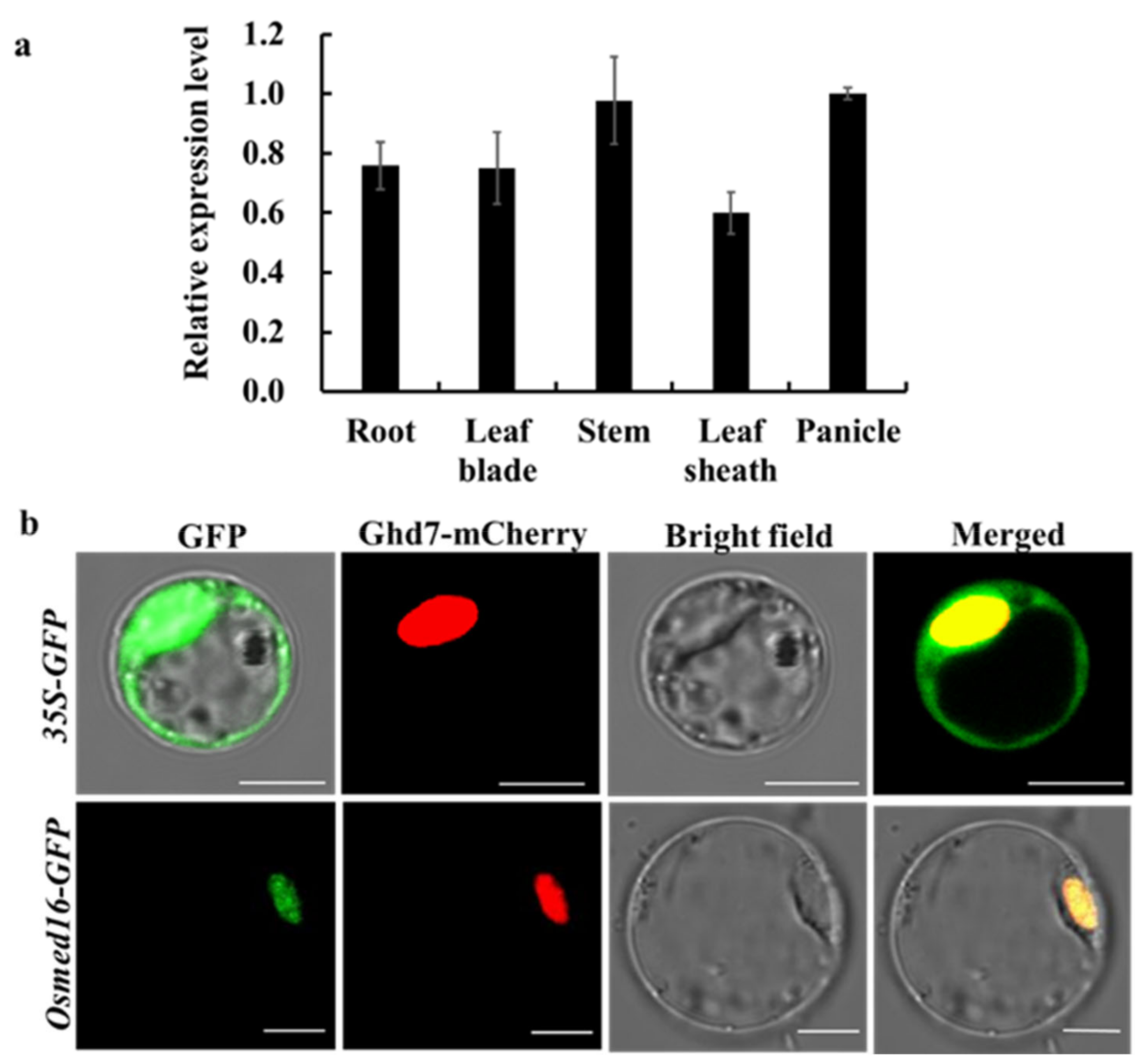
3.2. OsMed16 mRNA Expression Pattern and Protein Subcellular Localization
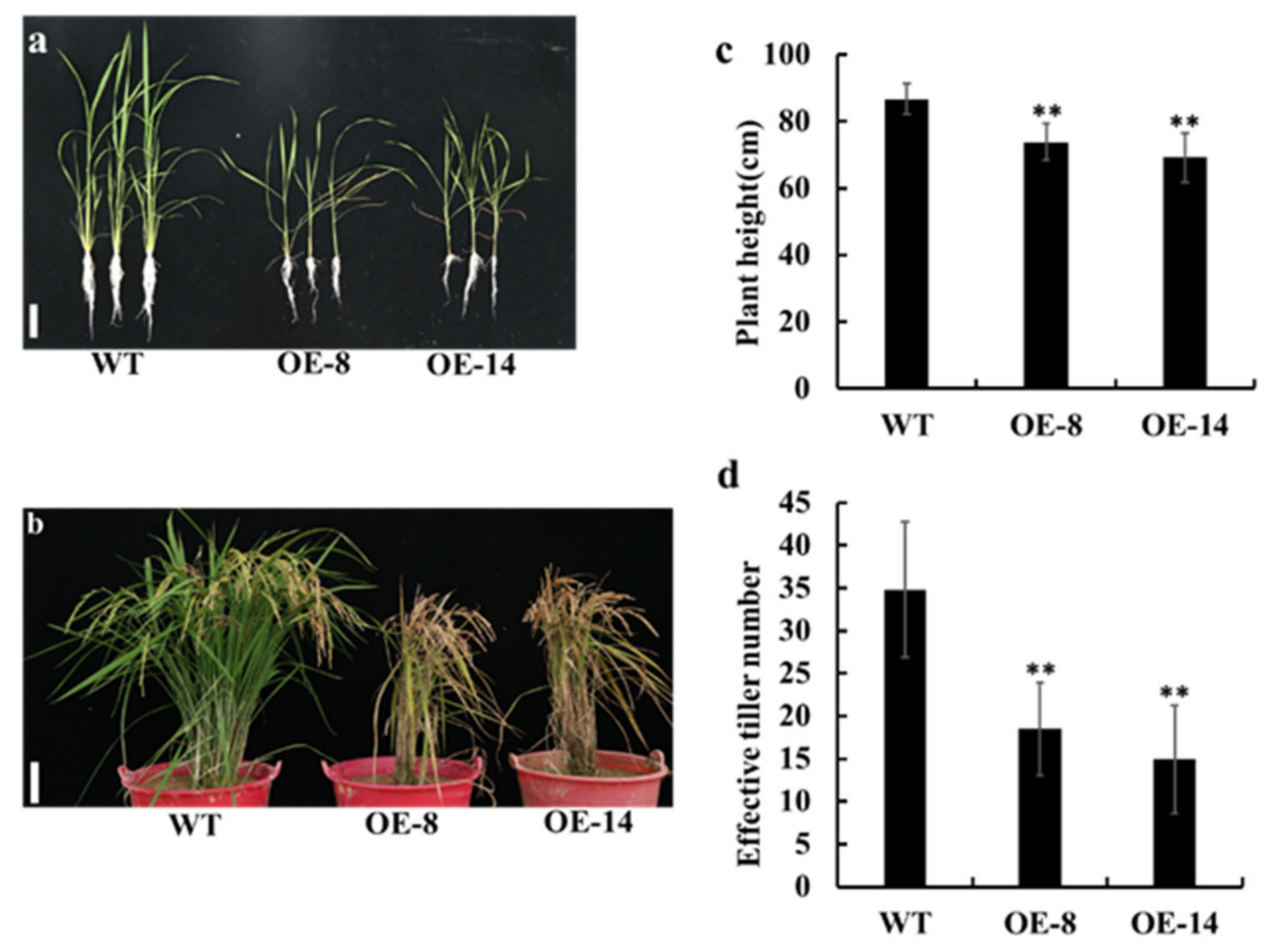
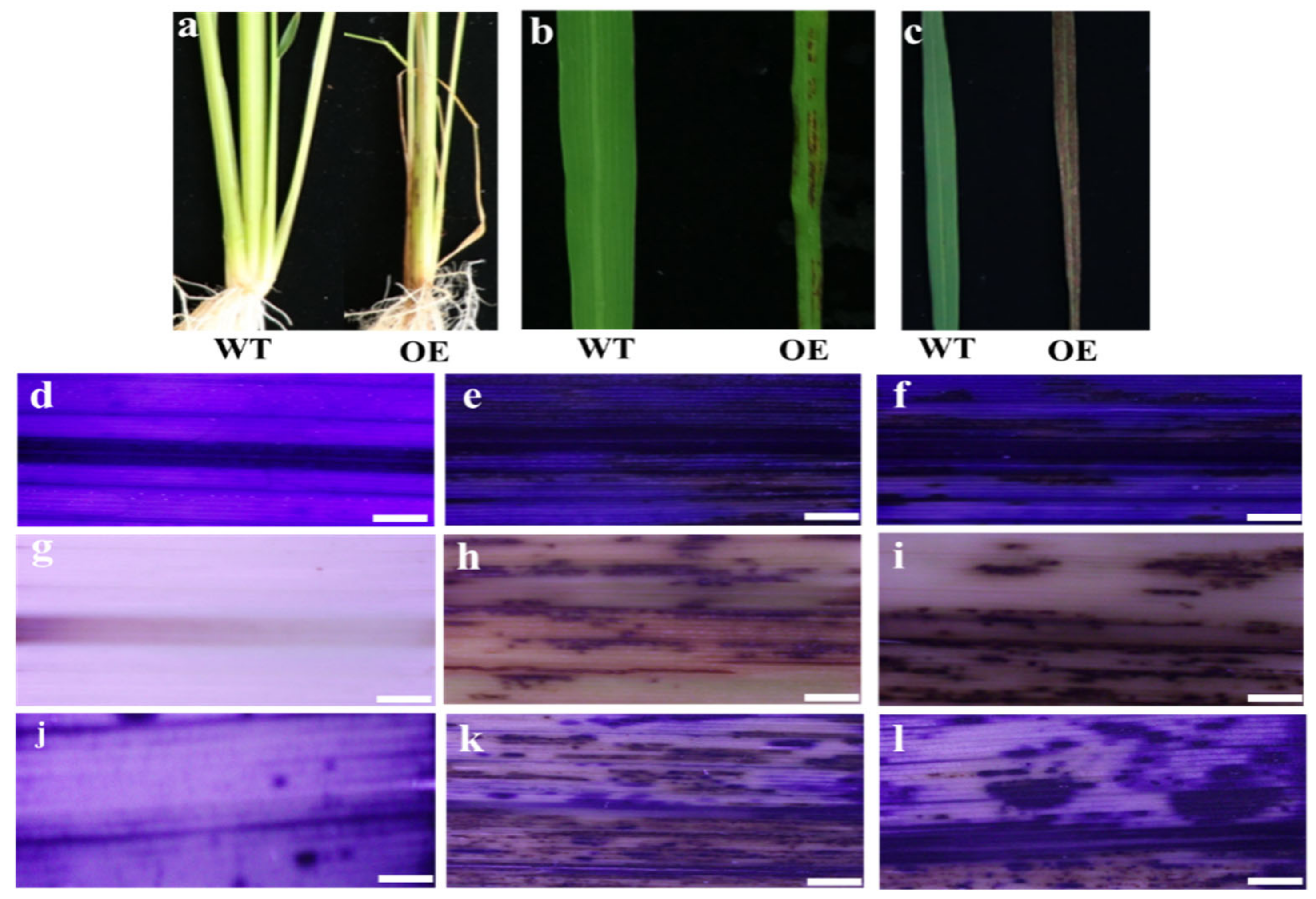
3.3. Overexpression of OsMed16 Caused the Inhibition of Growth of Rice and Spontaneous Cell Death
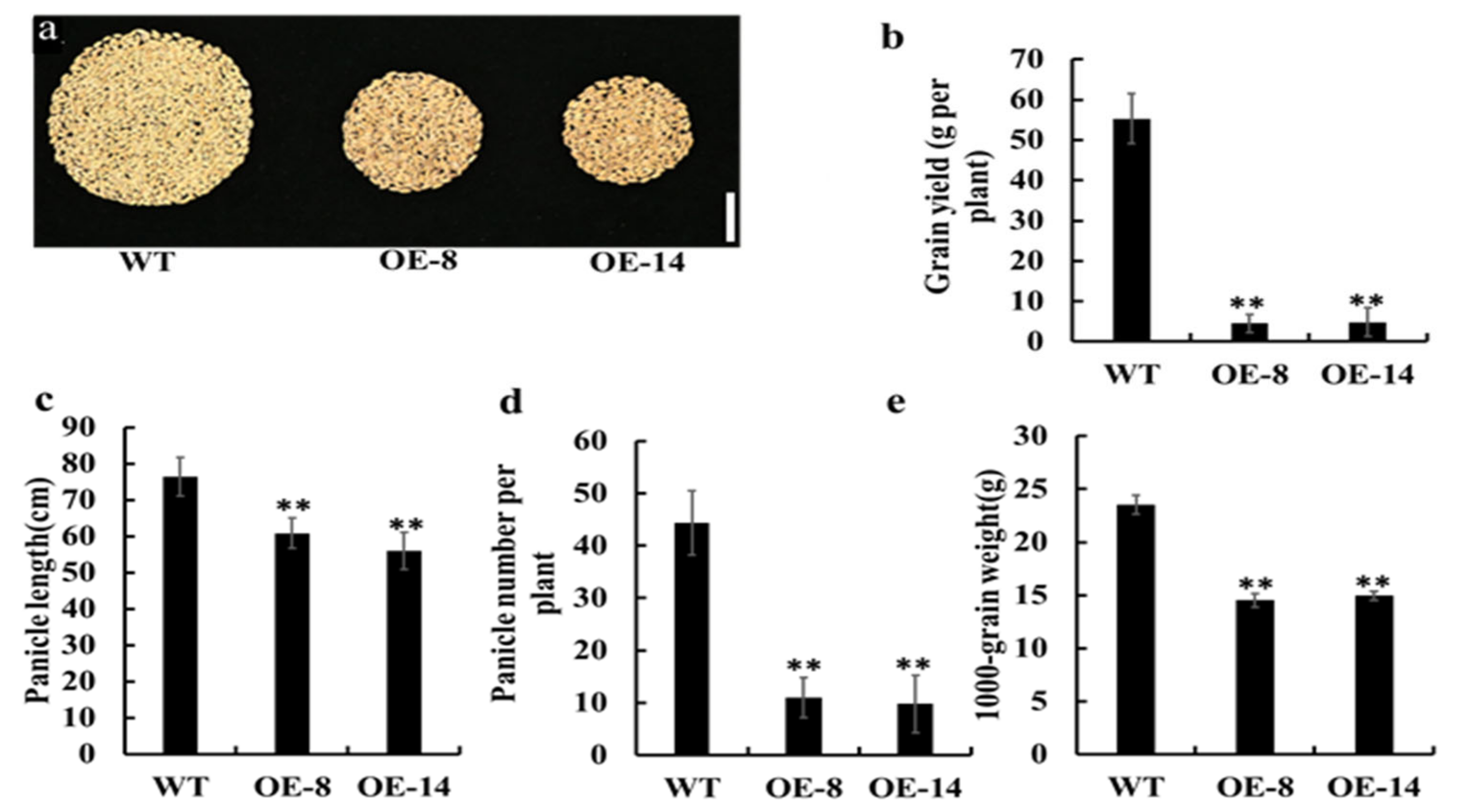
3.4. Overexpression of OsMed16 Reduced the Yield of Rice Grains
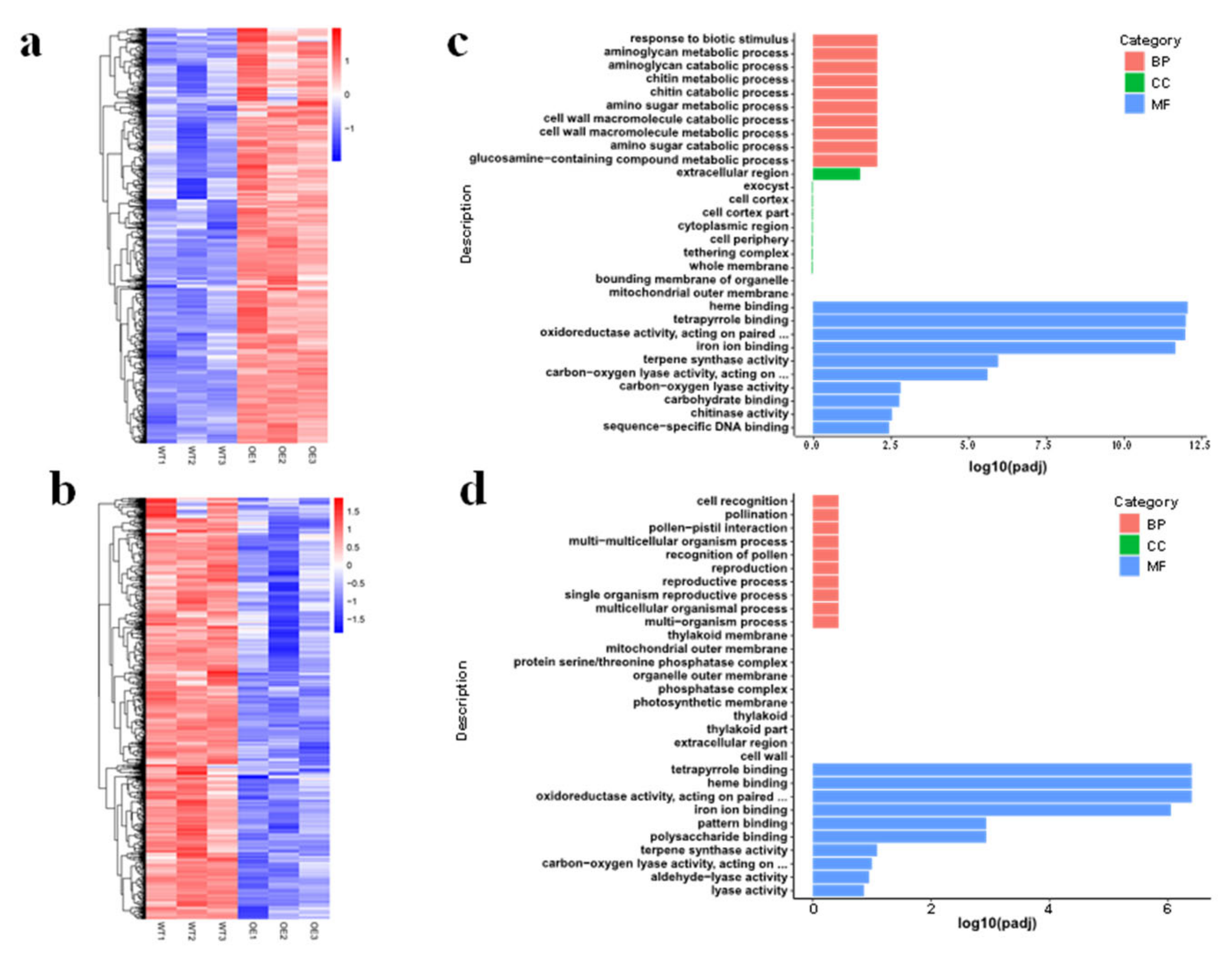
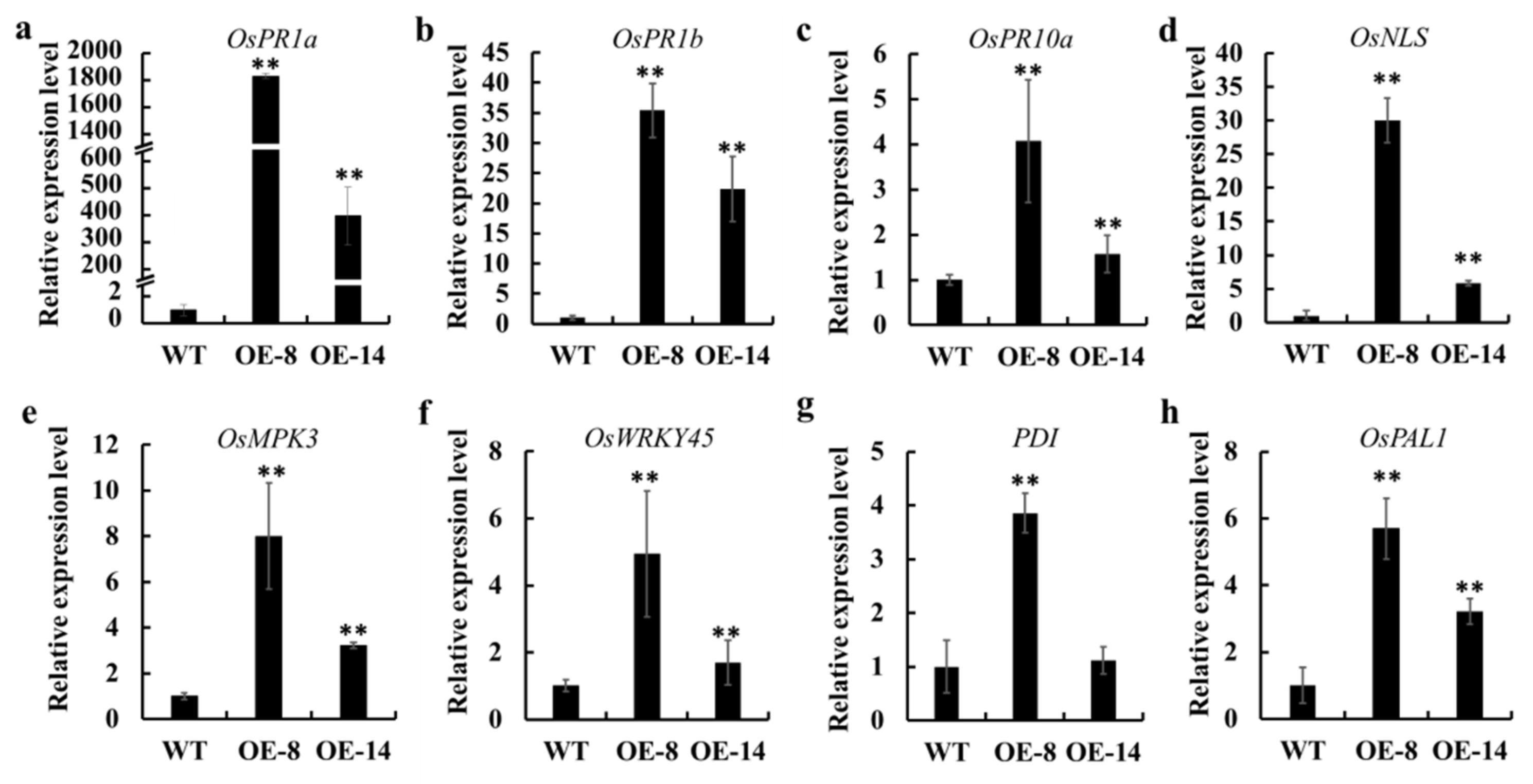
3.5. Transcriptome Changes in OsMed16-OE Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martinez, E. Multi-protein complexes in eukaryotic gene transcription. Plant Mol. Biol. 2002, 50, 925–947. [Google Scholar] [CrossRef]
- Orphanides, G.; Reinberg, D. A unified theory of gene expression. Cell 2002, 108, 439–451. [Google Scholar] [CrossRef]
- Zhai, Q.; Li, C. The plant Mediator complex and its role in jasmonate signaling. J. Exp. Bot. 2019, 70, 3415–3424. [Google Scholar] [CrossRef] [PubMed]
- Jeronimo, C.; Robert, F. The Mediator complex: At the nexus of RNA polymerase II transcription. Trends. Cell Biol. 2017, 27, 765–783. [Google Scholar] [CrossRef]
- Chadick, J.Z.; Asturias, F.J. Structure of eukaryotic Mediator complexes. Trends. Biochem. Sci. 2005, 30, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.D.; Oldenbroek, M.; Boyer, T.G. Mediator kinase module and human tumorigenesis. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 393–426. [Google Scholar]
- Kelleher, R.J.; Flanagan, P.M.; Kornberg, R.D. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell 1990, 61, 1209–1215. [Google Scholar] [CrossRef]
- Fondell, J.D.; Ge, H.; Roeder, R.G. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. USA 1996, 93, 8329–8333. [Google Scholar] [CrossRef]
