Metagenomics Assessment of Soil Fertilization on the Chemotaxis and Disease Suppressive Genes Abundance in the Maize Rhizosphere
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description, Samples Collection, and DNA Extraction
2.2. Sequencing of the Community DNA
2.3. Sequence Processing, Annotation and Statistical Analysis
3. Results
3.1. Metagenomics Dataset
3.2. Abundant Bacterial Phyla in the Samples
3.3. Chemotaxis Genes Present at the Maize Rhizosphere
3.4. Antimicrobial and Siderophore Genes Contributing to Disease Suppressive Soil
3.5. Diversity (α and β) Estimation of the Chemotaxis and the Disease Suppressive Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parkinson, J.S.; Hazelbauer, G.L.; Falke, J.J. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 2015, 23, 257–266. [Google Scholar] [CrossRef]
- Sourjik, V.; Wingreen, N.S. Responding to chemical gradients: Bacterial chemotaxis. Curr. Opin. Cell Biol. 2012, 24, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Wuichet, K.; Zhulin, I.B. Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal. 2010, 3, ra50. [Google Scholar] [CrossRef]
- Schweinitzer, T.; Josenhans, C. Bacterial energy taxis: A global strategy? Arch. Microbiol. 2010, 192, 507–520. [Google Scholar] [CrossRef]
- Zhu, Y.-G.; Su, J.-Q.; Cao, Z.; Xue, K.; Quensen, J.; Guo, G.-X.; Yang, Y.-F.; Zhou, J.; Chu, H.-Y.; Tiedje, J.M. A buried Neolithic paddy soil reveals loss of microbial functional diversity after modern rice cultivation. Sci. Bull. 2016, 61, 1052–1060. [Google Scholar] [CrossRef]
- Tejada, M.; Gonzalez, J.; García-Martínez, A.; Parrado, J. Effects of different green manures on soil biological properties and maize yield. Bioresour. Technol. 2008, 99, 1758–1767. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, R.; Kundu, S.; Prakash, V.; Gupta, H. Sustainability under combined application of mineral and organic fertilizers in a rainfed soybean–wheat system of the Indian Himalayas. Eur. J. Agron. 2008, 28, 33–46. [Google Scholar] [CrossRef]
