Genetic Regulation of Biomarkers as Stress Proxies in Dairy Cows
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethic Statement
2.2. Data and Sample Collection
2.3. Blood, Milk and Hair Assays
2.4. Analysis of Phenotypes
2.5. Genotyping and Quality Controls
2.6. Single-SNP Genome-Wide Association Study
2.7. Haplotype Genome-Wide Association Study
2.8. Genome-Wide Association Meta-Analysis
2.9. Analysis of Candidate for Putative Causative Variants
3. Results
3.1. Blood Assays and Outlier Analyses
3.2. Single-SNP GWAS Results
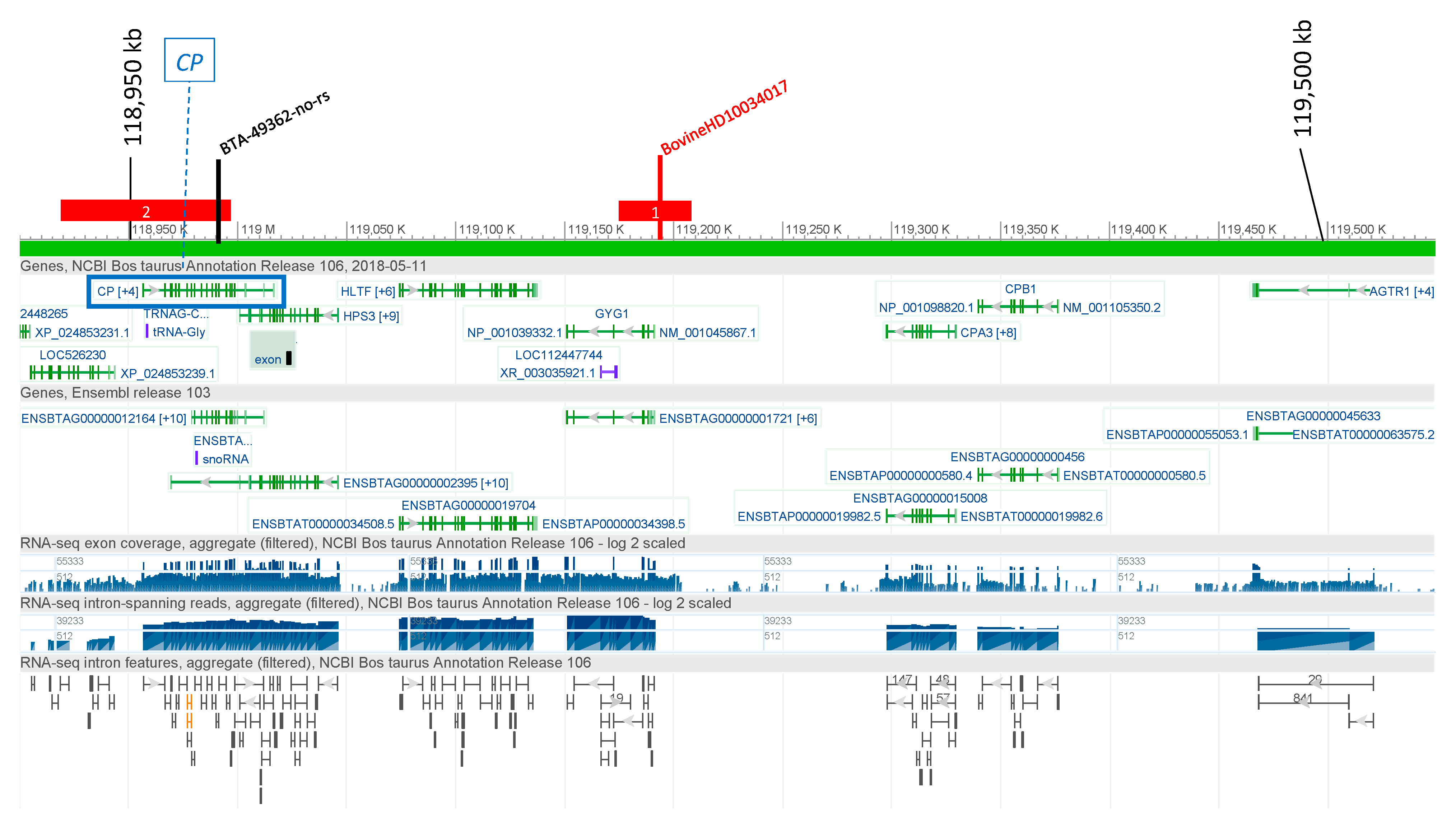
3.2.1. Ceruloplasmin (CP)
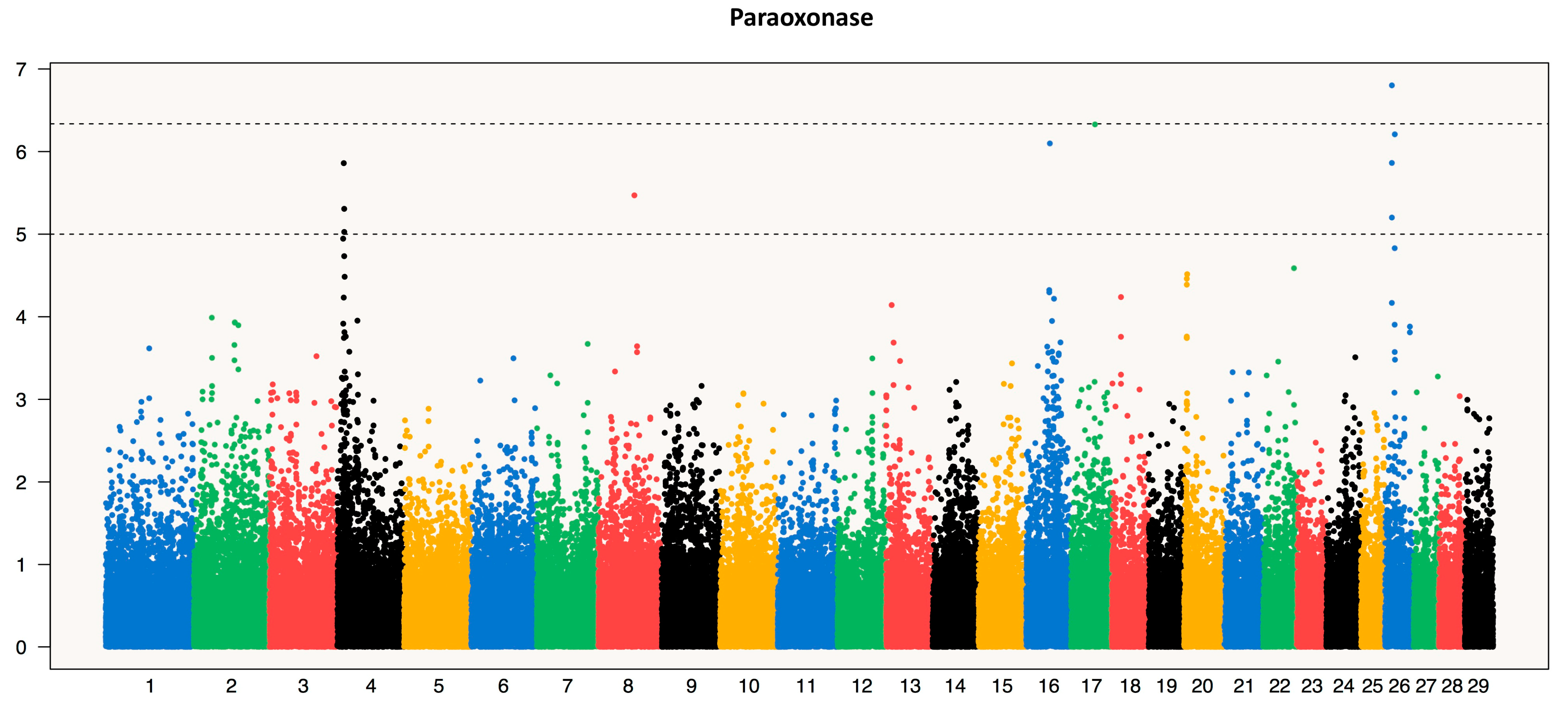
3.2.2. Paraoxonase (PON)
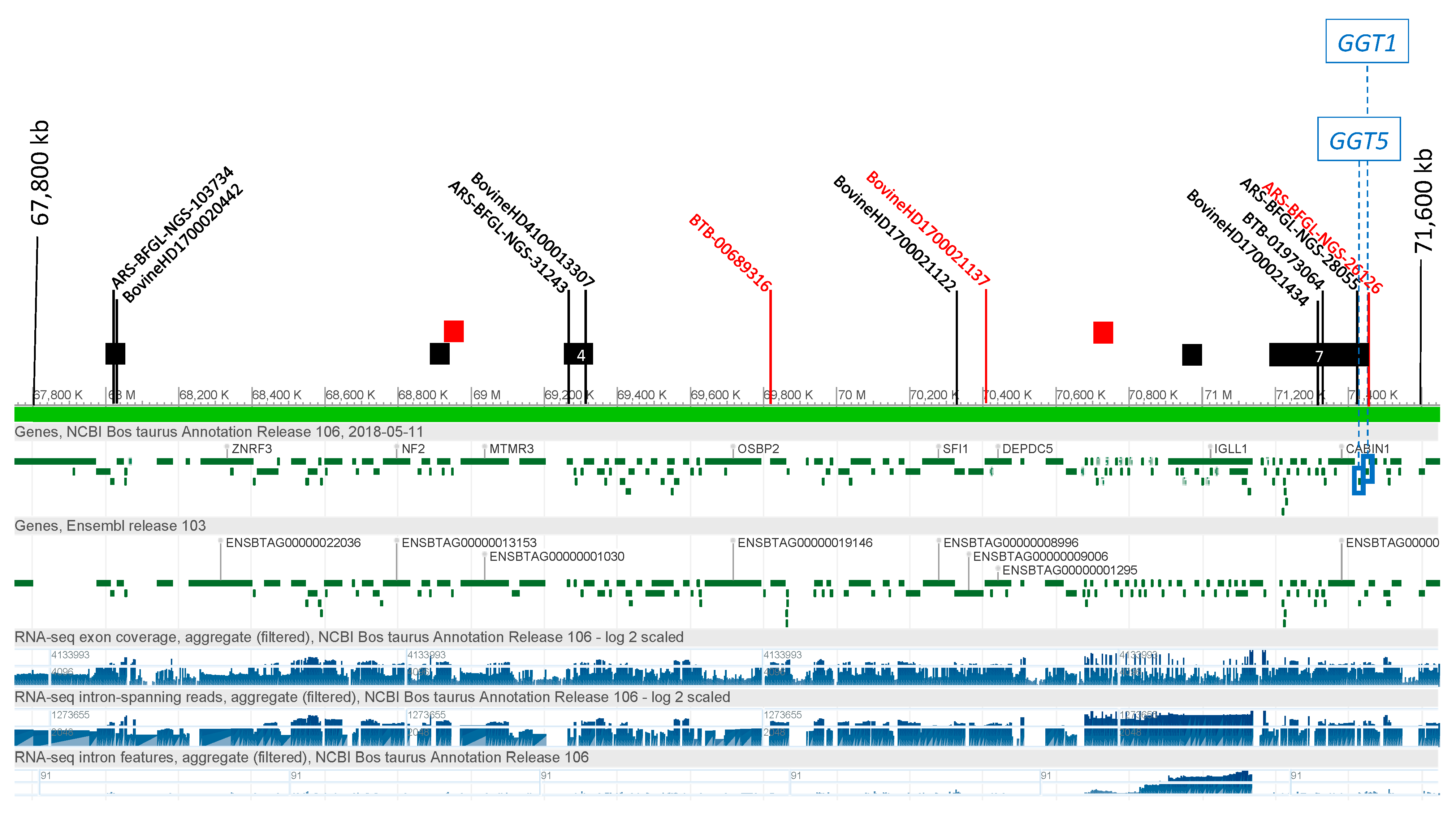
3.2.3. γ Glutamyl Transferase (GGT)
3.3. Genome-Wide Association Meta-analysis
3.4. Candidate Causal Variant Identification
4. Discussion
4.1. GWAS Analyses
4.2. Search for Candidate Causative Variants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pachauri, R.K.; Meyer, L.A. IPCC, 2014: Climate Change 2014: Synthesis Report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; IPCC: Geneve, Switzerland, 2014. [Google Scholar]
- Collier, R.; Renquist, B.; Xiao, Y. A 100-Year Review: Stress physiology including heat stress. J. Dairy Sci. 2017, 100, 10367–10380. [Google Scholar] [CrossRef]
- Zachut, M.; Marcela, Š.; Almeida, A.M.; De Gabai, G.; Mobasheri, A.; Hernández-Castellano, L.E. Biomarkers of fitness and welfare in dairy cattle: Healthy productivity. J. Dairy Res. 2020, 87, 4–13. [Google Scholar] [CrossRef]
- Kantanen, J.; Lã¸vendahl, P.; Strandberg, E.; Eythorsdottir, E.; Li, M.-H.; Kettunen-Prã¦Bel, A.; Berg, P.; Meuwissen, T.; Løvendahl, P. Utilization of farm animal genetic resources in a changing agro-ecological environment in the Nordic countries. Front. Genet. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Berghof, T.V.L.; Poppe, M.; Mulder, H.A. Opportunities to Improve Resilience in Animal Breeding Programs. Front. Genet. 2019, 9, 692. [Google Scholar] [CrossRef] [PubMed]
- Polsky, L.; Von Keyserlingk, M.A. Invited review: Effects of heat stress on dairy cattle welfare. J. Dairy Sci. 2017, 100, 8645–8657. [Google Scholar] [CrossRef] [PubMed]
- Yun, C.-H.; Wynn, P.; Ha, J.K. Stress, acute phase proteins and immune modulation in calves. Anim. Prod. Sci. 2014, 54, 1561–1568. [Google Scholar] [CrossRef]
- Jordan, E. Effects of Heat Stress on Reproduction. J. Dairy Sci. 2003, 86, E104–E114. [Google Scholar] [CrossRef]
- West, J.W. Effects of Heat-Stress on Production in Dairy Cattle. J. Dairy Sci. 2003, 86, 2131–2144. [Google Scholar] [CrossRef]
- Lykkesfeldt, J.; Svendsen, O. Oxidants and antioxidants in disease: Oxidative stress in farm animals. Vet. J. 2007, 173, 502–511. [Google Scholar] [CrossRef] [PubMed]
- McEwen, B.S. Stress, Adaptation, and Disease: Allostasis and Allostatic Load. Ann. N. Y. Acad. Sci. 1998, 840, 33–44. [Google Scholar] [CrossRef]
- Wood, S.K.; Walker, H.E.; Valentino, R.J.; Bhatnagar, S. Individual Differences in Reactivity to Social Stress Predict Susceptibility and Resilience to a Depressive Phenotype: Role of Corticotropin-Releasing Factor. Endocrinology 2010, 151, 1795–1805. [Google Scholar] [CrossRef] [PubMed]
- Nasca, C.; Zelli, D.; Bigio, B.; Piccinin, S.; Scaccianoce, S.; Nisticò, R.; McEwen, B.S. Stress dynamically regulates behavior and glutamatergic gene expression in hippocampus by opening a window of epigenetic plasticity. Proc. Natl. Acad. Sci. USA 2015, 112, 14960–14965. [Google Scholar] [CrossRef] [PubMed]
