High Levels of Genetic Variation in MHC-Linked Microsatellite Markers from Native Chicken Breeds
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and DNA Extraction
2.2. Ms Marker Identification and Primer Design
2.3. PCR Amplification and Genotyping
2.4. Population Genetics Statistics
MS Marker Diversity within and between Populations
2.5. Haplotype Construction
3. Results
3.1. MHC-B MS Marker Polymorphisms
3.2. MHC Diversity within and between Populations
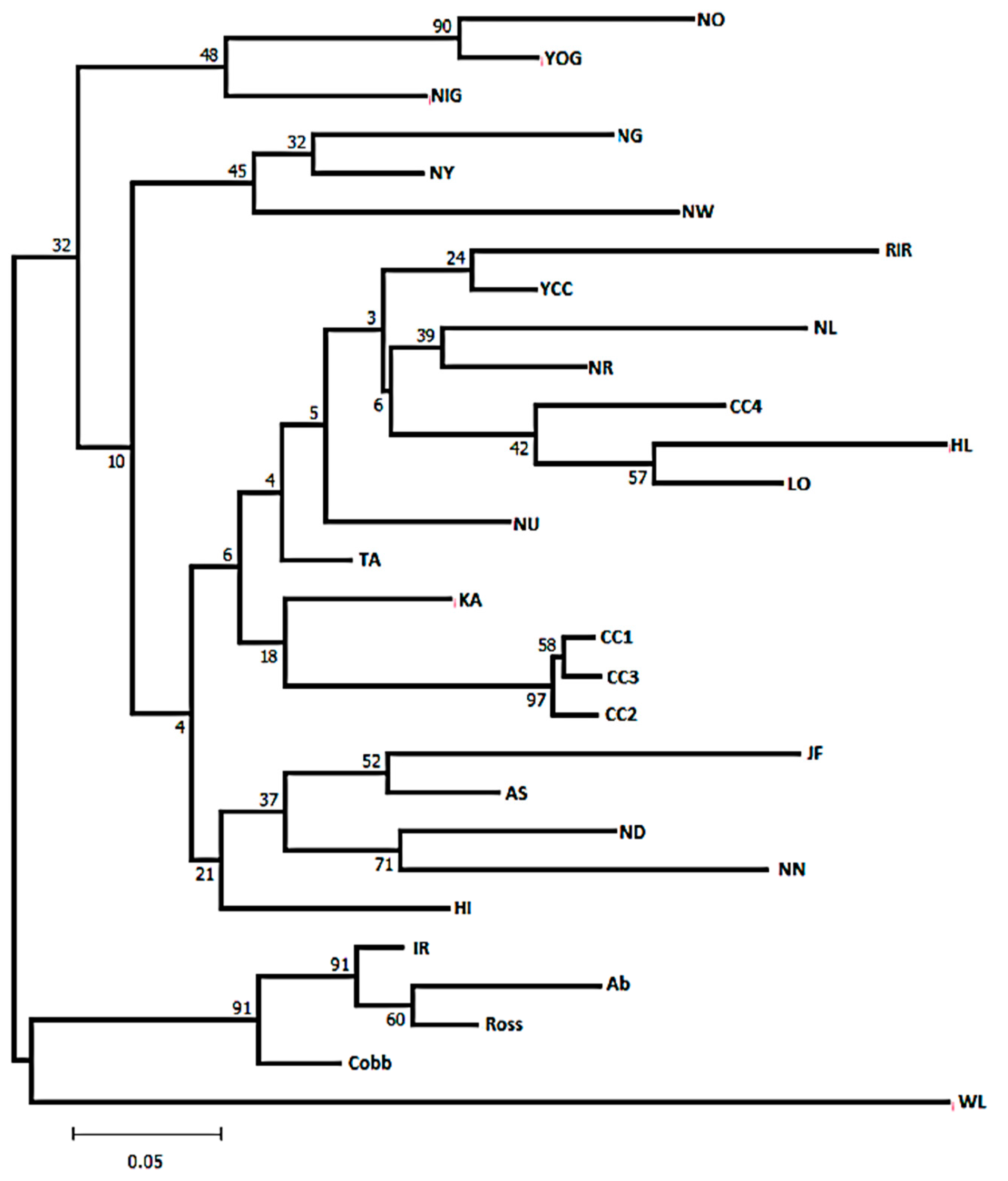
Population Structure and Genetic Distances
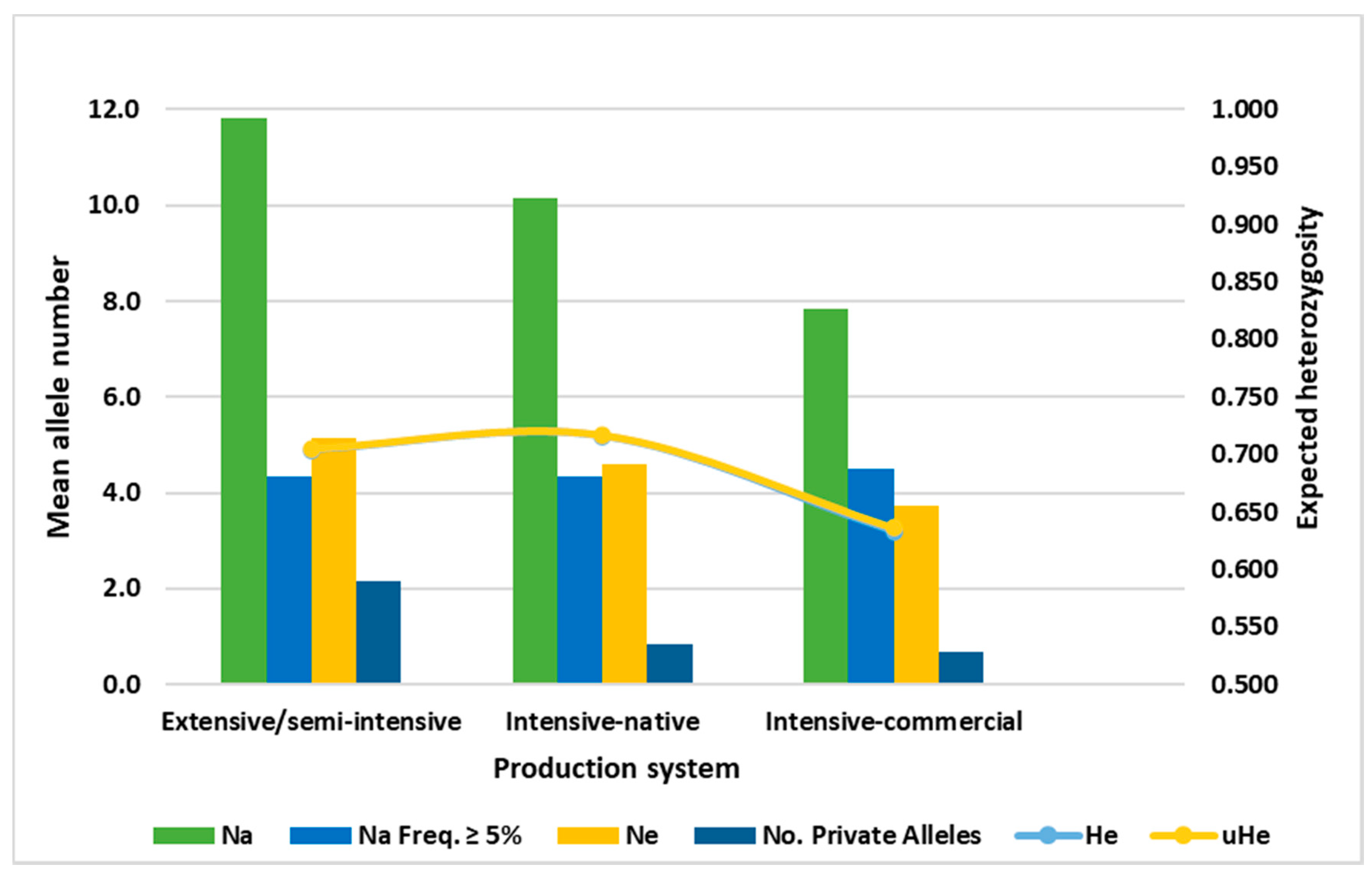
3.3. Allele Distribution by Production Systems
3.4. Haplotype Diversity
4. Discussion
4.1. Polymorphism of MHC-B Microsatellite Loci
4.2. Difference in MHC Diversity and Frequency among Populations
4.3. Differences in MHC Diversity among Production Systems
4.4. Haplotype Diversity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sommer, S. The importance of immune gene variability MHC in evolutionary ecology and conservation. Front. Zool. 2005, 2, 1–18. [Google Scholar] [CrossRef]
- Kaufman, J.; Jacob, J.; Shaw, J.; Walker, B.; Milne, S.; Beck, S.; Salomonsen, J. Gene organization determines evolution of function in the chicken MHC. Immunol. Rev. 1999, 167, 101–117. [Google Scholar] [CrossRef]
- Kaufman, J. The Avian MHC. In Avian Immunology, 2nd ed.; Schat, K.A., Kaspers, B., Kaiser, P., Eds.; Academic Press: Cambridge, MA, USA, 2014; pp. 149–167. [Google Scholar] [CrossRef]
- Piertney, S.B.; Oliver, M.K. The evolutionary ecology of the major histocompatibility complex. Heredity 2006, 96, 7–21. [Google Scholar] [CrossRef]
- Minias, P.; Pikus, E.; Whittingham, L.A.; Dunn, P.O. A global analysis of selection at the avian MHC. Evolution 2018, 72, 1278–1293. [Google Scholar] [CrossRef]