- Ajilogba, C.F.; Babalola, O.O.; Ahmad, F. Antagonistic effects of Bacillus species in biocontrol of tomato Fusarium wilt. Stud. Ethno Med. 2013, 7, 205–216. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef] [PubMed]
- Marschner, P.; Crowley, D.; Yang, C.H. Development of specific rhizosphere bacterial communities in relation to plant species, nutrition and soil type. Plant Soil 2004, 261, 199–208. [Google Scholar] [CrossRef]
- Babalola, O.O.; Glick, B.R. The use of microbial inoculants in African agriculture: Current practice and future prospects. J. Food Agric. Environ. 2012, 10, 540–549. [Google Scholar]
- Enebe, M.C.; Babalola, O.O. The impact of microbes in the orchestration of plants’ resistance to biotic stress: A disease management approach. Appl. Microbiol. Biotechnol. 2019, 103, 9–25. [Google Scholar] [CrossRef]
- Adegboye, M.F.; Babalola, O.O. Evaluation of antibiotic biosynthetic potential of actinomycete isolates to produce antimicrobial agents. Microbiol. Res. J. Int. 2015, 243–254. [Google Scholar] [CrossRef]
- Arseneault, T.; Goyer, C.; Filion, M. Pseudomonas fluorescens LBUM223 increases potato yield and reduces common scab symptoms in the field. Phytopathology 2015, 105, 1311–1317. [Google Scholar] [CrossRef]
- Latz, E.; Eisenhauer, N.; Rall, B.C.; Allan, E.; Roscher, C.; Scheu, S.; Jousset, A. Plant diversity improves protection against soil-borne pathogens by fostering antagonistic bacterial communities. J. Ecol. 2012, 100, 597–604. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; Mazzola, M. Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu. Rev. Phytopathol. 2012, 50, 403–424. [Google Scholar] [CrossRef]
- Ajayi, A.; Onibokun, E.; George, F.; Atolagbe, O. Isolation and characterization of chitinolytic bacteria for chitinase production from the African Catfish, Clarias gariepinus (Burchell, 1822). Res. J. Microbiol. 2016, 11, 119–125. [Google Scholar] [CrossRef]
- Adegboye, M.F.; Babalola, O.O. Taxonomy and ecology of antibiotic producing actinomycetes. Afr. J. Agric. Res. 2012, 7, 2255–2261. [Google Scholar]