- Jiang, Y.W.; Veschambre, P.; Erdjument-Bromage, H.; Tempst, P.; Conaway, J.W.; Conaway, R.C.; Kornberg, R.D. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc. Natl. Acad. Sci. USA 1998, 95, 8538–8543. [Google Scholar] [CrossRef]
- Kwon, J.Y.; Park, J.M.; Gim, B.S.; Han, S.J.; Lee, J.; Kim, Y.J. Caenorhabditis elegans mediator complexes are required for developmental-specific transcriptional activation. Proc. Natl. Acad. Sci. USA. 1999, 96, 14990–14995. [Google Scholar] [CrossRef]
- Bäckström, S.; Elfving, N.; Nilsson, R.; Wingsle, G.; Björklund, S. Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell. 2007, 26, 717–729. [Google Scholar] [CrossRef]
- Yang, Y.; Li, L.; Qu, L.J. Plant Mediator complex and its critical functions in transcription regulation. J. Integr. Plant Biol. 2016, 58, 106–118. [Google Scholar] [CrossRef]
- Mathur, S.; Vyas, S.; Kapoor, S.; Tyagi, A.K. The Mediator complex in plants: Structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress. Plant Physiol. 2011, 157, 1609–1627. [Google Scholar] [CrossRef] [PubMed]
- Elfving, N.; Davoine, C.; Benlloch, R.; Blomberg, J.; Brännström, K.; Müller, D.; Nilsson, A.; Ulfstedt, M.; Ronne, H.; Wingsle, G.; et al. The Arabidopsis thaliana Med25 mediator subunit integrates environmental cues to control plant development. Proc. Natl. Acad. Sci. USA 2011, 108, 8245–8250. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Li, Y. Control of final organ size by Mediator complex subunit25 in Arabidopsis thaliana. Development 2011, 38, 4545–4554. [Google Scholar] [CrossRef] [PubMed]
- Iñigo, S.; Alvarez, M.J.; Strasser, B.; Califano, A.; Cerdán, P.D. PFT1, the MED25 subunit of the plant Mediator complex, promotes flowering through CONSTANS dependent and independent mechanisms in Arabidopsis. Plant J. 2012, 69, 601–612. [Google Scholar] [CrossRef]
