Annotation and Molecular Characterisation of the TaIRO3 and TaHRZ Iron Homeostasis Genes in Bread Wheat (Triticum aestivum L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Validation of TaIRO3 and TaHRZ Genes
2.2. Phylogenetic Analyses
2.3. Bread Wheat Tissue Sampling and Quantitative Reverse Transcription-PCR (qRT-PCR) Analyses of the TaIRO3 and TaHRZ Genes
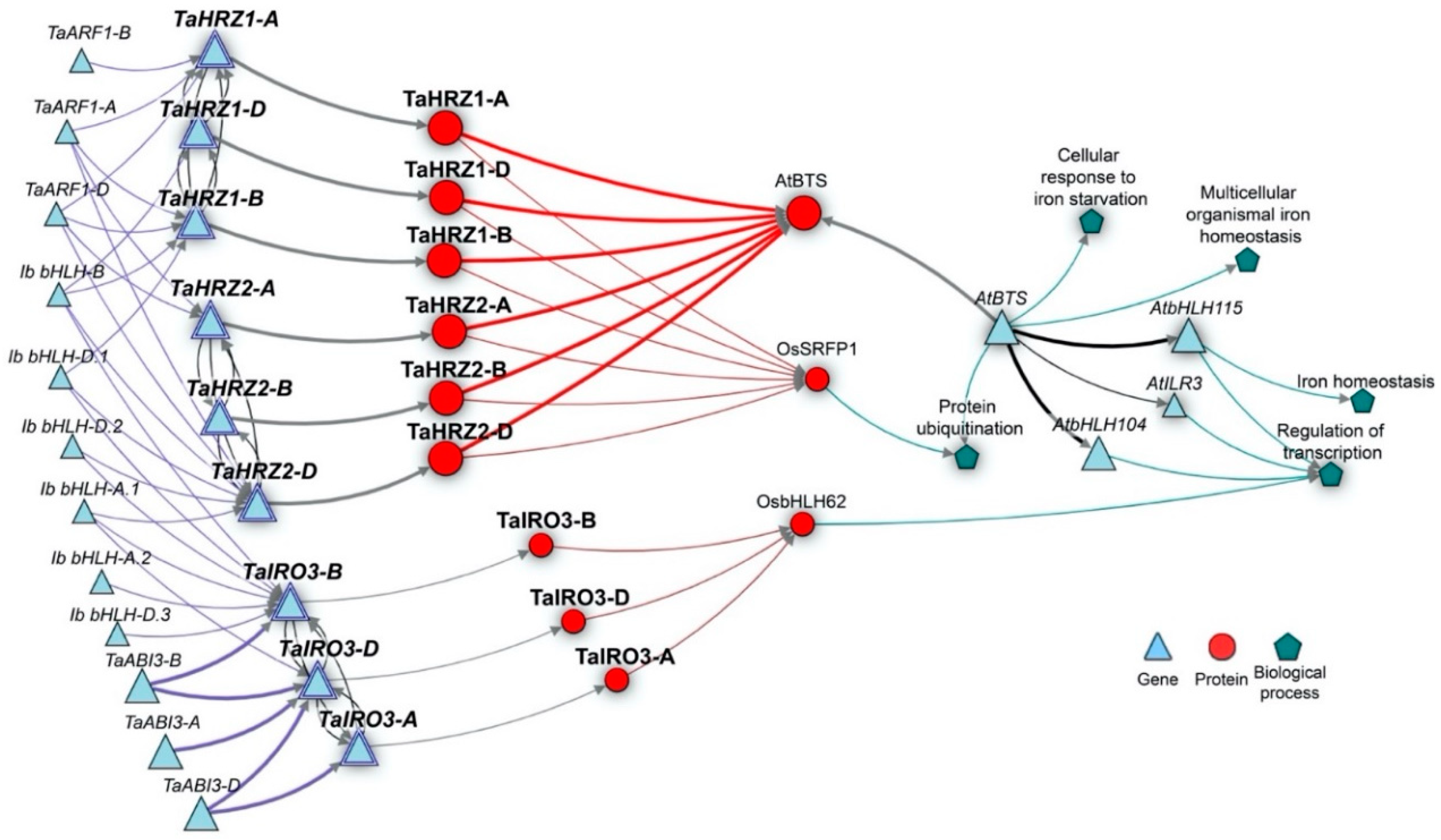
2.4. Gene Network Construction
2.5. Statistical Analysis
3. Results
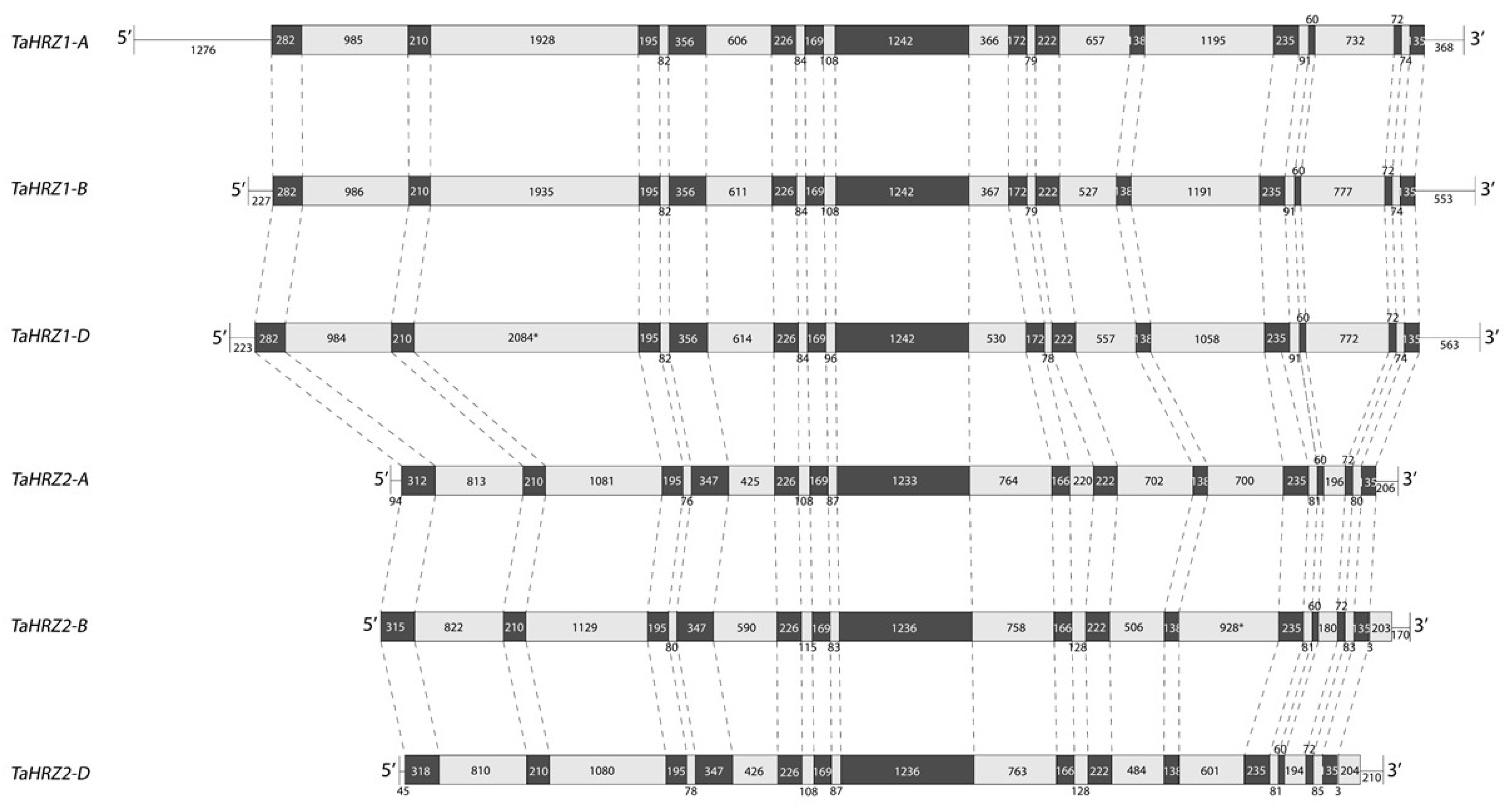
3.1. Three TaIRO3 Genes Are Located on Chromosomal Group 2, and Six TaHRZ Genes Are Located on Chromosomal Groups 1 and 3
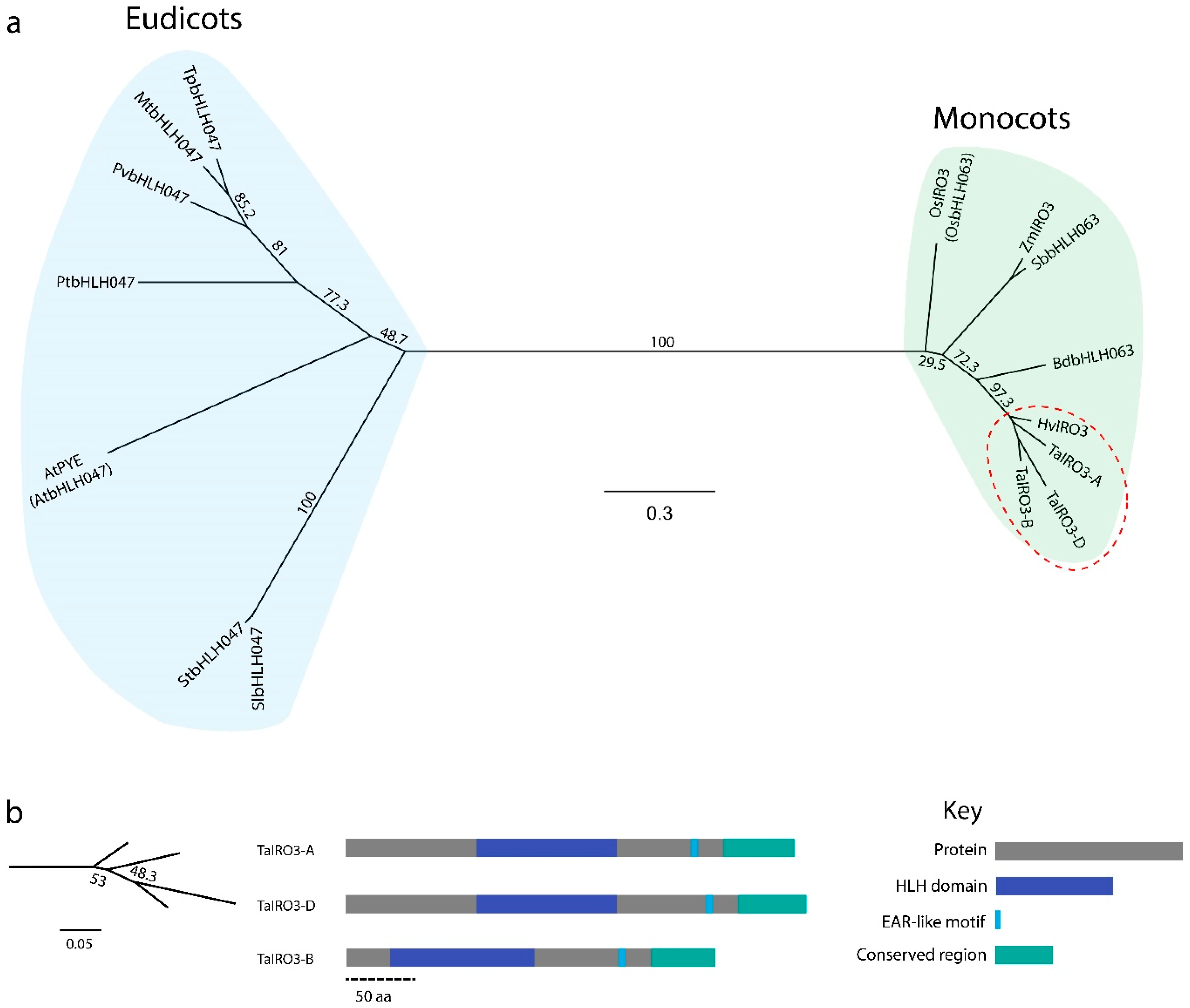
3.2. IRO3 Proteins Are Conserved within Graminoids and Separate between Monocots and Eudicots
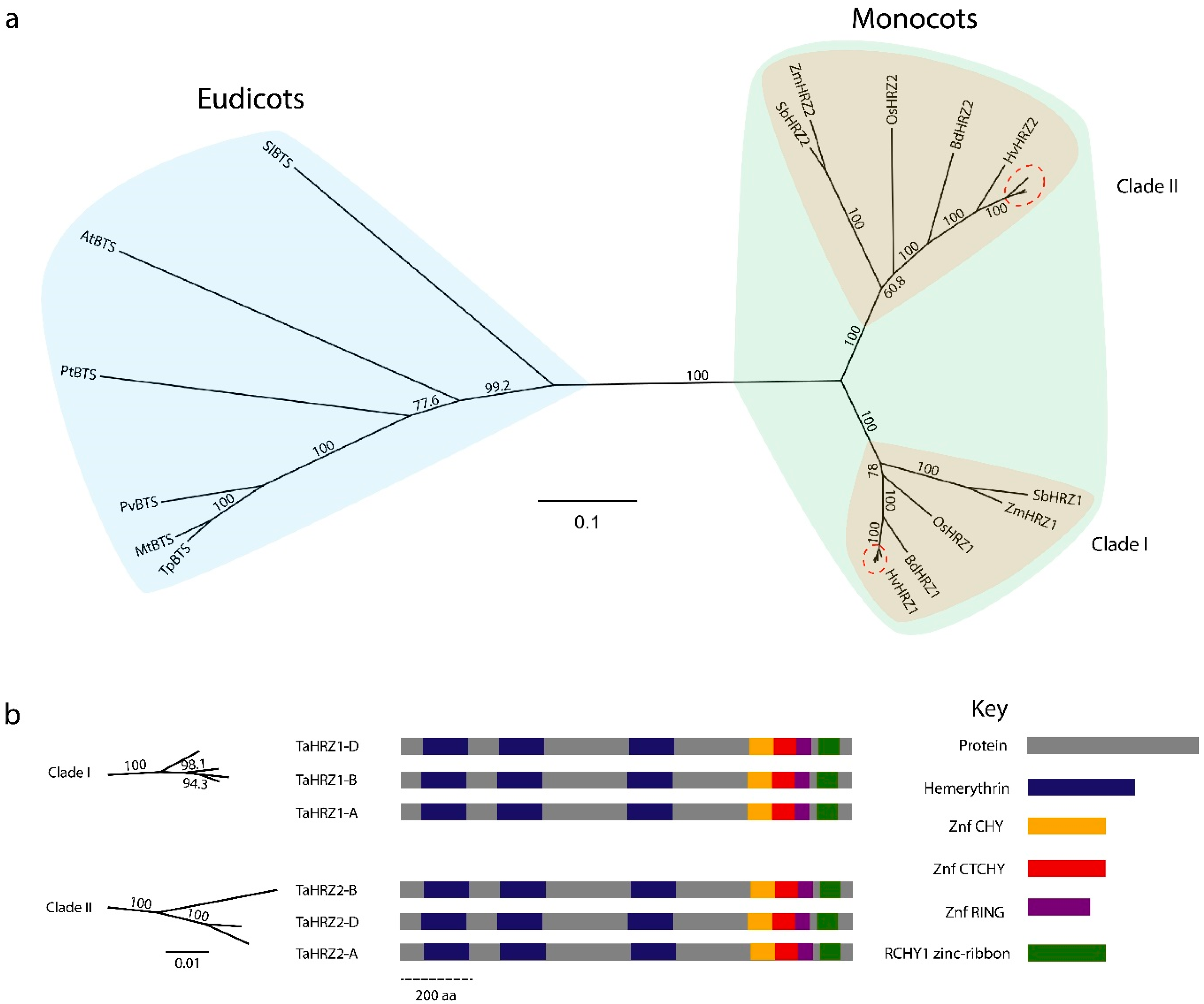
3.3. HRZ Proteins Are Highly Conserved and Separate into Two Clades in Graminaceous Species
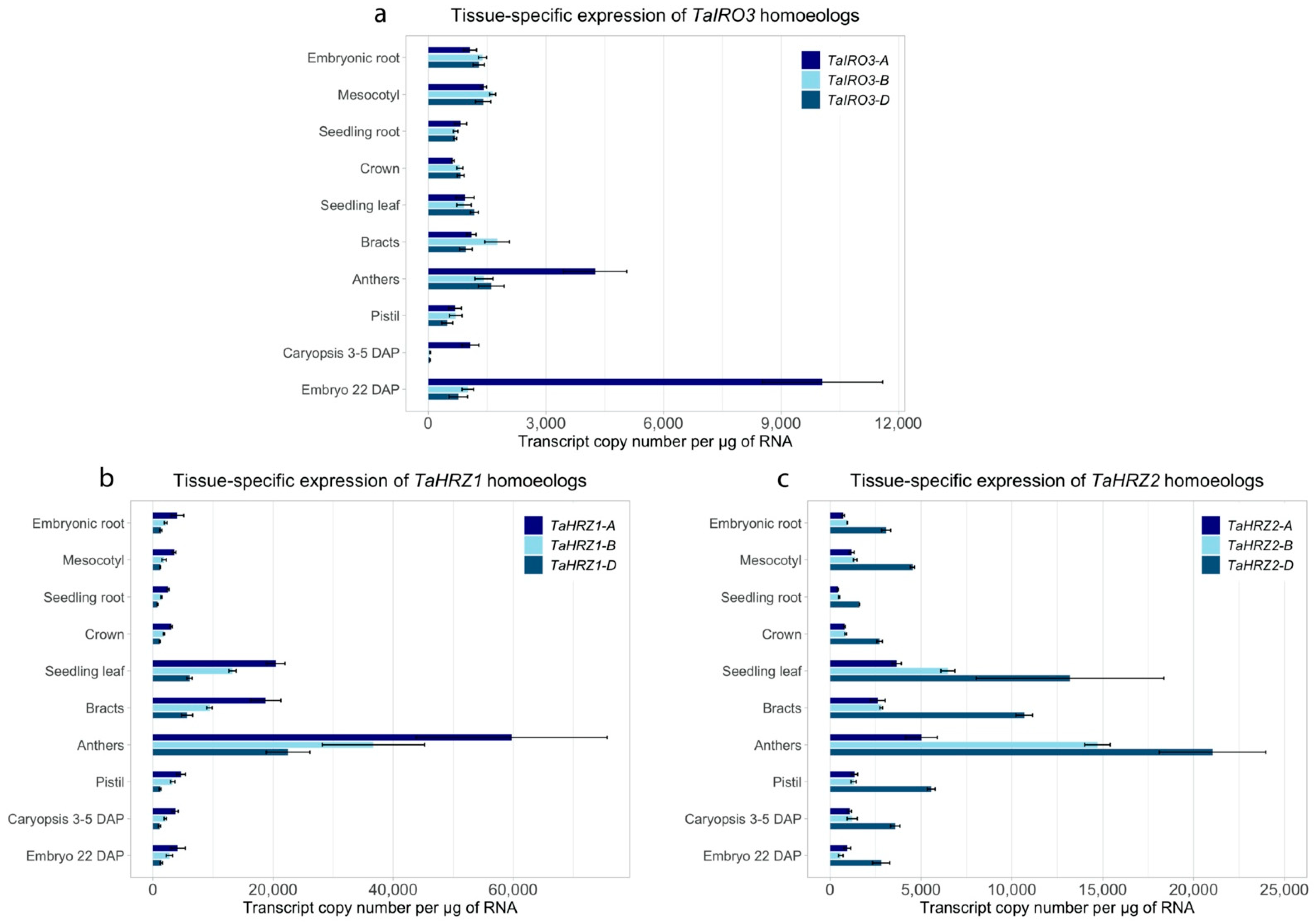
3.4. The TaIRO3, TaHRZ1, and TaHRZ2 Homoeologs Have Distinct Expression Patterns in Bread Wheat Tissues and Are Upregulated in Response to Fe Deficiency
3.5. The TaIRO3 and TaHRZ Genes Are Associated with Regulatory Components of Fe Homeostasis in Arabidopsis and Rice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kroh, G.E.; Pilon, M. Regulation of Iron Homeostasis and Use in Chloroplasts. Int. J. Mol. Sci. 2020, 21, 3395. [Google Scholar] [CrossRef]
- Ravet, K.; Pilon, M. Copper and Iron Homeostasis in Plants: The Challenges of Oxidative Stress. Antioxid. Redox Signal. 2013, 19, 919–932. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nishizawa, N.K. Iron Uptake, Translocation, and Regulation in Higher Plants. Annu. Rev. Plant Biol. 2012, 63, 131–152. [Google Scholar] [CrossRef]
- Connorton, J.M.; Balk, J.; Rodríguez-Celma, J. Iron homeostasis in plants—A brief overview. Metallomics 2017, 9, 813–823. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nozoye, T.; Nishizawa, N.K. Iron transport and its regulation in plants. Free Radic. Biol. Med. 2019, 133, 11–20. [Google Scholar] [CrossRef]