- Enebe, M.C.; Babalola, O.O. Soil fertilization affects the abundance and distribution of carbon and nitrogen cycling genes in the maize rhizosphere. AMB Express 2021, 11, 1–10. [Google Scholar] [CrossRef]
- Adebayo, A.R.; Kutu, F.R.; Sebetha, E.T. Data on root system architecture of water efficient maize as affected by different nitrogen fertilizer rates and plant density. Data Brief 2020, 30, 105561. [Google Scholar] [CrossRef]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A. The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008, 9, 386. [Google Scholar] [CrossRef]
- Kent, W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef]
- Wilke, A.; Harrison, T.; Wilkening, J.; Field, D.; Glass, E.M.; Kyrpides, N.; Mavrommatis, K.; Meyer, F. The M5nr: A novel non-redundant database containing protein sequences and annotations from multiple sources and associated tools. BMC Bioinform. 2012, 13, 141. [Google Scholar] [CrossRef]
- Enebe, M.C.; Babalola, O.O. Effects of inorganic and organic treatments on the microbial community of maize rhizosphere by a shotgun metagenomics approach. Ann. Microbiol. 2020, 70, 1–10. [Google Scholar] [CrossRef]
- Marschner, P.; Kandeler, E.; Marschner, B. Structure and function of the soil microbial community in a long-term fertilizer experiment. Soil Biol. Biochem. 2003, 35, 453–461. [Google Scholar] [CrossRef]
- Lee, S.B.; Lee, C.H.; Jung, K.Y.; Do Park, K.; Lee, D.; Kim, P.J. Changes of soil organic carbon and its fractions in relation to soil physical properties in a long-term fertilized paddy. Soil Tillage Res. 2009, 104, 227–232. [Google Scholar] [CrossRef]
- Enebe, M.C.; Babalola, O.O. The influence of plant growth-promoting rhizobacteria in plant tolerance to abiotic stress: A survival strategy. Appl. Microbiol. Biotechnol. 2018, 102, 7821–7835. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, R.L.; Pieterse, C.M.; Bakker, P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Beier, S.; Bertilsson, S. Bacterial chitin degradation—mechanisms and ecophysiological strategies. Front. Microbiol. 2013, 4, 149. [Google Scholar] [CrossRef]