- Terenina, E.; Cavigelli, S.; Mormède, P.; Zhao, W.; Parks, C.; Lu, L.; Zhao, W.; Parks, C.; Lu, L.; Jones, B.; et al. Genetic Factors Mediate the Impact of Chronic Stress and Subsequent Response to Novel Acute Stress. Front. Neurosci. 2019, 13, 1–12. [Google Scholar] [CrossRef]
- Altieri, M.A.; Nicholls, C.I.; Henao, A.; Lana, M.A. Agroecology and the design of climate change-resilient farming systems. Agron. Sustain. Dev. 2015, 35, 869–890. [Google Scholar] [CrossRef]
- Döring, T.F.; Vieweger, A.; Pautasso, M.; Vaarst, M.; Finckh, M.R.; Wolfe, M.S. Resilience as a universal criterion of health. J. Sci. Food Agric. 2014, 95, 455–465. [Google Scholar] [CrossRef]
- Kasper, C.; Ribeiro, D.; De Almeida, A.M.; Larzul, C.; Liaubet, L.; Murani, E. Omics Application in Animal Science—A Special Emphasis on Stress Response and Damaging Behaviour in Pigs. Genes 2020, 11, 920. [Google Scholar] [CrossRef]
- Carabaño, M.J.; Ramón, M.; Menéndez-Buxadera, A.; Molina, A.; Díaz, C. Selecting for heat tolerance. Anim. Front. 2019, 9, 62–68. [Google Scholar] [CrossRef]
- Carabaño, M.J.; Ramón, M.; Díaz, C.; Molina, A.; Serradilla, J.M. Breeding and genetics symposium: Breeding for resilience to heat stress effects in dairy ruminants. A comprehensive review. J. Anim. Sci. 2017, 95, 1813–1826. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Jiang, C. Inflammation: The Common Pathway of Stress-Related Diseases. Front. Hum. Neurosci. 2017, 11, 1–11. [Google Scholar] [CrossRef]
- Min, L.; Zheng, N.; Zhao, S.; Cheng, J.; Yang, Y.; Zhang, Y.; Yang, H.; Wang, J. Long-term heat stress induces the inflammatory response in dairy cows revealed by plasma proteome analysis. Biochem. Biophys. Res. Commun. 2016, 471, 296–302. [Google Scholar] [CrossRef]
- Kumar, B.; Manuja, A.; Palok, A. Stress and its impact on farm animals. Front. Biosci. 2012, E4, 1759–1767. [Google Scholar] [CrossRef]
- Padgett, D.A.; Glaser, R. How stress influences the immune response. Trends Immunol. 2003, 24, 444–448. [Google Scholar] [CrossRef]
- Rohleder, N. Stimulation of Systemic Low-Grade Inflammation by Psychosocial Stress. Psychosom. Med. 2014, 76, 181–189. [Google Scholar] [CrossRef]
- Eckersall, P.; Bell, R. Acute phase proteins: Biomarkers of infection and inflammation in veterinary medicine. Vet. J. 2010, 185, 23–27. [Google Scholar] [CrossRef]
- Sordillo, L.M.; Contreras, G.A.; Aitken, S.L. Metabolic factors affecting the inflammatory response of periparturient dairy cows. Anim. Health Res. Rev. 2009, 10, 53–63. [Google Scholar] [CrossRef]
- Putman, A.; Brown, J.; Gandy, J.; Wisnieski, L.; Sordillo, L. Changes in biomarkers of nutrient metabolism, inflammation, and oxidative stress in dairy cows during the transition into the early dry period. J. Dairy Sci. 2018, 101, 9350–9359. [Google Scholar] [CrossRef] [PubMed]
- Alharthi, A.; Zhou, Z.; Lopreiato, V.; Trevisi, E.; Loor, J.J. Body condition score prior to parturition is associated with plasma and adipose tissue biomarkers of lipid metabolism and inflammation in Holstein cows. J. Anim. Sci. Biotechnol. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gruys, E.; Toussaint, M.; Niewold, T.; Koopmans, S. Acute phase reaction and acute phase proteins. J. Zhejiang Univ. A 2005, 6, 1045–1056. [Google Scholar] [CrossRef] [PubMed]
- Bertoni, G.; Trevisi, E. Use of the Liver Activity Index and Other Metabolic Variables in the Assessment of Metabolic Health in Dairy Herds. Vet. Clin. N. Am. Food Anim. Pract. 2013, 29, 413–431. [Google Scholar] [CrossRef]