- Gao, F.; Robe, K.; Gaymard, F.; Izquierdo, E.; Dubos, C. The Transcriptional Control of Iron Homeostasis in Plants: A Tale of bHLH Transcription Factors? Front. Plant Sci. 2019, 10, 6. [Google Scholar] [CrossRef]
- Riaz, N.; Guerinot, M.L. All together now: Regulation of the iron deficiency response. J. Exp. Bot. 2021, 72, 2045–2055. [Google Scholar] [CrossRef]
- Gao, F.; Dubos, C. Transcriptional integration of plant responses to iron availability. J. Exp. Bot. 2021, 72, 2056–2070. [Google Scholar] [CrossRef]
- Kobayashi, T.; Itai, R.N.; Aung, M.S.; Senoura, T.; Nakanishi, H.; Nishizawa, N.K. The rice transcription factor IDEF1 directly binds to iron and other divalent metals for sensing cellular iron status. Plant J. 2011, 69, 81–91. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nagasaka, S.; Senoura, T.; Itai, R.N.; Nakanishi, H.; Nishizawa, N.K. Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nat. Commun. 2013, 4, 2792. [Google Scholar] [CrossRef]
- Selote, D.; Samira, R.; Matthiadis, A.; Gillikin, J.W.; Long, T.A. Iron-Binding E3 Ligase Mediates Iron Response in Plants by Targeting Basic Helix-Loop-Helix Transcription Factors. Plant Physiol. 2015, 167, 273–286. [Google Scholar] [CrossRef]
- Stone, S.L. The role of ubiquitin and the 26S proteasome in plant abiotic stress signaling. Front. Plant Sci. 2014, 5, 135. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Yao, X.; Liang, G.; Yu, D. POSITIVE REGULATOR OF IRON HOMEOSTASIS1, OsPRI1, Facilitates Iron Homeostasis. Plant Physiol. 2017, 175, 543–554. [Google Scholar] [CrossRef]