- Schwensow, N.; Fietz, J.; Dausmann, K.H.; Sommer, S. Neutral versus adaptive genetic variation in parasite resistance: Importance of major histocompatibility complex supertypes in a free-ranging primate. Heredity 2007, 99, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Collin, H.; Burri, R.; Comtesse, F.; Fumagalli, L. Combining molecular evolution and environmental genomics to unravel adaptive processes of MHC class IIB diversity in european minnows phoxinus phoxinus. Ecol. Evol. 2013, 3, 2568–2585. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liu, Z.; Ren, L.; Wei, Z.; Wang, P.; Li, N.; Zhao, Y. Immunoglobulin genes and diversity: What we have learned from domestic animals. J. Anim. Sci. Biotechnol. 2012, 3, 1–5. [Google Scholar] [CrossRef]
- Hedrick, P.W. Pathogen resistance and genetic variation at MHC loci. Evolution 2002, 56, 1902–1908. [Google Scholar] [CrossRef] [PubMed]
- Banat, G.R.; Tkalcigrc, S.; Dzielawa, J.A.; Jackwood, M.W.; Saggese, M.D.; Yates, L.; Kopulos, R.; Briles, W.E.; Collisson, E.W. Association of the chicken MHC B haplotypes with resistance to avian coronavirus. Dev. Comp. Immunol. 2013, 39, 430–437. [Google Scholar] [CrossRef]
- Miller, M.M.; Taylor, R.L., Jr. Brief review of the chicken Major Histocompatibility Complex: The genes, their distribution on chromosome 16, and their contributions to disease resistance distribution on chromosome 16, and their contributions to disease resistance. Poult. Sci. 2016, 95, 375–392. [Google Scholar] [CrossRef] [PubMed]
- Bettridge, J.M.; Psifidi, A.; Terfa, Z.G.; Desta, T.T.; Lozano-Jaramillo, M.; Dessie, T.; Kaiser, P.; Wigley, P.; Hanotte, O.; Christley, R.M. The role of local adaptation in sustainable production of village chickens. Nat. Sustain. 2018, 1, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Walugembe, M.; Bertolini, F.; Dematawewa, C.M.B.; Reis, M.P.; Elbeltagy, A.R.; Schmidt, C.J.; Lamont, S.J.; Rothschild, M.F. Detection of selection signatures among Brazilian, Sri Lankan, and Egyptian chicken populations under different environmental conditions. Front. Genet. 2019, 9, 737. [Google Scholar] [CrossRef] [PubMed]