- Knight, H.; Mugford, S.G.; Ulker, B.; Gao, D.; Thorlby, G.; Knight, M.R. Identification of SFR6, a key component in cold acclimation acting post-translationally on CBF function. Plant J. 2009, 58, 97–108. [Google Scholar] [CrossRef]
- Huang, J.; Sun, Y.; Orduna, A.R.; Jetter, R.; Li, X. The Mediator kinase module acts as a positive regulator of salicylic acid accumulation and systemic acquired resistance. Plant J. 2019, 98, 842–852. [Google Scholar] [CrossRef]
- Dwivedi, N.; Maji, S.; Waseem, M.; Thakur, P.; Kumar, V.; Parida, S.K.; Thakur, J.K. The Mediator subunit OsMED15a is a transcriptional co-regulator of seed size/weight–modulating genes in rice. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 194432. [Google Scholar] [CrossRef]
- Malik, N.; Ranjan, R.; Parida, S.K.; Agarwal, P.; Tyagi, A.K. Mediator subunit OsMED14_1 plays an important role in rice development. Plant J. 2020, 101, 1411–1429. [Google Scholar] [CrossRef] [PubMed]
- Wathugala, D.L.; Richards, S.A.; Knight, H.; Knight, M.R. OsSFR6 is a functional rice orthologue of SENSITIVE TO FREEZING-6 and can act as a regulator of COR gene expression, osmotic stress and freezing tolerance in Arabidopsis. New Phytol. 2011, 191, 984–995. [Google Scholar] [CrossRef]
- Yamaji, N.; Ma, J.F. Spatial distribution and temporal variation of the rice silicon transporter Lsi1. Plant Physiol. 2007, 143, 1306–1313. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A robust CRISPR/Cas9 system for convenient, High-Efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, N.; Takemoto, Y.; Miyaji, T.; Mitani-Ueno, N.; Yoshida, K.T.; Ma, J.F. Erratum: Reducing phosphorus accumulation in rice grains with an impaired transporter in the node. Nature 2017, 543, 136. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ma, Q.; Yi, R.; Li, L.; Liang, Z.; Zeng, T.; Zhang, Y.; Huang, H.; Zhang, X.; Yin, X.; Cai, Z.; et al. GsMATE encoding a multidrug and toxic compound extrusion transporter enhances aluminum tolerance in Arabidopsis thaliana. BMC Plant Biol. 2018, 18, 212. [Google Scholar] [CrossRef]