- Kobayashi, T.; Ozu, A.; Kobayashi, S.; An, G.; Jeon, J.-S.; Nishizawa, N.K. OsbHLH058 and OsbHLH059 transcription factors positively regulate iron deficiency responses in rice. Plant Mol. Biol. 2019, 101, 471–486. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Pu, M.; Xu, P.; Liang, G.; Yu, D. Oryza sativa POSITIVE REGULATOR OF IRON DEFICIENCY RESPONSE 2 (OsPRI2) and OsPRI3 are involved in the maintenance of Fe homeostasis. Plant Cell Environ. 2020, 43, 261–274. [Google Scholar] [CrossRef]
- Ogo, Y.; Itai, R.N.; Kobayashi, T.; Aung, M.S.; Nakanishi, H.; Nishizawa, N.K. OsIRO2 is responsible for iron utilization in rice and improves growth and yield in calcareous soil. Plant Mol. Biol. 2011, 75, 593–605. [Google Scholar] [CrossRef]
- Long, T.A.; Tsukagoshi, H.; Busch, W.; Lahner, B.; Salt, D.E.; Benfey, P.N. The bHLH Transcription Factor POPEYE Regulates Response to Iron Deficiency in Arabidopsis Roots. Plant Cell 2010, 22, 2219–2236. [Google Scholar] [CrossRef]
- Zheng, L.; Ying, Y.; Wang, L.; Wang, F.; Whelan, J.; Shou, H. Identification of a novel iron regulated basic helix-loop-helix protein involved in Fe homeostasis in Oryza sativa. BMC Plant Biol. 2010, 10, 166. [Google Scholar] [CrossRef]
- Wang, W.; Ye, J.; Ma, Y.; Wang, T.; Shou, H.; Zheng, L. OsIRO3 Plays an Essential Role in Iron Deficiency Responses and Regulates Iron Homeostasis in Rice. Plants 2020, 9, 1095. [Google Scholar] [CrossRef]
- Wang, F.; Itai, R.N.; Nozoye, T.; Kobayashi, T.; Nishizawa, N.K.; Nakanishi, H. The bHLH protein OsIRO3 is critical for plant survival and iron (Fe) homeostasis in rice (Oryza sativa L.) under Fe-deficient conditions. Soil Sci. Plant Nutr. 2020, 66, 579–592. [Google Scholar] [CrossRef]
- ABARES, Agricultural Forecasts and Outlook: September 2020. 2020. Available online: https://doi.org/10.25814/5f3caa10eee79 (accessed on 8 October 2020).
- Chen, Y.; Barak, P. Iron Nutrition of Plants in Calcareous Soils. Adv. Agron. 1982, 35, 217–240. [Google Scholar] [CrossRef]
- Lindsay, W.L. Soil and plant relationships associated with iron deficiency with emphasis on nutrient interactions. J. Plant Nutr. 1984, 7, 489–500. [Google Scholar] [CrossRef]