- Fleck, A. Clinical and nutritional aspects of changes in acute-phase proteins during inflammation. In Proceedings of the Nutrition Society; CABI Publishing: Wallingford, UK, 1989; Volume 48, pp. 347–354. [Google Scholar]
- Bomba, L.; Nicolazzi, E.L.; Milanesi, M.; Negrini, R.; Mancini, G.; Biscarini, F.; Stella, A.; Valentini, A.; Ajmone-Marsan, P. Relative extended haplotype homozygosity signals across breeds reveal dairy and beef specific signatures of selection. Genet. Sel. Evol. 2015, 47, 1–14. [Google Scholar] [CrossRef]
- Marras, G.; Gaspa, G.; Sorbolini, S.; DiMauro, C.; Ajmone-Marsan, P.; Valentini, A.; Williams, J.L.; Macciotta, N.P.P. Analysis of runs of homozygosity and their relationship with inbreeding in five cattle breeds farmed in Italy. Anim. Genet. 2014, 46, 110–121. [Google Scholar] [CrossRef]
- Lopreiato, V.; Minuti, A.; Trimboli, F.; Britti, D.; Morittu, V.; Cappelli, F.P.; Loor, J.; Trevisi, E. Immunometabolic status and productive performance differences between periparturient Simmental and Holstein dairy cows in response to pegbovigrastim. J. Dairy Sci. 2019, 102, 9312–9327. [Google Scholar] [CrossRef]
- Associazione Allevatori Friuli Venezia Giulia (Codroipo). Available online: www.aafvg.it (accessed on 2 April 2021).
- Bionaz, M.; Trevisi, E.; Calamari, L.; Librandi, F.; Ferrari, A.; Bertoni, G. Plasma Paraoxonase, Health, Inflammatory Conditions, and Liver Function in Transition Dairy Cows. J. Dairy Sci. 2007, 90, 1740–1750. [Google Scholar] [CrossRef]
- Sgorlon, S.; Mattiello, A.; Ronutti, L.; Sandri, M.; Stefanon, B. Concentration of elements in the hair of growing and adult dogs. Ital. J. Anim. Sci. 2019, 18, 1126–1134. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org (accessed on 2 April 2021).
- Rosen, B.D.; Bickhart, D.M.; Schnabel, R.D.; Koren, S.; Elsik, C.G.; Tseng, E.; Rowan, T.N.; Low, W.Y.; Zimin, A.; Couldrey, C.; et al. De novo assembly of the cattle reference genome with single-molecule sequencing. GigaScience 2020, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 1–16. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Browning, S.R.; Browning, B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007, 81, 1084–1097. [Google Scholar] [CrossRef] [PubMed]
- Browning, B.L.; Browning, S.R. Genotype Imputation with Millions of Reference Samples. Am. J. Hum. Genet. 2016, 98, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A Tool for Genome-wide Complex Trait Analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Yang, J.; Zaitlen, N.A.; Goddard, M.E.; Visscher, P.M.; Price, A.L. Advantages and pitfalls in the application of mixed-model association methods. Nat. Genet. 2014, 46, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Shim, H.; Chasman, D.I.; Smith, J.D.; Mora, S.; Ridker, P.M.; Nickerson, D.A.; Krauss, R.M.; Stephens, M. A Multivariate Genome-Wide Association Analysis of 10 LDL Subfractions, and Their Response to Statin Treatment, in 1868 Caucasians. PLOS ONE 2015, 10, e0120758. [Google Scholar] [CrossRef] [PubMed]
- Loh, P.-R.; Danecek, P.; Palamara, P.F.; Fuchsberger, C.; A Reshef, Y.; Finucane, H.K.; Schoenherr, S.; Forer, S.S.L.; McCarthy, S.; Abecasis, C.F.G.R.; et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 2016, 48, 1443–1448. [Google Scholar] [CrossRef] [PubMed]
- Utsunomiya, Y.T.; Milanesi, M.; Barbato, M.; Utsunomiya, A.T.H.; Sölkner, J.; Ajmone-Marsan, P.; Garcia, J.F. Unsupervised detection of ancestry tracks with the GHap r package. Methods Ecol. Evol. 2020, 11, 1448–1454. [Google Scholar] [CrossRef]