- Thomas, F.; Hehemann, J.-H.; Rebuffet, E.; Czjzek, M.; Michel, G. Environmental and gut bacteroidetes: The food connection. Front. Microbiol. 2011, 2, 93. [Google Scholar] [CrossRef]
- Chamizo, S.; Mugnai, G.; Rossi, F.; Certini, G.; De Philippis, R. Cyanobacteria inoculation improves soil stability and fertility on different textured soils: Gaining insights for applicability in soil restoration. Front. Environ. Sci. 2018, 6, 49. [Google Scholar] [CrossRef]
- Borneman, J.; Skroch, P.W.; O’Sullivan, K.M.; Palus, J.A.; Rumjanek, N.G.; Jansen, J.L.; Nienhuis, J.; Triplett, E.W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl. Environ. Microbiol. 1996, 62, 1935–1943. [Google Scholar] [CrossRef]
- Smit, E.; Leeflang, P.; Gommans, S.; van den Broek, J.; van Mil, S.; Wernars, K. Diversity and seasonal fluctuations of the dominant members of the bacterial soil community in a wheat field as determined by cultivation and molecular methods. Appl. Environ. Microbiol. 2001, 67, 2284–2291. [Google Scholar] [CrossRef]
- Valinsky, L.; Della Vedova, G.; Scupham, A.J.; Alvey, S.; Figueroa, A.; Yin, B.; Hartin, R.J.; Chrobak, M.; Crowley, D.E.; Jiang, T. Analysis of bacterial community composition by oligonucleotide fingerprinting of rRNA genes. Appl. Environ. Microbiol. 2002, 68, 3243–3250. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Mhete, M.; Eze, P.N.; Rahube, T.O.; Akinyemi, F.O. Soil properties influence bacterial abundance and diversity under different land-use regimes in semi-arid environments. Sci. Afr. 2020, 7, e00246. [Google Scholar] [CrossRef]
- Shivlata, L.; Tulasi, S. Thermophilic and alkaliphilic Actinobacteria: Biology and potential applications. Front. Microbiol. 2015, 6, 1014. [Google Scholar] [CrossRef]
- Martínez-Alonso, M.; Escolano Sánchez, J.; Montesinos Seguí, E.; Gaju, N. Diversity of the bacterial community in the surface soil of a pear orchard based on 16S rRNA gene analysis. Int. Microbiol. 2010, 13, 123–134. [Google Scholar] [PubMed]