- Adamski, N.M.; Borrill, P.; Brinton, J.; A Harrington, S.; Marchal, C.; Bentley, A.R.; Bovill, W.D.; Cattivelli, L.; Cockram, J.; Contreras-Moreira, B.; et al. A roadmap for gene functional characterisation in crops with large genomes: Lessons from polyploid wheat. eLife 2020, 9, e55646. [Google Scholar] [CrossRef]
- The International Wheat Genome Sequencing Consortium (IWGSC); Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [PubMed]
- Bolser, D.M.; Kerhornou, A.; Walts, B.; Kersey, P. Triticeae Resources in Ensembl Plants. Plant Cell Physiol. 2015, 56, e3. [Google Scholar] [CrossRef] [PubMed]
- Bonneau, J.; O’Brien, M.; Plett, D.C.; Johnson, A.A.T. Genetic Pathways Important for Iron Nutrition and Biofortification of Bread Wheat. Annu. Plant Rev. Online 2019, 237–272. [Google Scholar] [CrossRef]
- Wang, M.; Gong, J.; Bhullar, N.K. Iron deficiency triggered transcriptome changes in bread wheat. Comput. Struct. Biotechnol. J. 2020, 18, 2709–2722. [Google Scholar] [CrossRef]
- Beasley, J.T.; Bonneau, J.P.; Johnson, A.A.T. Characterisation of the nicotianamine aminotransferase and deoxymugineic acid synthase genes essential to Strategy II iron uptake in bread wheat (Triticum aestivum L.). PLoS ONE 2017, 12, e0177061. [Google Scholar] [CrossRef]
- Bonneau, J.; Baumann, U.; Beasley, J.; Julien, B.; Johnson, A.A.T. Identification and molecular characterization of the nicotianamine synthase gene family in bread wheat. Plant Biotechnol. J. 2016, 14, 2228–2239. [Google Scholar] [CrossRef]
- Sharma, S.; Kaur, G.; Kumar, A.; Meena, V.; Kaur, J.; Pandey, A.K. Overlapping transcriptional expression response of wheat zinc-induced facilitator-like transporters emphasize important role during Fe and Zn stress. BMC Mol. Biol. 2019, 20, 1–17. [Google Scholar] [CrossRef]