- Seroussi, E.; Knytl, M.; Pitel, F.; Elleder, D.; Krylov, V.; Leroux, S.; Morisson, M.; Yosefi, S.; Miyara, S.; Ganesan, S.; et al. Avian Expression Patterns and Genomic Mapping Implicate Leptin in Digestion and TNF in Immunity, Suggesting That Their Interacting Adipokine Role Has Been Acquired Only in Mammals. Int. J. Mol. Sci. 2019, 20, 4489. [Google Scholar] [CrossRef]
- Lwelamira, J.; Kifaro, G.C.; Gwakisa, P.S.; Msoffe, P.L.M. Association of LEI0258 microsatellite alleles with antibody response against newcastle disease virus vaccine and body weight in two Tanzania chicken ecotypes. Afr. J. Biotechnol. 2008, 7, 714–720. [Google Scholar]
- Matsuzaka, Y.; Makino, S.; Nakajima, K.; Tomizawa, M.; Oka, A.; Bahram, S.; Kulski, J.K.; Tamiya, G.; Inoko, H. New polymorphic microsatellite markers in the human MHC class III region. Tissue Antigens 2001, 57, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Gourraud, P.A.; Mano, S.; Barnetche, T.; Carrington, M.; Inoko, H.; Cambon-Thomsen, A. Integration of microsatellite characteristics in the MHC region: A literature and sequence based analysis. Tissue Antigens 2004, 64, 543–555. [Google Scholar] [CrossRef]
- Tanaka, M.; Ando, A.; Renard, C.; Chardon, P.; Domuki, M.; Okumura, N.; Awata, T.; Uenishi, H. Development of dense microsatellite markers in the entire SLA region and evalustion of their polymorphism in pocine breeds. Immunogenetics 2005, 57, 690–696. [Google Scholar] [CrossRef]
- Crooijmans, R.P.M.A.; Dijkhof, R.J.M.; van der Poel, J.J.; Groenen, M.A.M. New microsatellite markers in chicken optimised for automated fluorescent genotyping. Anim. Genet. 1997, 28, 427–437. [Google Scholar] [CrossRef]
- McConnell, S.K.J.; Dawson, D.A.; Wardle, A.; Burke, T. The isolation and mapping of 19 tetranucleotide microsatellite markers in the chicken. Anim. Genet. 1999, 30, 183–189. [Google Scholar] [CrossRef]