- Zhang, Y.; Su, J.; Duan, S.; Ao, Y.; Dai, J.; Liu, J.; Wang, P.; Li, Y.; Liu, B.; Feng, D.; et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 2011, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Chen, J.; Zeng, L.; Goh, M.; Leung, H.; Khush, G.S.; Wang, G.L. Characterizing rice lesion mimic mutants and identifying a mutant with broad spectrum resistance to rice blast and bacterial blight. Mol. Plant Microbe Interact. 2000, 13, 869–876. [Google Scholar] [CrossRef]
- Thordal-Christensen, H.; Zhang, Z.; Wei, Y.; Collinge, D.B. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley—powdery mildew interaction. Plant J. 1997, 11, 1187–1194. [Google Scholar] [CrossRef]
- Qiao, Y.; Jiang, W.; Lee, J.; Park, B.; Choi, M.S.; Piao, R.; Woo, M.O.; Roh, J.H.; Han, L.; Paek, N.C.; et al. SPL28 encodes a clathrinassociated adaptor protein complex 1, medium subunit micro 1 (AP1M1) and is responsible for spotted leaf and early senescence in rice (Oryza sativa). New Phytol. 2010, 185, 258–274. [Google Scholar] [CrossRef]
- Shen, C.X.; Li, D.; He, R.H.; Fang, Z.; Xia, Y.M.; Gao, J.; Shen, H.; Cao, M.L. Comparative transcriptome analysis of RNA-Seq data for cold-tolerant and cold-sensitive rice genotypes under cold stress. J. Plant Biol. 2014, 57, 337–348. [Google Scholar] [CrossRef]
- Bourbon, H.M.; Aguilera, A.; Ansari, A.Z.; Asturias, F.J.; Berk, A.J.; Bjorklund, S.; Blackwell, T.K.; Borggrefe, T.; Carey, M.; Carlson, M.; et al. A unified nomenclature for protein subunits of Mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell 2004, 14, 553–557. [Google Scholar] [CrossRef]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef]