- Sharma, S.; Kaur, G.; Kumar, A.; Meena, V.; Ram, H.; Kaur, J.; Pandey, A.K. Gene Expression Pattern of Vacuolar-Iron Transporter-Like (VTL) Genes in Hexaploid Wheat during Metal Stress. Plants 2020, 9, 229. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Kawakami, Y.; Bhullar, N.K. Molecular Analysis of Iron Deficiency Response in Hexaploid Wheat. Front. Sustain. Food Syst. 2019, 3, 67. [Google Scholar] [CrossRef]
- Kaur, G.; Shukla, V.; Kumar, A.; Kaur, M.; Goel, P.; Singh, P.; Shukla, A.; Meena, V.; Kaur, J.; Singh, J.; et al. Integrative analysis of hexaploid wheat roots identifies signature components during iron starvation. J. Exp. Bot. 2019, 70, 6141–6161. [Google Scholar] [CrossRef]
- Ramirez-Gonzalez, R.H.; Borrill, P.; Lang, D.; Harrington, S.A.; Brinton, J.; Venturini, L.; Davey, M.; Jacobs, J.; Van Ex, F.; Pasha, A.; et al. The transcriptional landscape of polyploid wheat. Science 2018, 361, eaar6089. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; De La Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Alaux, M.; International Wheat Genome Sequencing Consortium; Rogers, J.; Letellier, T.; Flores, R.; Alfama, F.; Pommier, C.; Mohellibi, N.; Durand, S.; Kimmel, E.; et al. Linking the International Wheat Genome Sequencing Consortium bread wheat reference genome sequence to wheat genetic and phenomic data. Genome Biol. 2018, 19, 111. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.L.; Contreras-Moreira, B.; De Silva, N.; Maslen, G.; Akanni, W.; Allen, J.; Alvarez-Jarreta, J.; Barba, M.; Bolser, D.M.; Cambell, L.; et al. Ensembl Genomes 2020—enabling non-vertebrate genomic research. Nucleic Acids Res. 2020, 48, D689–D695. [Google Scholar] [CrossRef]
- Boguski, M.S.; Lowe, T.M.; Tolstoshev, C.M. dbEST—Database for “expressed sequence tags”. Nat. Genet. 1993, 4, 332–333. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.S.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Chilton, J.M.; Grüning, B.; Johnson, J.E.; Soranzo, N. NCBI BLAST+ integrated into Galaxy. GigaScience 2015, 4, 39. [Google Scholar] [CrossRef]