- Buitenhuis, A.J.; Rodenburg, T.B.; Van Hierden, Y.M.; Siwek, M.; Cornelissen, S.J.B.; Nieuwland, M.G.B.; Crooijmans, R.P.M.A.; Groenen, M.A.M.; Koene, P.; Korte, S.M.; et al. Mapping quantitative trait loci affecting feather pecking behavior and stress response in laying hens. Poult. Sci. 2003, 82, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Fulton, J.E.; Juul-Madsen, H.R.; Ashwell, C.M.; McCarron, A.M.; Arthur, J.A.; O’Sullivan, N.P.; Taylor, R.L. Molecular genotype identification of the Gallus gallus major histocompatibility complex. Immunogenetics 2006, 58, 407–421. [Google Scholar] [CrossRef]
- Sironi, L.; Williams, J.L.; Stella, A.; Minozzi, G.; Moreno, A.; Ramelli, P.; Han, J.; Weigend, S.; Wan, J.; Lombardi, G.; et al. Genomic study of the response of chicken to highly pathogenic avian influenza virus. BMC Proc. 2011, 5, S25. [Google Scholar] [CrossRef]
- Chazara, O.; Juul-Madsen, H.; Chang, C.S.; Tixier-Boichard, M.; Bed’hom, B. Correlation in chicken between the marker LEI0258 alleles and Major Histocompatibility Complex sequences. BMC Proc. 2011, 4, S29. [Google Scholar] [CrossRef]
- Chazara, O.; Chang, C.S.; Bruneau, N.; Benabdeljelil, K.; Fotsa, J.C.; Kayang, B.B.; Loukou, N.E.; Osei-Amponsah, R.; Yapi-Gnaore, V.; Youssao, I.A.K.; et al. Diversity and evolution of the highly polymorphic tandem repeat LEI0258 in the chicken MHC-B region. Immunogenetics 2013, 65, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Lian, L.; Qu, L.; Zheng, J.; Yang, N. Abundant polymorphisms at the microsatellite locus LEI0258 in indigenous chickens. Poult. Sci. 2013, 92, 3113–3119. [Google Scholar] [CrossRef]
- Nikbakht, G.; Esmailnejad, A. Chicken major histocompatibility complex polymorphism and its association with production traits. Immunogenetics 2015, 67, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, P.; Liu, R.; Zheng, M.; Sun, Y.; Wu, D.; Hu, Y.; Wen, J.; Zhao, G. The identification of loci for immune traits in chickens using a genome-wide association study. PLoS ONE 2015, 30, e0117269. [Google Scholar] [CrossRef]
- Rashid, M.A.; Manjula, P.; Farque, S.; Bhuiyan, A.K.F.H.; Seo, D.; Alam, J.; Lee, J.H.; Bhuiyan, M.S.A. Genetic diversity and population structure of indigenous chicken of Bangladesh using microsatellite markers. Asian Autralas J. Anim. Sci. 2020, 11, 1732–1742. [Google Scholar] [CrossRef] [PubMed]