- Xue, W.; Xing, Y.; Weng, X.; Zhao, Y.; Tang, W.; Wang, L.; Zhou, H.; Yu, S.; Xu, C.; Li, X.; et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Khanna-Chopra, R. Leaf senescence and abiotic stresses share reactive oxygen species-mediated chloroplast degradation. Protoplasma 2012, 249, 469–481. [Google Scholar] [CrossRef]
- Li, W.; Shao, M.; Yang, J.; Zhong, W.; Okada, K.; Yamane, H.; Qian, G.; Liu, F. Oscyp71Z2 involves diterpenoid phytoalexin biosynthesis that contributes to bacterial blight resistance in rice. Plant Sci. 2013, 207, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, F.; Wang, J.; Fan, F.; Zhu, J.; Yang, J.; Liu, F.; Zhong, W. Overexpressing CYP71Z2 enhances resistance to bacterial blight by suppressing auxin biosynthesis in rice. PLoS ONE 2015, 10, e0119867. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, S.; Maekawa, M.; Arite, T.; Onishi, K.; Takamure, I.; Kyozuka, J. Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol. 2005, 46, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Takeda, Y.; Koshiba, T.; Tobimatsu, Y.; Suzuki, S.; Murakami, S.; Yamamura, M.; Rahman, M.M.; Takano, T.; Hattori, T.; Sakamoto, M.; et al. Regulation of CONIFERALDEHYDE 5-HYDROXYLASE expression to modulate cell wall lignin structure in rice. Planta 2017, 246, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.; Xin, Y.; Tan, Y.; Hu, X.; Bai, J.; Liu, Z.Y.; Yu, Y.; Li, L.; Peng, C.; Fan, T.; et al. Natural variation in the HAN1 gene confers chilling tolerance in rice and allowed adaptation to a temperate climate. Proc. Natl. Acad. Sci. USA 2019, 116, 3494–3501. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Q.; Wu, J.; Zheng, X.; Zheng, S.; Sun, X.; Qiu, Q.; Lu, T. Gene knockout study reveals that cytosolic ascorbate peroxidase 2 (OsAPX2) plays a critical role in growth and reproduction in rice under drought, salt and cold stresses. PLoS ONE 2013, 8, e57472. [Google Scholar] [CrossRef]