- Berardini, T.Z.; Reiser, L.; Li, D.; Mezheritsky, Y.; Muller, R.; Strait, E.; Huala, E. The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 2015, 53, 474–485. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Schreiber, A.W.; Sutton, T.; A Caldo, R.; Kalashyan, E.; Lovell, B.; Mayo, G.; Muehlbauer, G.J.; Druka, A.; Waugh, R.; Wise, R.P.; et al. Comparative transcriptomics in the Triticeae. BMC Genom. 2009, 10, 285. [Google Scholar] [CrossRef] [PubMed]
- Sears, E.R. The Aneuploids of Common Wheat; University of Missouri, College of Agriculture, Agricultural Experiment Station: Columbia, MO, USA, 1954; Volume 572, pp. 1–58. [Google Scholar]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Borrill, P.; Ramirez-Gonzalez, R.; Uauy, C. expVIP: A Customizable RNA-seq Data Analysis and Visualization Platform. Plant Physiol. 2016, 170, 2172–2186. [Google Scholar] [CrossRef]
- Harrington, S.A.; Backhaus, A.E.; Singh, A.; Hassani-Pak, K.; Uauy, C. The Wheat GENIE3 Network Provides Biologically-Relevant Information in Polyploid Wheat. G3 Genes Genomes Genet. 2020, 10, 3675–3686. [Google Scholar] [CrossRef]
- Pires, N.; Dolan, L. Origin and Diversification of Basic-Helix-Loop-Helix Proteins in Plants. Mol. Biol. Evol. 2009, 27, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis Basic/Helix-Loop-Helix Transcription Factor Family [W]. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef]
- Buck, M.J.; Atchley, W.R. Phylogenetic Analysis of Plant Basic Helix-Loop-Helix Proteins. J. Mol. Evol. 2003, 56, 742–750. [Google Scholar] [CrossRef]
- Kagale, S.; Rozwadowski, K. EAR motif-mediated transcriptional repression in plants: An underlying mechanism for epigenetic regulation of gene expression. Epigenetics 2011, 6, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lei, R.; Pu, M.; Cai, Y.; Lu, C.; Li, Z.; Liang, G. bHLH11 negatively regulates Fe homeostasis by its EAR motifs recruiting corepressors in Arabidopsis. BioRxiv 2020. [Google Scholar] [CrossRef]