- Nemergut, D.R.; Townsend, A.R.; Sattin, S.R.; Freeman, K.R.; Fierer, N.; Neff, J.C.; Bowman, W.D.; Schadt, C.W.; Weintraub, M.N.; Schmidt, S.K. The effects of chronic nitrogen fertilization on alpine tundra soil microbial communities: Implications for carbon and nitrogen cycling. Environ. Microbiol. 2008, 10, 3093–3105. [Google Scholar] [CrossRef]
- Kuramae, E.E.; Gamper, H.A.; Yergeau, E.; Piceno, Y.M.; Brodie, E.L.; DeSantis, T.Z.; Andersen, G.L.; Van Veen, J.A.; Kowalchuk, G.A. Microbial secondary succession in a chronosequence of chalk grasslands. ISME J. 2010, 4, 711–715. [Google Scholar] [CrossRef] [PubMed]
- Perez, C.; Dill-Macky, R.; Kinkel, L.L. Management of soil microbial communities to enhance populations of Fusarium graminearum-antagonists in soil. Plant Soil 2008, 302, 53–69. [Google Scholar] [CrossRef]
- Huang, J.; Li, H.; Yuan, H. Effect of organic amendments on Verticillium wilt of cotton. Crop. Prot. 2006, 25, 1167–1173. [Google Scholar] [CrossRef]
- Ahmad, F.; Babalola, O.O.; Siddiqui, M.A. Integrated Approach for Management of Nematodes in Chickpea. J. Pure Appl. Microbiol. 2012, 6, 1063–1068. [Google Scholar]
- Zeng, J.; Liu, X.; Song, L.; Lin, X.; Zhang, H.; Shen, C.; Chu, H. Nitrogen fertilization directly affects soil bacterial diversity and indirectly affects bacterial community composition. Soil Biol. Biochem. 2016, 92, 41–49. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; Van Der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Bouwmeester, H.J.; Roux, C.; Lopez-Raez, J.A.; Becard, G. Rhizosphere communication of plants, parasitic plants and AM fungi. Trends Plant Sci. 2007, 12, 224–230. [Google Scholar] [CrossRef]
- Neal, A.L.; Ahmad, S.; Gordon-Weeks, R.; Ton, J. Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS ONE 2012, 7, e35498. [Google Scholar] [CrossRef]
- Zhang, Y.; Shen, H.; He, X.; Thomas, B.W.; Lupwayi, N.Z.; Hao, X.; Thomas, M.C.; Shi, X. Fertilization shapes bacterial community structure by alteration of soil pH. Front. Microbiol. 2017, 8, 1325. [Google Scholar] [CrossRef]