- Manjula, P.; Fulton, J.E.; Seo, D.; Lee, J.H. Major histocompatibility complex B (MHC-B) variability in Korean native chicken. Poult. Sci. 2020, 99, 4704–4713. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem Repeats Database. 2016. Available online: Tandem.bu.edu/cgi-bin/trdb/trdb.exe (accessed on 14 May 2019).
- Matschiner, M.; Salzburger, W. TANDEM: Integrating automated allele binning into genetics and genomics workflows. Bioinformatics 2009, 25, 1982–1983. [Google Scholar] [CrossRef]
- Hosomichi, K.; Miller, M.M.; Goto, R.M.; Wang, Y.; Suzuki, S.; Kulski, J.K.; Nishibori, M.; Inoko, H.; Hanzawa, K.; Shiina, T. Contribution of mutation, recombination, and gene conversion to chicken MHC-B haplotype diversity. J. Immunol. 2008, 181, 3393–3399. [Google Scholar] [CrossRef] [PubMed]
- Shiina, T.; Briles, W.E.; Goto, R.M.; Hosomichi, K.; Yanagiya, K.; Shimizu, S.; Inoko, H.; Miller, M.M. Extended gene map reveals Tripartite Motif, C-Type Lectin, and Ig superfamily type genes within a subregion of the chicken MHC-B affecting infectious disease. J. Immunol. 2007, 178, 7162–7172. [Google Scholar] [CrossRef]
- Raymond, M.J.J.H. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 19, 1512. [Google Scholar] [CrossRef]
- Kalinowsky, S.T. Counting alleles with rarefaction: Private alleles and hierarchical sampling designs. Conserv. Gen. 2004, 5, 539–543. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Nei, M.; Tajima, F.; Tateno, Y. Accuracy of estimated phylogenetic tree from molecular data. J. Mol. Evol. 1983, 19, 153–170. [Google Scholar] [CrossRef] [PubMed]
- Takezaki, N.; Nei, M.; Tamura, K. POPTREEW: Web Version of POPTREE for Constructing Population Trees from Allele Frequency Data and Computing Other Population Statistics. 2014. Available online: http://www.med.kagawau.ac.jp/~genomelb/takezaki/poptreew/index.html (accessed on 15 November 2020).