- Utsunomiya, Y.T.; Milanesi, M.; Utsunomiya, A.T.H.; Ajmone-Marsan, P.; Garcia, J.F. GHap: An R package for genome-wide haplotyping. Bioinformatics 2016, 32, 2861–2862. [Google Scholar] [CrossRef]
- Willer, C.J.; Li, Y.; Abecasis, G.R. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010, 26, 2190–2191. [Google Scholar] [CrossRef]
- Stouffer, S.A.; Suchman, E.A.; Devinney, L.C.; Star, S.A.; Williams, R.M.; Lumsdaine, A.H.; Brewster, S.M.; Cottrell, L.S. The American Soldier: Combat and its aftermath. Ann. Am. Acad. Pol. Soc. Sci. 1949, 265, 173–175. [Google Scholar] [CrossRef]
- Daetwyler, H.D.; Capitan, A.; Pausch, H.; Stothard, P.; Van Binsbergen, R.; Brøndum, R.F.; Liao, X.; Djari, A.; Rodriguez, S.C.; Grohs, C.; et al. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Genet. 2014, 46, 858–865. [Google Scholar] [CrossRef]
- Sargolzaei, M.; Schenkel, F.; Jansen, G.; Schaeffer, L. Extent of Linkage Disequilibrium in Holstein Cattle in North America. J. Dairy Sci. 2008, 91, 2106–2117. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 1–14. [Google Scholar] [CrossRef]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 4, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Huang, C. LASAGNA-Search 2.0: Integrated transcription factor binding site search and visualisation in a browser. Bioinformatics 2014, 30, 1923–1925. [Google Scholar] [CrossRef]
- Trevisi, E.; Amadori, M.; Cogrossi, S.; Razzuoli, E.; Bertoni, G. Metabolic stress and inflammatory response in high-yielding, periparturient dairy cows. Res. Vet. Sci. 2012, 93, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, A.C.; Daetwyler, H.D.; Chamberlain, A.J.; Ponce, C.H.; Sargolzaei, M.; Schenkel, F.S.; Sahana, G.; Govignon-Gion, A.; Boitard, S.; Dolezal, M.; et al. Meta-analysis of genome-wide association studies for cattle stature identifies common genes that regulate body size in mammals. Nat. Genet. 2018, 50, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Osorio, J.; Trevisi, E.; Ji, P.; Drackley, J.; Luchini, D.; Bertoni, G.; Loor, J. Biomarkers of inflammation, metabolism, and oxidative stress in blood, liver, and milk reveal a better immunometabolic status in peripartal cows supplemented with Smartamine M or MetaSmart. J. Dairy Sci. 2014, 97, 7437–7450. [Google Scholar] [CrossRef]
- Kaya, S.; Merhan, O.; Kacar, C.; Colak, A.; Bozukluhan, K. Determination of ceruloplasmin, some other acute phase proteins, and biochemical parameters in cows with endometritis. Vet. World 2016, 9, 1056–1062. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, I.I.; Gould, T.D. The Endophenotype Concept in Psychiatry: Etymology and Strategic Intentions. Am. J. Psychiatry 2003, 160, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Lay, S.L.; Simard, G.; Martinez, M.C.; Andriantsitohaina, R. Oxidative Stress and Metabolic Pathologies: From an Adipocentric Point of View. Oxid. Med. Cell Longev. 2014, 2014, 1–18. [Google Scholar] [CrossRef]
- Salim, S. Oxidative Stress and Psychological Disorders. Curr. Neuropharmacol. 2014, 12, 140–147. [Google Scholar] [CrossRef]
- Slavich, G.M.; Irwin, M.R. From stress to inflammation and major depressive disorder: A social signal transduction theory of depression. Psychol. Bull. 2014, 140, 774–815. [Google Scholar] [CrossRef]
- Haroon, E.; Raison, C.L.; Miller, A.H. Psychoneuroimmunology Meets Neuropsychopharmacology: Translational Implications of the Impact of Inflammation on Behavior. Neuropsychopharmacology 2011, 37, 137–162. [Google Scholar] [CrossRef] [PubMed]