- Geisseler, D.; Scow, K.M. Long-term effects of mineral fertilizers on soil microorganisms—A review. Soil Biol. Biochem. 2014, 75, 54–63. [Google Scholar] [CrossRef]
- Zhulin, I.B. The Superfamily of Chemotaxis Transducers: From Physiology to Genomics and Back. Sci. Direct 2001, 45, 157–198. [Google Scholar]
- Chao, X.; Muff, T.J.; Park, S.-Y.; Zhang, S.; Pollard, A.M.; Ordal, G.W.; Bilwes, A.M.; Crane, B.R. A receptor-modifying deamidase in complex with a signaling phosphatase reveals reciprocal regulation. Cell 2006, 124, 561–571. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Igo, M.M.; Ninfa, A.J.; Stock, J.B.; Silhavy, T.J. Phosphorylation and dephosphorylation of a bacterial transcriptional activator by a transmembrane receptor. Genes Dev. 1989, 3, 1725–1734. [Google Scholar] [CrossRef] [PubMed]
- Zschiedrich, C.P.; Keidel, V.; Szurmant, H. Molecular mechanisms of two-component signal transduction. J. Mol. Biol. 2016, 428, 3752–3775. [Google Scholar] [CrossRef] [PubMed]
- Güvener, Z.T.; Harwood, C.S. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol. Microbiol. 2007, 66, 1459–1473. [Google Scholar] [CrossRef] [PubMed]
- Sourjik, V. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol. 2004, 12, 569–576. [Google Scholar] [CrossRef]
- Porter, S.L.; Wadhams, G.H.; Armitage, J.P. Signal processing in complex chemotaxis pathways. Nat. Rev. Microbiol. 2011, 9, 153–165. [Google Scholar] [CrossRef]
- Stader, J.; Matsumura, P.; Vacante, D.; Dean, G.; Macnab, R. Nucleotide sequence of the Escherichia coli motB gene and site-limited incorporation of its product into the cytoplasmic membrane. J. Bacteriol. 1986, 166, 244–252. [Google Scholar] [CrossRef]
- Zhao, X.; Norris, S.J.; Liu, J. Molecular architecture of the bacterial flagellar motor in cells. Biochemistry 2014, 53, 4323–4333. [Google Scholar] [CrossRef]
- Matilla, M.A.; Krell, T. Chemoreceptor-based signal sensing. Curr. Opin. Biotechnol. 2017, 45, 8–14. [Google Scholar] [CrossRef]