- Du, Y.; Chu, H.; Chu, I.K.; Lo, C. CYP93G2 is a flavanone 2-hydroxylase required for C-glycosylflavone biosynthesis in rice. Plant Physiol. 2010, 154, 324–333. [Google Scholar] [CrossRef]
- Zhou, Z.; Pang, Z.; Li, G.; Lin, C.; Wang, J.; Lv, Q.; He, C.; Zhu, L. Endoplasmic reticulum membrane-bound MoSec62 is involved in the suppression of rice immunity and is essential for the pathogenicity of Magnaporthe oryzae. Mol. Plant Pathol. 2016, 17, 1211–1222. [Google Scholar] [CrossRef]
- Sun, L.; Yang, D.L.; Kong, Y.; Chen, Y.; Li, X.Z.; Zeng, L.J.; Li, Q.; Wang, E.T.; He, Z.H. Sugar homeostasis mediated by cell wall invertase GRAIN INCOMPLETE FILLING 1 (GIF1) plays a role in pre-existing and induced defence in rice. Mol. Plant Pathol. 2014, 15, 161–173. [Google Scholar] [CrossRef]
- Delteil, A.; Blein, M.; Faivre-Rampant, O.; Guellim, A.; Estevan, J.; Hirsch, J.; Bevitori, R.; Michel, C.; Morel, J.B. Building a mutant resource for the study of disease resistance in rice reveals the pivotal role of several genes involved in defence. Mol. Plant Pathol. 2012, 13, 72–82. [Google Scholar] [CrossRef]
- Johal, G.S.; Hulbert, S.H.; Briggs, S.P. Disease lesion mimic of maize: A model for cell death in plants. Bioessays 1995, 17, 685–692. [Google Scholar] [CrossRef]
- Dietrich, R.A.; Delaney, T.P.; Uknes, S.J.; Ward, E.R.; Ryals, J.A.; Dangl, J.L. Arabidopsis mutants simulating disease resistance response. Cell 1994, 77, 565–577. [Google Scholar] [CrossRef]