- Bielli, P.; Calabrese, L. Structure to function relationships in ceruloplasmin: A ’moonlighting’ protein. Cell. Mol. Life Sci. 2002, 59, 1413–1427. [Google Scholar] [CrossRef] [PubMed]
- Floris, G.; Medda, R.; Padiglia, A.; Musci, G. The physiopathological significance of ceruloplasmin. Biochem. Pharmacol. 2000, 60, 1735–1741. [Google Scholar] [CrossRef]
- Cerone, S.I.; Sansinanea, A.S.; Streitenberger, S.A.; Garcia, M.C.; Auza, N.J. Cytochrome c Oxidase, Cu,Zn-Superoxide Dismutase, and Ceruloplasmin Activities in Copper-Deficient Bovines. Biol. Trace Element Res. 2000, 73, 269–278. [Google Scholar] [CrossRef]
- Bakhautdin, B.; Febbraio, M.; Goksoy, E.; A De La Motte, C.; Gulen, M.F.; Childers, E.P.; Hazen, S.L.; Li, X.; Fox, P.L. Protective role of macrophage-derived ceruloplasmin in inflammatory bowel disease. Gut 2012, 62, 209–219. [Google Scholar] [CrossRef]
- Texel, S.J.; Camandola, S.; Ladenheim, B.; Rothman, S.M.; Mughal, M.R.; Unger, E.L.; Cadet, J.L.; Mattson, M.P. Ceruloplasmin deficiency results in an anxiety phenotype involving deficits in hippocampal iron, serotonin, and BDNF. J. Neurochem. 2011, 120, 125–134. [Google Scholar] [CrossRef]
- Hussein, H.A.; Staufenbiel, R.; Müller, A.E.; El-Sebaie, A.; Abd-El-Salam, M. Ceruloplasmin activity in Holstein dairy cows: Effects of lactation stages and anticoagulants. Comp. Haematol. Int. 2012, 21, 705–710. [Google Scholar] [CrossRef]
- Seo, J.; Osorio, J.S.; Schmitt, E.; Corrêa, M.N.; Bertoni, G.; Trevisi, E.; Loor, J.J. Hepatic purinergic signaling gene network expression and its relationship with inflammation and oxidative stress biomarkers in blood from peripartal dairy cattle. J Dairy Sci. 2014, 97, 861–873. [Google Scholar] [CrossRef]
- Garelnabi, M.; Litvinov, D.; Mahini, H. Antioxidant and anti-inflammatory role of paraoxonase 1: Implication in arteriosclerosis diseases. N. Am. J. Med. Sci. 2012, 4, 523–532. [Google Scholar] [CrossRef]
- Moreira, E.G.; Boll, K.M.; Correia, D.G.; Soares, J.F.; Rigobello, C.; Maes, M.; Maesd, M. Why Should Psychiatrists and Neuroscientists Worry about Paraoxonase 1? Curr. Neuropharmacol. 2019, 17, 1004–1020. [Google Scholar] [CrossRef]
- Aviram, M.; Rosenblat, M. Paraoxonases 1, 2, and 3, oxidative stress, and macrophage foam cell formation during atherosclerosis development. Free. Radic. Biol. Med. 2004, 37, 1304–1316. [Google Scholar] [CrossRef]
- Silveira, P.A.; Schwegler, E.; Montagner, P.; Krause, A.R.; Acosta, D.A.; Halfen, J.; Garlet, T.; Barros, C.C.; Corrêa, M.N.; Schneider, A. Characterization of single nucleotide polymorphisms in the promoter region of the bovine paraoxonase 1 (PON1) gene affecting serum enzyme activity in dairy cows. Vet. J. 2015, 205, 101–103. [Google Scholar] [CrossRef]
- Bertoni, G.; Trevisi, E.; Han, X.; Bionaz, M. Effects of Inflammatory Conditions on Liver Activity in Puerperium Period and Consequences for Performance in Dairy Cows. J. Dairy Sci. 2008, 91, 3300–3310. [Google Scholar] [CrossRef]
- Rodríguez-Jiménez, S.; Haerr, K.; Trevisi, E.; Loor, J.; Cardoso, F.; Osorio, J. Prepartal standing behavior as a parameter for early detection of postpartal subclinical ketosis associated with inflammation and liver function biomarkers in peripartal dairy cows. J. Dairy Sci. 2018, 101, 8224–8235. [Google Scholar] [CrossRef]