- Antunez-Lamas, M.; Cabrera, E.; Lopez-Solanilla, E.; Solano, R.; González-Melendi, P.; Chico, J.M.; Toth, I.; Birch, P.; Pritchard, L.; Liu, H. Bacterial chemoattraction towards jasmonate plays a role in the entry of Dickeya dadantii through wounded tissues. Mol. Microbiol. 2009, 74, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Hegde, M.; Englert, D.L.; Schrock, S.; Cohn, W.B.; Vogt, C.; Wood, T.K.; Manson, M.D.; Jayaraman, A. Chemotaxis to the quorum-sensing signal AI-2 requires the Tsr chemoreceptor and the periplasmic LsrB AI-2-binding protein. J. Bacteriol. 2011, 193, 768–773. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Kato, J.; Kuroda, A.; Ikeda, T.; Takiguchi, N.; Ohtake, H. Identification and characterization of two chemotactic transducers for inorganic phosphate in Pseudomonas aeruginosa. J. Bacteriol. 2000, 182, 3400–3404. [Google Scholar] [CrossRef] [PubMed]
- Esuola, C.O.; Babalola, O.O.; Heine, T.; Schwabe, R.; Schlömann, M.; Tischler, D. Identification and characterization of a FAD-dependent putrescine N-hydroxylase (GorA) from Gordonia rubripertincta CWB2. J. Mol. Catal. B Enzym. 2016, 134, 378–389. [Google Scholar] [CrossRef]
- Ahmadi, M.K.; Fawaz, S.; Jones, C.H.; Zhang, G.; Pfeifer, B.A. Total biosynthesis and diverse applications of the nonribosomal peptide-polyketide siderophore yersiniabactin. Appl. Environ. Microbiol. 2015, 81, 5290–5298. [Google Scholar] [CrossRef] [PubMed]
- Sritharan, M. Iron homeostasis in Mycobacterium tuberculosis: Mechanistic insights into siderophore-mediated iron uptake. J. Bacteriol. 2016, 198, 2399–2409. [Google Scholar] [CrossRef]
- Perry, R.D.; Balbo, P.B.; Jones, H.A.; Fetherston, J.D.; DeMoll, E. Yersiniabactin from Yersinia pestis: Biochemical characterization of the siderophore and its role in iron transport and regulation. Microbiology 1999, 145, 1181–1190. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H.; Nonomiya, T.; Usami, M.; Ohta, T.; Ōmura, S. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc. Natl. Acad. Sci. USA 1999, 96, 9509–9514. [Google Scholar] [CrossRef]