- Büschges, R.; Hollricher, K.; Panstruga, R.; Simons, G.; Wolter, M.; Frijters, A.; Van Daelen, R.; van der Lee, T.; Diergaarde, P.; Groenendijk, J.; et al. The barley Mlo gene: A novel control element of plant pathogen resistance. Cell 1997, 88, 695–705. [Google Scholar] [CrossRef]
- Gray, J.; Close, P.S.; Briggs, S.P.; Johal, G.S. A novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell 1997, 89, 25–31. [Google Scholar] [CrossRef]
- Badigannavar, A.M.; Kale, D.M.; Eapen, S.; Murty, G.S.S. Inheritance of disease lesion mimic leaf trait in groundnut. J. Hered. 2002, 93, 50–52. [Google Scholar] [CrossRef] [PubMed]
- Malamy, J.; Carr, J.P.; Klessig, D.F.; Raskin, I. Salicylic acid: A likely endogenous signal in the resistance response of tobacco to viral infection. Science 1990, 250, 1002–1004. [Google Scholar] [CrossRef]
- Takahashi, A.; Kawasaki, T.; Henmi, K.; Shi, I.K.; Kodama, O.; Satoh, H.; Shimamoto, K. Lesion mimic mutants of rice with alterations in early signaling events of defense. Plant J. 1999, 17, 535–545. [Google Scholar] [CrossRef]
- Matin, M.N.; Pandeya, D.; Baek, K.H.; Lee, D.S.; Lee, J.; Kang, H.D.; Kang, S.G. Phenotypic and genotypic analysis of rice lesion mimic mutants. Plant Pathol. J. 2010, 26, 159–169. [Google Scholar] [CrossRef]
- Toufighi, K.; Brady, S.M.; Austin, R.; Ly, E.; Provart, N.J. The Botany Array Resource: E-Northerns, Expression Angling, and promoter analyses. Plant J. 2005, 43, 153–163. [Google Scholar] [CrossRef]
- Dhawan, R.; Luo, H.; Foerster, A.M.; AbuQamar, S.; Du, H.N.; Briggs, S.D.; Scheid, O.M.; Mengiste, T. HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 2009, 21, 1000–1019. [Google Scholar] [CrossRef]
- Ou, B.; Yin, K.Q.; Liu, S.N.; Yang, Y.; Gu, T.; Hui, J.M.; Zhang, L.; Miao, J.; Kondou, Y.; Matsui, M.; et al. A high-throughput screening system for Arabidopsis transcription factors and its application to Med25-dependent transcriptional regulation. Mol. Plant 2011, 4, 546–555. [Google Scholar] [CrossRef] [PubMed]