- Spielmann, J.; Vert, G. The many facets of protein ubiquitination and degradation in plant root iron-deficiency responses. J. Exp. Bot. 2021, 72, 2071–2082. [Google Scholar] [CrossRef]
- Sheriff, S.; Hendrickson, W.A.; Smith, J.L. Structure of myohemerythrin in the azidomet state at 1.7 1.3 Å resolution. J. Mol. Biol. 1987, 197, 273–296. [Google Scholar] [CrossRef]
- Rodríguez-Celma, J.; Chou, H.; Kobayashi, T.; Long, T.A.; Balk, J. Hemerythrin E3 Ubiquitin Ligases as Negative Regulators of Iron Homeostasis in Plants. Front. Plant Sci. 2019, 10, 98. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, S.; Khokhar, W.; Jabre, I.; Reddy, A.S.N.; Byrne, L.J.; Wilson, C.M.; Syed, N.H. Alternative Splicing and Protein Diversity: Plants Versus Animals. Front. Plant Sci. 2019, 10, 708. [Google Scholar] [CrossRef]
- Dong, C.; He, F.; Berkowitz, O.; Liu, J.; Cao, P.; Tang, M.; Shi, H.; Wang, W.; Li, Q.; Shen, Z.; et al. Alternative Splicing Plays a Critical Role in Maintaining Mineral Nutrient Homeostasis in Rice (Oryza sativa). Plant Cell 2018, 30, 2267–2285. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Du, B.; Liu, D.; Qi, X. Splicing factor SR34b mutation reduces cadmium tolerance in Arabidopsis by regulating iron-regulated transporter 1 gene. Biochem. Biophys. Res. Commun. 2014, 455, 312–317. [Google Scholar] [CrossRef]
- Schuler, M.; Rellán-Álvarez, R.; Fink-Straube, C.; Abadía, J.; Bauer, P. Nicotianamine Functions in the Phloem-Based Transport of Iron to Sink Organs, in Pollen Development and Pollen Tube Growth in Arabidopsis. Plant Cell 2012, 24, 2380–2400. [Google Scholar] [CrossRef]
- Pfeifer, M.; Kugler, K.G.; Sandve, S.R.; Zhan, B.; Rudi, H.; Hvidsten, T.R.; Mayer, K.F.X.; Olsen, O.-A. International Wheat Genome Sequencing Consortium Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 2014, 345, 1250091. [Google Scholar] [CrossRef] [PubMed]