- Molamane, D.K.; Simianer, H.; Weigend, A.; Reimer, C.; Schmitt, A.O.; Weigend, S. The SYNBREED chicken diversity panel: A global resource to assess chicken diversity at high genomic resolution. BMC Genom. 2019, 20, 345. [Google Scholar] [CrossRef]
- Kirk, H.; Freeland, J.R. Applications and implications of neutral versus non-neutral markers in molecular ecology. Int. J. Mol. Sci. 2011, 12, 3966–3988. [Google Scholar] [CrossRef]
- Mwambene, P.L.; Kyallo, M.; Machuka, E.; Githae, D.; Pelle, R. Genetic diversity of 10 indigenous chicken ecotypes from Southern Highlands of Tanzania based on Major Histocompatibility Complex-linked microsatellite LEI0258 marker typing. Poult. Sci. 2019, 98, 2734–2746. [Google Scholar] [CrossRef]
- Chapuis, M.; Estoup, A. Microsatellite Null Alleles and Estimation of Population Differentiation. Mol. Bio. Evol. 2007, 24, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Gray, I.C. From linkage peak to curlprit gene: Following up linkage analysis of complex phenotypes with population based association studies. In Bioinformatics for Geneticists; Barnes, M.C., Gray, I.C., Eds.; John Wiley & Sons: West Sussex, UK, 2003; Volume 8, pp. 166–178. [Google Scholar]
- Silva, P.; Liyanage, R.P.; Senadheera, R.; Dematawewa, C.M.B. Monograph on Indigenous Chicken in Sri Lanka; UNEP-GEF-ILRI FAnGR Asia Project; University of Peradeniya: Kandi, Sri Lanka, 2016; pp. 1–74. [Google Scholar]
- Manjula, P.; Fulton, J.E.; Seo, D.; Lee, J.H. Comparison of Major histocompatibility complex B variability in Sri Lankan indigenous chickens with five global chicken populations using MHC-B SNP panel. Anim. Gent. 2021; under review. [Google Scholar]
- Manjula, P.; Bed’Hom, B.; Hoque, M.R.; Cho, S.; Seo, D.; Chazara, O.; Lee, S.H.; Lee, J.H. Genetic diversity of MHC-B in 12 chicken populations in Korea revealed by single nucleotide polymorphisms. Immunogenetics 2020, 72, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Jayasena, D.D.; Jo, C.; Lee, J.H. The breeding history and commercial development of the Korean native chicken. World’s Poult. Sci. J. 2017, 73, 163–174. [Google Scholar] [CrossRef]