- Çevik, V.; Kidd, B.N.; Zhang, P.; Hill, C.; Kiddle, S.; Denby, K.J.; Holub, E.B.; Cahill, D.M.; Manners, J.M.; Schenk, P.M.; et al. MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol. 2012, 160, 541–555. [Google Scholar] [CrossRef]
- Chen, R.; Jiang, H.; Li, L.; Zhai, Q.; Qi, L.; Zhou, W.; Liu, X.; Li, H.; Zheng, W.; Sun, J.; et al. The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 2012, 24, 2898–2916. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, G.; Lucob-Agustin, N.; Kashihara, K.; Fujii, Y.; Inukai, Y.; Gomi, K. Rice MEDIATOR25, OsMED25, is an essential subunit for jasmonate-mediated root development and OsMYC2-mediated leaf senescence. Plant Sci. 2021, 36, 110853. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, J.; Yang, G.; Xin, Y.; Wang, Z.; Yan, W.; Chen, Z.; Tang, X.; Xia, J. Overexpression of OsMed16 Inhibits the Growth of Rice and Causes Spontaneous Cell Death. Genes 2021, 12, 656. https://doi.org/10.3390/genes12050656
Jiang J, Yang G, Xin Y, Wang Z, Yan W, Chen Z, Tang X, Xia J. Overexpression of OsMed16 Inhibits the Growth of Rice and Causes Spontaneous Cell Death. Genes. 2021; 12(5):656. https://doi.org/10.3390/genes12050656
Chicago/Turabian StyleJiang, Jie, Guangzhe Yang, Yafeng Xin, Zhigang Wang, Wei Yan, Zhufeng Chen, Xiaoyan Tang, and Jixing Xia. 2021. "Overexpression of OsMed16 Inhibits the Growth of Rice and Causes Spontaneous Cell Death" Genes 12, no. 5: 656. https://doi.org/10.3390/genes12050656
APA StyleJiang, J., Yang, G., Xin, Y., Wang, Z., Yan, W., Chen, Z., Tang, X., & Xia, J. (2021). Overexpression of OsMed16 Inhibits the Growth of Rice and Causes Spontaneous Cell Death. Genes, 12(5), 656. https://doi.org/10.3390/genes12050656