- Choulet, F.; Alberti, A.; Theil, S.; Glover, N.; Barbe, V.; Daron, J.; Pingault, L.; Sourdille, P.; Couloux, A.; Paux, E.; et al. Structural and functional partitioning of bread wheat chromosome 3B. Science 2014, 345, 1249721. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Ogo, Y.; Itai, R.N.; Nakanishi, H.; Takahashi, M.; Mori, S.; Nishizawa, N.K. The transcription factor IDEF1 regulates the response to and tolerance of iron deficiency in plants. Proc. Natl. Acad. Sci. USA 2007, 104, 19150–19155. [Google Scholar] [CrossRef] [PubMed]
- McElver, J.; Tzafrir, I.; Aux, G.; Rogers, R.; Ashby, C.; Smith, K.; Thomas, C.; Schetter, A.; Zhou, Q.; A Cushman, M.; et al. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 2001, 159, 1751–1763. [Google Scholar] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carey-Fung, O.; Beasley, J.T.; Johnson, A.A.T. Annotation and Molecular Characterisation of the TaIRO3 and TaHRZ Iron Homeostasis Genes in Bread Wheat (Triticum aestivum L.). Genes 2021, 12, 653. https://doi.org/10.3390/genes12050653
Carey-Fung O, Beasley JT, Johnson AAT. Annotation and Molecular Characterisation of the TaIRO3 and TaHRZ Iron Homeostasis Genes in Bread Wheat (Triticum aestivum L.). Genes. 2021; 12(5):653. https://doi.org/10.3390/genes12050653
Chicago/Turabian StyleCarey-Fung, Oscar, Jesse T. Beasley, and Alexander A. T. Johnson. 2021. "Annotation and Molecular Characterisation of the TaIRO3 and TaHRZ Iron Homeostasis Genes in Bread Wheat (Triticum aestivum L.)" Genes 12, no. 5: 653. https://doi.org/10.3390/genes12050653
APA StyleCarey-Fung, O., Beasley, J. T., & Johnson, A. A. T. (2021). Annotation and Molecular Characterisation of the TaIRO3 and TaHRZ Iron Homeostasis Genes in Bread Wheat (Triticum aestivum L.). Genes, 12(5), 653. https://doi.org/10.3390/genes12050653