- Levasseur, R.; Barrios, R.; Elefteriou, F.; Glass, D.A.; Lieberman, M.W.; Karsenty, G. Reversible Skeletal Abnormalities in γ-Glutamyl Transpeptidase-Deficient Mice. Endocrinology 2003, 144, 2761–2764. [Google Scholar] [CrossRef][Green Version]
- Coon, T.A.; McKelvey, A.C.; Lear, T.; Rajbhandari, S.; Dunn, S.R.; Connelly, W.; Zhao, J.Y.; Han, S.; Liu, Y.; Weathington, N.M.; et al. The proinflammatory role of HECTD2 in innate immunity and experimental lung injury. Sci. Transl. Med. 2015, 7, 295ra109. [Google Scholar] [CrossRef]
- Simino, J.; Shi, G.; Bis, J.C.; Chasman, D.I.; Ehret, G.B.; Gu, X.; Guo, X.; Hwang, S.-J.; Sijbrands, E.; Smith, A.V.; et al. Gene-Age Interactions in Blood Pressure Regulation: A Large-Scale Investigation with the CHARGE, Global BPgen, and ICBP Consortia. Am. J. Hum. Genet. 2014, 95, 24–38. [Google Scholar] [CrossRef]
- Liu, Z.; Naranjo, A.; Thiele, C.J. CASZ1b, the Short Isoform of CASZ1 Gene, Coexpresses with CASZ1a during Neurogenesis and Suppresses Neuroblastoma Cell Growth. PLoS ONE 2011, 6, e18557. [Google Scholar] [CrossRef]
- Bhaskaran, N.; Liu, Z.; Saravanamuthu, S.S.; Yan, C.; Hu, Y.; Dong, L.; Zelenka, P.; Zheng, L.; Bletsos, V.; Harris, R.; et al. Identification of Casz1 as a Regulatory Protein Controlling T Helper Cell Differentiation, Inflammation, and Immunity. Front. Immunol. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Okumoto, K.; El Shermely, M.; Natsui, M.; Kosako, H.; Natsuyama, R.; Marutani, T.; Fujiki, Y. The peroxisome counteracts oxidative stresses by suppressing catalase import via Pex14 phosphorylation. eLife 2020, 9, 1–25. [Google Scholar] [CrossRef]
- Rubin, C.-J.; Zody, M.C.; Eriksson, J.; Meadows, J.R.S.; Sherwood, E.; Webster, M.T.; Jiang, L.; Ingman, M.; Sharpe, T.; Ka, S.; et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 2010, 464, 587–591. [Google Scholar] [CrossRef]
- Besnard, A.; Galan-Rodriguez, B.; Vanhoutte, P.; Caboche, J. Umrs-inserm C-C. Elk-1 a transcription factor with multiple facets in the brain. Front. Neurosci. 2011, 5, 1–11. [Google Scholar] [CrossRef]
- Kno, B.; Nordheim, A. Functional versatility of transcription factors in the nervous system: The SRF paradigm. Cell Press. 2009, 32, 432–442. [Google Scholar]
- Gomez-Ferreria, M.A.; Rey-Campos, J. Functional Domains of FOXJ2. J. Mol. Biol. 2003, 329, 631–644. [Google Scholar] [CrossRef]
- Silveira, P.A.S.; Butler, W.; LaCount, S.; Overton, T.; Barros, C.C.; Schneider, A. Polymorphisms in the anti-oxidant paraoxonase-1 (PON1) gene associated with fertility of postpartum dairy cows. Theriogenology 2019, 125, 302–309. [Google Scholar] [CrossRef]
- Tsarovina, K.; Reiff, T.; Stubbusch, J.; Kurek, D.; Grosveld, F.G.; Parlato, R. The Gata3 Transcription Factor Is Required for the Survival of Embryonic and Adult Sympathetic Neurons. J. Neurosci. 2010, 30, 10833–10843. [Google Scholar] [CrossRef]
- Filosto, M.; Tonin, P.; Vattemi, G.; Savio, C.; Rizzuto, N.; Tomelleri, G. Transcription factors c-Jun/activator protein-1 and nuclear factor-kappa B in oxidative stress response in mitochondrial diseases. Neuropathol. Appl. Neurobiol. 2003, 29, 52–59. [Google Scholar] [CrossRef]
- Triner, D.; Castillo, C.; Hakim, J.B.; Xue, X.; Greenson, J.K.; Nuñez, G. Myc-Associated Zinc Finger Protein Regulates the Proiflammatory Response in Colitis and Colon Cancer via STAT3 signaling. Mol. Cell Biol. 2018, 38, 1–15. [Google Scholar] [CrossRef]
- Ryu, H.; Lee, J.; Zaman, K.; Kubilis, J.; Ferrante, R.J.; Ross, B.D.; Neve, R.; Ratan, R.R. Sp1 and Sp3 Are Oxidative Stress-Inducible, Antideath Transcription Factors in Cortical Neurons. J. Neurosci. 2003, 23, 3597–3606. [Google Scholar] [CrossRef]