- Van Pée, K.-H.; Zehner, S. Enzymology and Molecular Genetics of Biological Halogenation. In Natural Production of Organohalogen Compounds; Springer: Berlin/Heidelberg, Germany, 2003; pp. 171–199. [Google Scholar]
- Kovacevic, S.; Tobin, M.B.; Miller, J.R. The beta-lactam biosynthesis genes for isopenicillin N epimerase and deacetoxycephalosporin C synthetase are expressed from a single transcript in Streptomyces clavuligerus. J. Bacteriol. 1990, 172, 3952–3958. [Google Scholar] [CrossRef]
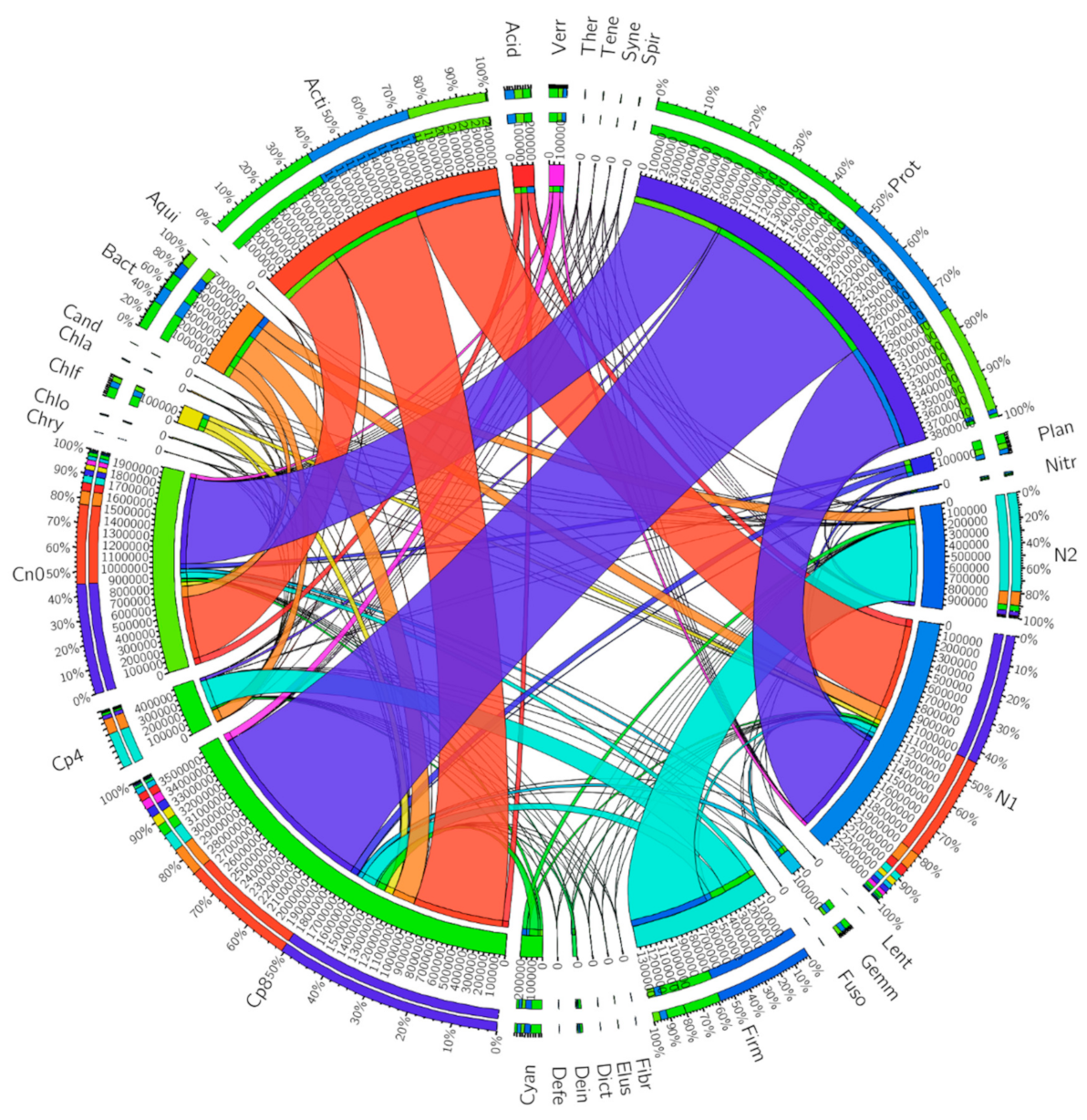
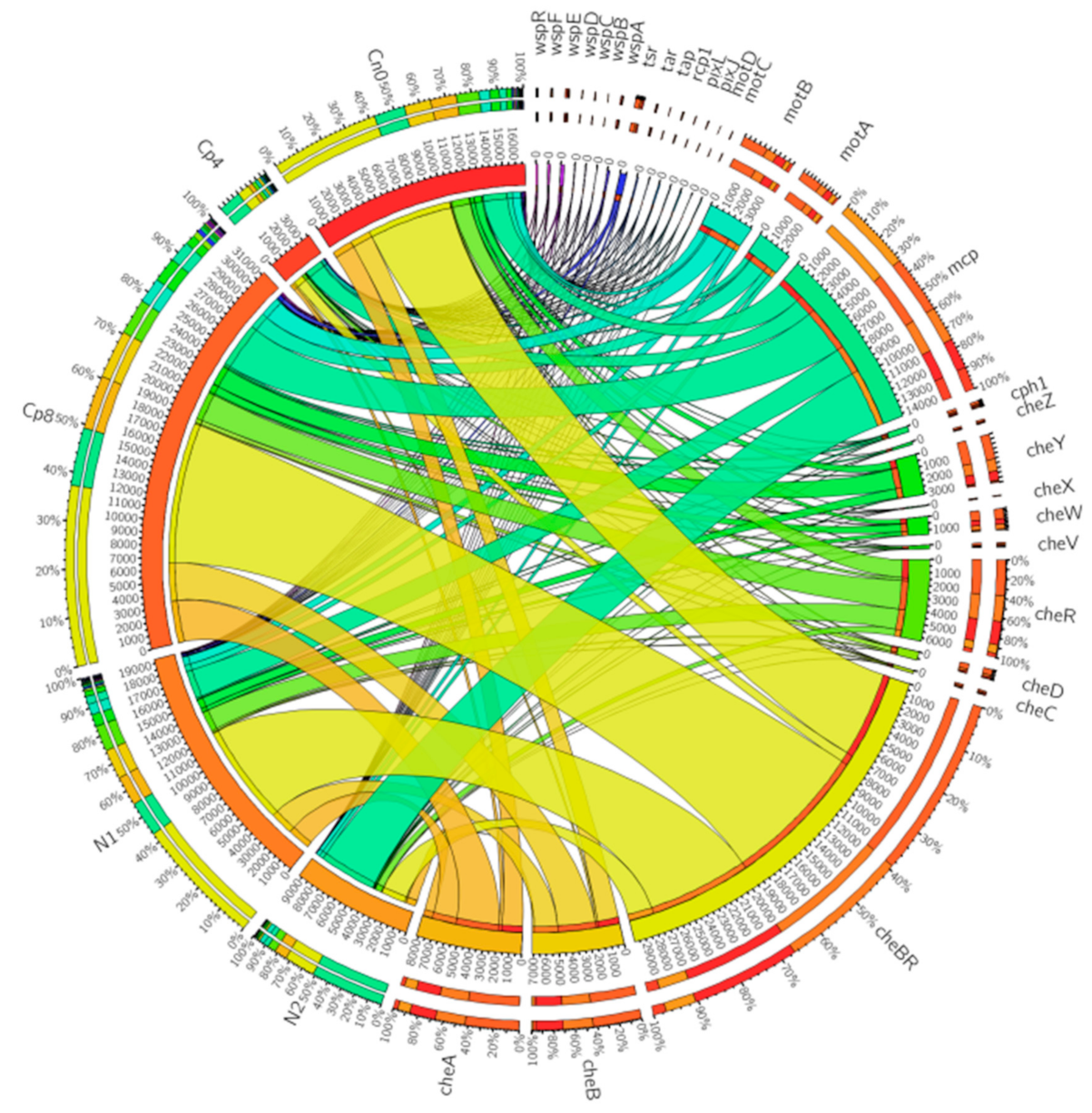

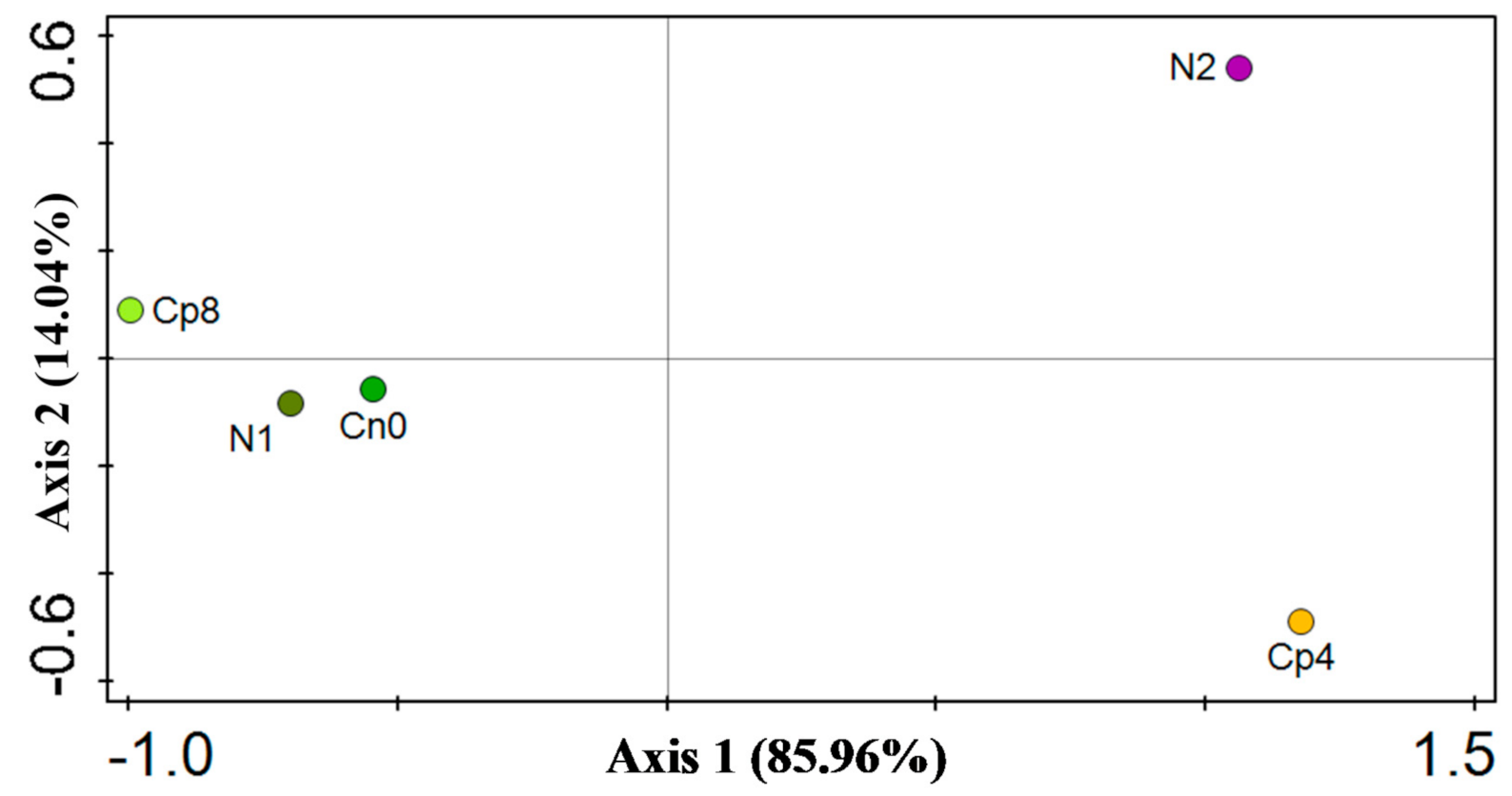
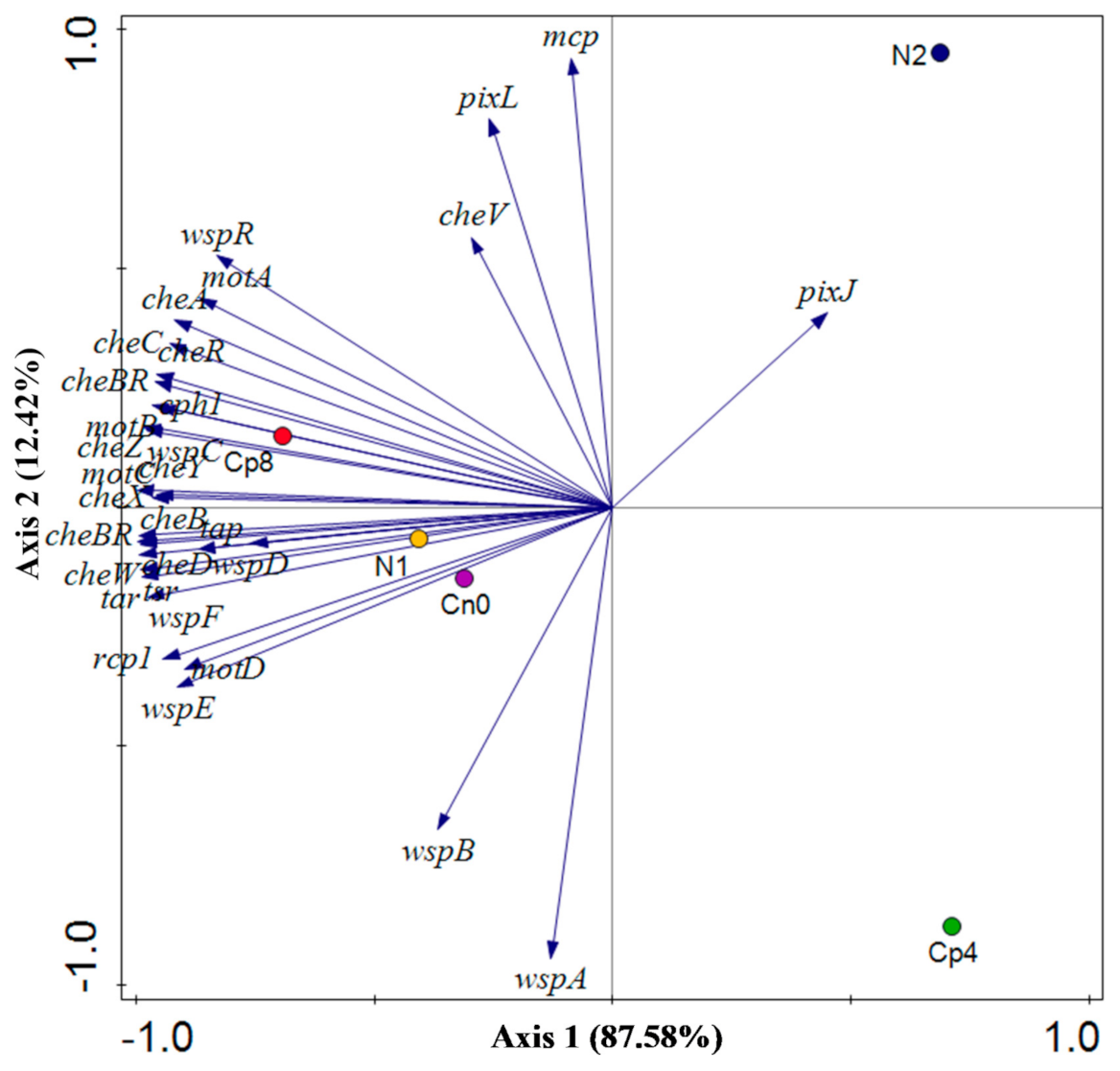
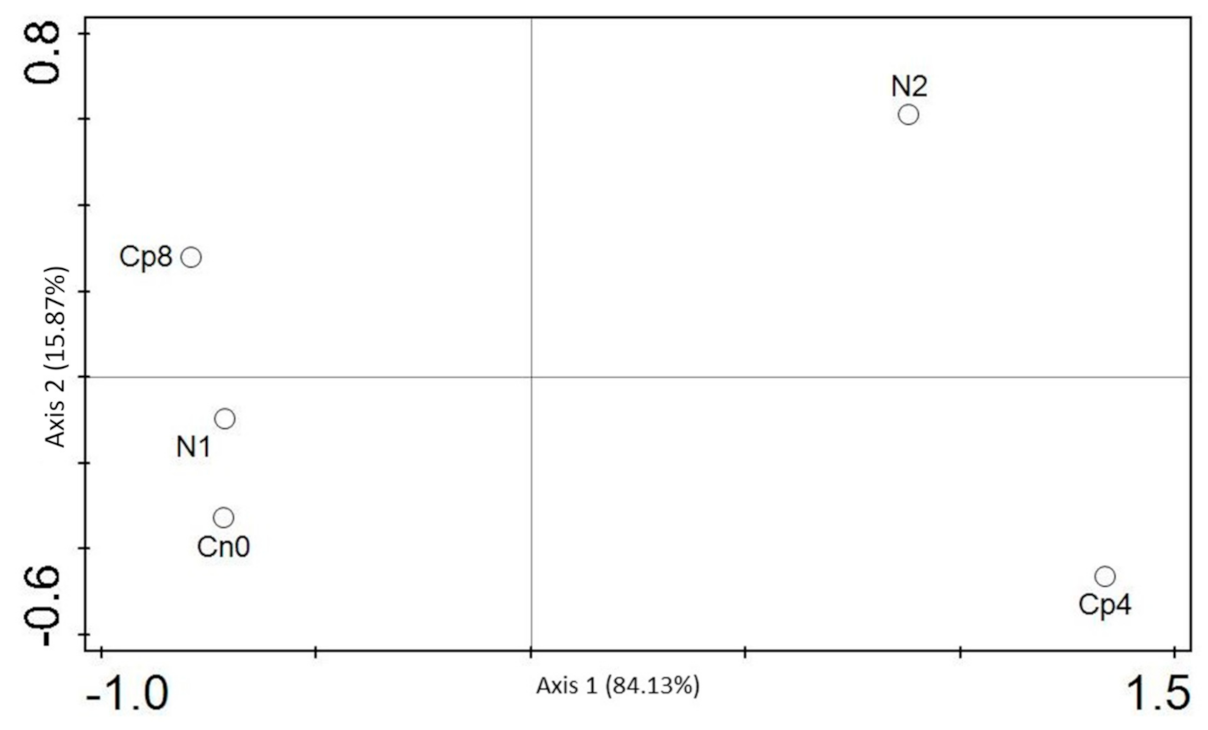
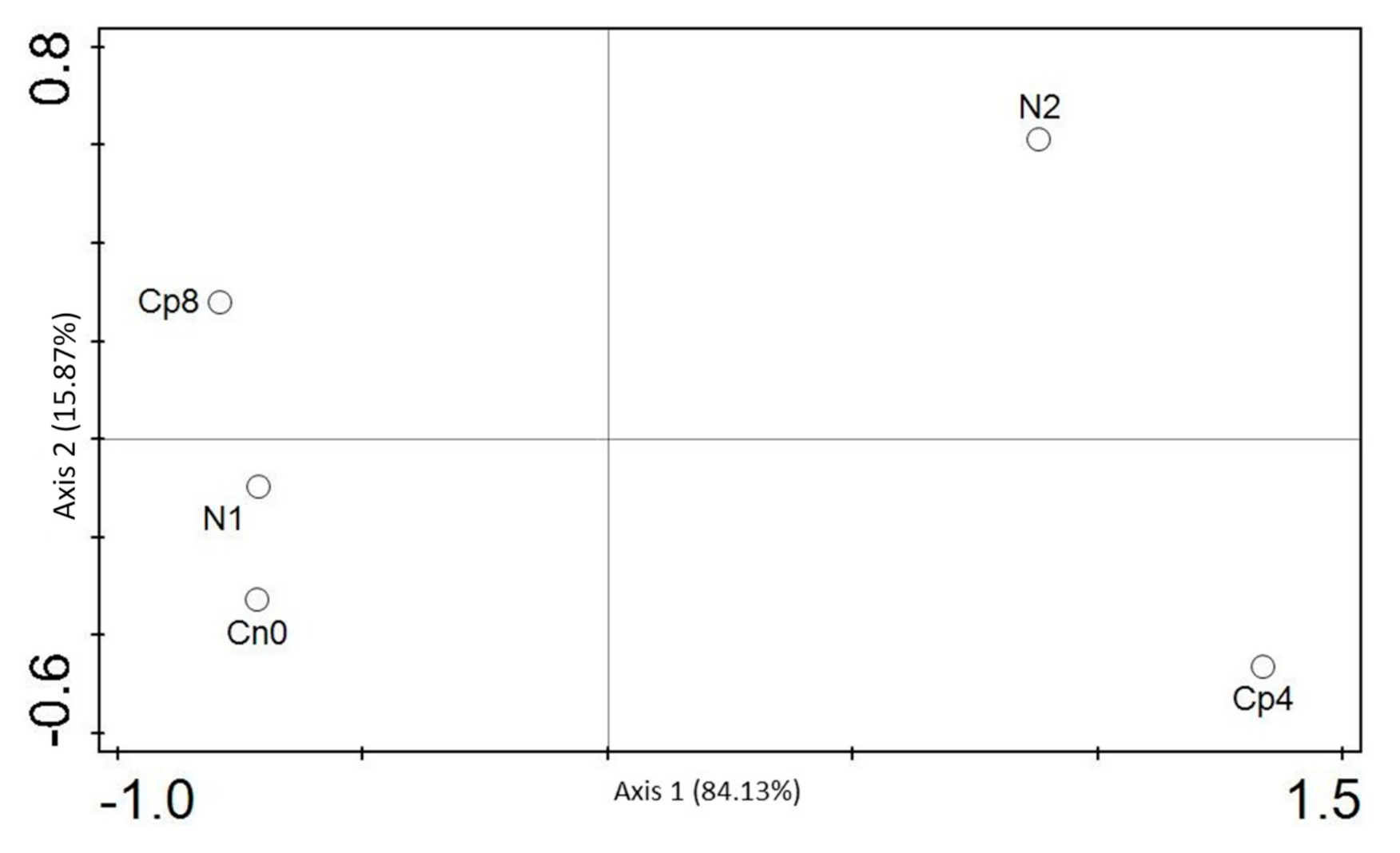
| Cp8 | Cp4 | N2 | N1 | Cn0 | p Value | |
|---|---|---|---|---|---|---|
| Simpson | 0.8377 | 0.8025 | 0.6948 | 0.8065 | 0.8139 | 5.7 × 10−4 |
| Shannon | 2.219 | 2.068 | 1.649 | 2.084 | 2.105 | |
| Evenness | 0.2967 | 0.2824 | 0.1928 | 0.2593 | 0.2648 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Enebe, M.C.; Babalola, O.O. Metagenomics Assessment of Soil Fertilization on the Chemotaxis and Disease Suppressive Genes Abundance in the Maize Rhizosphere. Genes 2021, 12, 535. https://doi.org/10.3390/genes12040535
Enebe MC, Babalola OO. Metagenomics Assessment of Soil Fertilization on the Chemotaxis and Disease Suppressive Genes Abundance in the Maize Rhizosphere. Genes. 2021; 12(4):535. https://doi.org/10.3390/genes12040535
Chicago/Turabian StyleEnebe, Matthew Chekwube, and Olubukola Oluranti Babalola. 2021. "Metagenomics Assessment of Soil Fertilization on the Chemotaxis and Disease Suppressive Genes Abundance in the Maize Rhizosphere" Genes 12, no. 4: 535. https://doi.org/10.3390/genes12040535
APA StyleEnebe, M. C., & Babalola, O. O. (2021). Metagenomics Assessment of Soil Fertilization on the Chemotaxis and Disease Suppressive Genes Abundance in the Maize Rhizosphere. Genes, 12(4), 535. https://doi.org/10.3390/genes12040535