- Nakashima, X.H.; Tsujimura, K.; Irie, K.; Ishizu, M.; Pan, M.; Kameda, T. Canonical TGF- B Signaling Negatively Regulates Neuronal Morphogenesis through TGIF/Smad Complex-Mediated CRMP2 Suppression. J. Neurosci. 2018, 38, 4791–4810. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, M.; Antoine, M.; Hébert, J.M. SMAD4 is essential for generating subtypes of neurons during cerebellar development. Dev. Biol. 2012, 365, 82–90. [Google Scholar] [CrossRef] [PubMed]

| Phenotype | Type | IH | IS | ||||
| n | Mean (SD) | Range | n | Mean (SD) | Range | ||
| Body Condition Score | Animal condition | 335 | 2.4 (0.39) | 1.05–3.75 | 307 | 3 (0.47) | 1.75–4 |
| Days in milk (DIM) | Animal condition | 335 | 167.38 (61.82) | 36–283 | 307 | 154.51 (79.03) | 15–404 |
| Somatic Cell Count | Mammary health | 335 | 4.85 (0.42) | 3.48–5.68 | 307 | 4.76 (0.47) | 3.6–5.7 |
| Milk yield | Mammary metabolism | 335 | 37.05 (9.22) | 11.9–62.5 | 306 | 26.76 (6.49) | 11.2–48.9 |
| Lactations | Animal condition | 335 | 1.99 (1.3) | 01-lug | 307 | 2.56 (1.62) | 01-set |
| Casein | Milk composition/ | 335 | 2.5 (0.21) | 1.94–3.15 | 306 | 2.73 (0.25) | 1.98–3.47 |
| Fat (milk) | Milk composition/Mammary metabolism | 335 | 3.57 (0.65) | 1.78–5.99 | 306 | 3.75 (0.65) | 1.94–7.53 |
| Protein (milk) | Milk composition/Mammary metabolism | 335 | 3.16 (0.28) | 2.43–4.03 | 307 | 3.48 (0.34) | 2.59–4.53 |
| Blood biomarker | Type | IH | IS | ||||
| n | Mean (SD) | Range | n | Mean (SD) | Range | ||
| Albumin | Liver function | 331 | 37.2 (3.21) | 17.25–47.44 | 297 | 37.51 (1.98) | 31.59–43.2 |
| Total bilirubin | Liver function | 332 | 0.7 (0.56) | 0.02–4.61 | 297 | 1.14 (0.75) | 0.03–4.29 |
| Total protein | Liver function | 331 | 78.61 (7.25) | 30.13–104 | 297 | 77.59 (4.55) | 67.9–91.29 |
| Globulin | Liver function/Immune response | 331 | 41.41 (7.11) | 12.88–78.06 | 297 | 40.08 (5.04) | 28.99–56.54 |
| Paraoxonase | Liver function/Lipoprotein metabolism | 326 | 106.75 (25.99) | 32.29–216.22 | 295 | 102.58 (23.74) | 47.38–197.44 |
| AST/GOT | Liver function/Protein metabolism | 330 | 105.5 (29.64) | 35.55–243.2 | 297 | 90.42 (24.45) | 57.34–233.68 |
| GGT | Liver function/ Protein metabolism | 331 | 33.73 (12.61) | 9.96–128.15 | 297 | 27.47 (6.57) | 14.7–64.64 |
| Cholesterol | Liver function/Energy metabolism | 331 | 6.28 (1.59) | 1.8–10.16 | 297 | 4.8 (1.1) | 2.37–10.45 |
| Glucose | Energy metabolism | 333 | 3.81 (0.42) | 2.66–5.06 | 297 | 3.89 (0.36) | 2.98–5.1 |
| NEFA | Energy metabolism/Lipid metabolism | 333 | 0.13 (0.08) | 0.04–0.73 | 297 | 0.11 (0.06) | 0.03–0.37 |
| BHB | Energy metabolism | 327 | 0.72 (0.24) | 0.19–1.71 | 294 | 0.7 (0.22) | 0.07–1.56 |
| Ceruloplasmin | Inflammatory response | 332 | 2.54 (0.6) | 1.13–4.74 | 294 | 2.44 (0.65) | 0.71–4.14 |
| Haptoglobin | Inflammatory response | 331 | 0.37 (0.32) | 0.02–2.24 | 297 | 0.33 (0.28) | 0.03–2.35 |
| Calcium | Mineral metabolism | 127 | 2.61 (0.23) | 1.3–2.92 | 0 | NA (NA) | NA |
| Zinc | Mineral metabolism/Immune function | 325 | 13.43 (3.19) | 5.18–32.7 | 295 | 12.76 (2.87) | 7.28–27.81 |
| Creatinine | Protein metabolism/Renal function | 327 | 87.37 (9.05) | 40.58–120 | 295 | 113.13 (12.26) | 77.42–158.62 |
| Urea | Protein metabolism | 331 | 5.34 (1.43) | 2.62–10.74 | 297 | 4.65 (1.13) | 1.67–8.68 |
| Milk biomarker | Type | IH | IS | ||||
| n | Mean (SD) | Range | n | Mean (SD) | Range | ||
| BHB | Energy metabolism | 316 | 0.16 (0.03) | 0.06–0.36 | 297 | 0.17 (0.05) | 0.03–0.3 |
| Cortisol | Immune system | 313 | 501.71 (312.18) | 41.74–1822.64 | 299 | 515.46 (263.55) | 60–1539.81 |
| Urea | Milk composition/Protein metabolism | 335 | 22.87 (8.29) | 9.4–51.4 | 307 | 19.97 (6.14) | 3.65–38.3 |
| Hair biomarker | Type | IH | IS | ||||
| n | Mean (SD) | Range | n | Mean (SD) | Range | ||
| Cortisol | Immune system | 309 | 3.53 (2.22) | 0.45–12.5 | 278 | 3.23 (1.63) | 0.84–10.66 |
| Gene | Chr | Pos | Alt | Ref | Distance_gene_start | TFBS.N | TFBS.name | rs | r2.SIM | r2.HOL |
|---|---|---|---|---|---|---|---|---|---|---|
| CP | 1 | 118,900,034 | T | G | 1347 | 2 | Elk-1(T00250)|SRF(T00764) | rs385773690 | 0.337 | <0.3 |
| CP | 1 | 118,900,683 | C | T | 698 | 1 | FOXJ2 (long isoform)(T04169) | rs381127256 | 0.320 | <0.3 |
| PON1 | 4 | 12,576,347 | G | A | 19 | 1 | GATA-3(T00311) | rs109606244 | <0.3 | <0.3 |
| PON1 | 4 | 12,576,418 | G | T | 90 | 1 | TBP(T00794) | rs377892116 | <0.3 | <0.3 |
| PON1 | 4 | 12,576,463 | G | A | 135 | 0 | rs109953053 | <0.3 | <0.3 | |
| PON1 | 4 | 12,576,634 | A | C | 306 | 2 | ER-α(T00261)|COUP-TF2(T00045) | rs110459801 | <0.3 | <0.3 |
| PON1 | 4 | 12,576,853 | T | C | 525 | 1 | c-Jun(T00133) | rs381274305 | <0.3 | <0.3 |
| PON1 | 4 | 12,576,916 | T | A | 588 | 0 | rs110270756 | <0.3 | <0.3 | |
| GGT5 | 17 | 71,454,646 | T | G | 1277 | 0 | rs109325809 | <0.3 | 0.597 | |
| GGT5 | 17 | 71,454,782 | A | G | 1413 | 0 | rs41854700 | <0.3 | 0.402 | |
| GGT5 | 17 | 71,455,325 | GCCC | G | 1956 | 1 | Smad4(T04292) | rs133286128 | <0.3 | 0.566 |
| GGT1 | 17 | 71,471,816 | T | G | 161 | 1 | ATF(T00051) | rs210579585 | <0.3 | 0.641 |
| GGT1 | 17 | 71,471,932 | A | G | 277 | 0 | rs208460991 | <0.3 | 0.628 | |
| GGT1 | 17 | 71,472,255 | C | T | 600 | 1 | VDR(T00885) | rs209913616 | <0.3 | 0.632 |
| GGT1 | 17 | 71,472,377 | G | C | 722 | 2 | MAZ(T00490)|Sp1(T00759) | rs210564197 | <0.3 | 0.647 |
| GGT1 | 17 | 71,472,410 | C | G | 755 | 0 | rs208475328 | <0.3 | 0.618 | |
| GGT1 | 17 | 71,472,863 | T | C | 1208 | 0 | rs209562610 | <0.3 | 0.541 | |
| GGT1 | 17 | 71,473,234 | C | G | 1579 | 2 | COUP-TF1(T00149)|ER-α(T00261) | rs41854716 | <0.3 | 0.401 |
| GGT11 | 17 | 71,473,281 | ACC | AC | 1626 | 0 | rs464903245 | <0.3 | 0.434 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Milanesi, M.; Passamonti, M.M.; Cappelli, K.; Minuti, A.; Palombo, V.; Sgorlon, S.; Capomaccio, S.; D’Andrea, M.; Trevisi, E.; Stefanon, B.; et al. Genetic Regulation of Biomarkers as Stress Proxies in Dairy Cows. Genes 2021, 12, 534. https://doi.org/10.3390/genes12040534
Milanesi M, Passamonti MM, Cappelli K, Minuti A, Palombo V, Sgorlon S, Capomaccio S, D’Andrea M, Trevisi E, Stefanon B, et al. Genetic Regulation of Biomarkers as Stress Proxies in Dairy Cows. Genes. 2021; 12(4):534. https://doi.org/10.3390/genes12040534
Chicago/Turabian StyleMilanesi, Marco, Matilde Maria Passamonti, Katia Cappelli, Andrea Minuti, Valentino Palombo, Sandy Sgorlon, Stefano Capomaccio, Mariasilvia D’Andrea, Erminio Trevisi, Bruno Stefanon, and et al. 2021. "Genetic Regulation of Biomarkers as Stress Proxies in Dairy Cows" Genes 12, no. 4: 534. https://doi.org/10.3390/genes12040534
APA StyleMilanesi, M., Passamonti, M. M., Cappelli, K., Minuti, A., Palombo, V., Sgorlon, S., Capomaccio, S., D’Andrea, M., Trevisi, E., Stefanon, B., Williams, J. L., & Ajmone-Marsan, P. (2021). Genetic Regulation of Biomarkers as Stress Proxies in Dairy Cows. Genes, 12(4), 534. https://doi.org/10.3390/genes12040534