- Greenbaum, G.; Templeton, A.R.; Zarmi, Y.; Bar-David, S. Allelic Richness following Population Founding Events—A Stochastic Modeling Framework Incorporating Gene Flow and Genetic Drift. PLoS ONE 2014, 9, e115203. [Google Scholar] [CrossRef] [PubMed]
- Petit, R.J.; El Mousadik, A.; Pons, O. Identifying populations for conservation on the basis of genetic markers. Conserv. Biol. 1998, 12, 844–855. [Google Scholar] [CrossRef]
- Caballero, A.; Garcia-Dorado, A. Allelic diversity and its implications for the rate of adaptation. Genetics 2013, 11, 158410. [Google Scholar] [CrossRef] [PubMed]
- Radwan, J.; Babik, W.; Kaufman, J.; Lenz, T.L.; Winternitz, J. Advances in the Evolutionary understanding of MHC polymorphism. Trends Genet. 2020, 36, 298–311. [Google Scholar] [CrossRef]
- Bodmer, W.F. Evolutionary significance of the HL-A system. Nature 1972, 237, 139–145. [Google Scholar] [CrossRef]
- Jaworska, J.; Molik, K.R.; Potocka, I.W.; Siemieniuch, M. Inter- and intrabreed diversity of the major histocompatibility complex MHC in primitive and draft horse breeds. PLoS ONE 2020, 15, e0228658. [Google Scholar] [CrossRef]
- Fulton, J.E.; Lund, A.R.; Mccarron, A.M.; Pinegar, K.N.; Korver, D.R.; Classen, H.L.; Aggrey, S.; Utterbach, C.; Anthony, N.B.; Berres, M.E. MHC variability in heritage breeds of chickens. Poult. Sci. 2016, 95, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Han, L.; Han, J.; Liu, J.; Jiang, Q.; Guo, D.; Qu, L. Establishment of six homozygous MHC-B haplotype populations associated with susceptibility to Marek’s disease in Chinese specific pathogen-free BWEL chickens. Infect. Genet. Evol. 2014, 29, 15–25. [Google Scholar] [CrossRef] [PubMed]
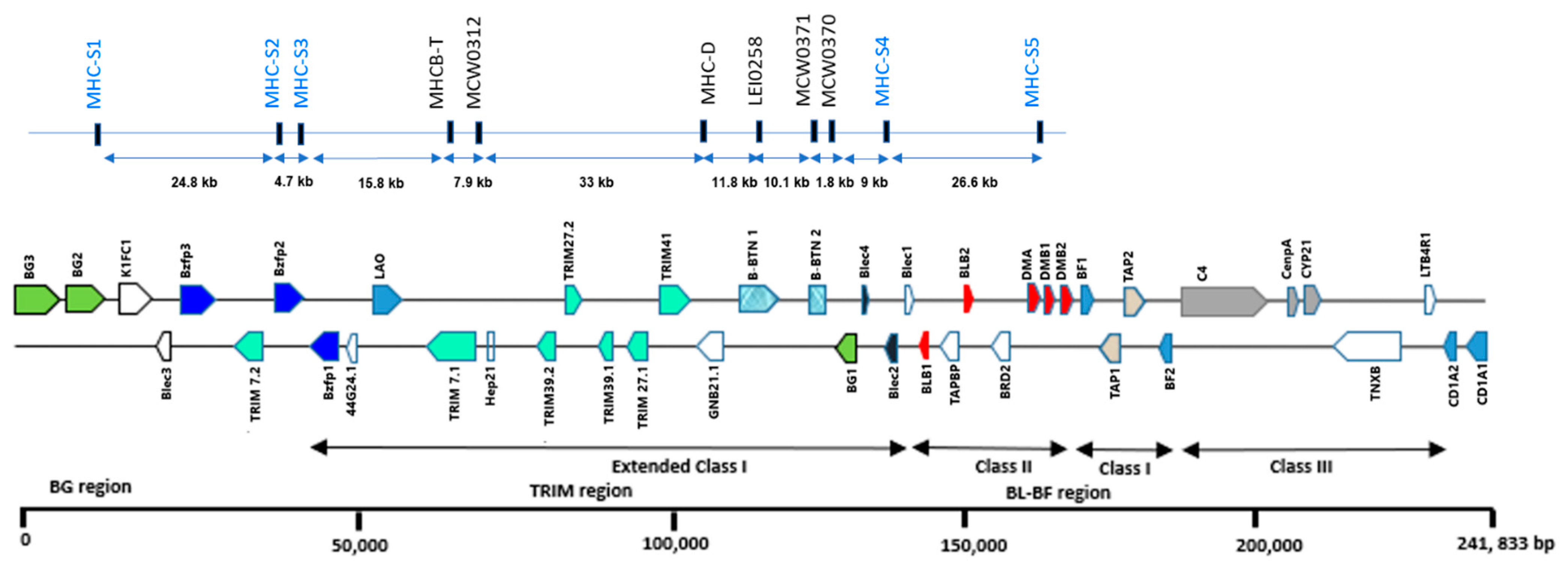
| Marker | Range | N | Na | Ho | He | PIC | FIS | F (null) | HWE |
|---|---|---|---|---|---|---|---|---|---|
| MHC-S1 a | 231–263 | 166 | 4 | 0.205 | 0.402 | 0.368 | 0.380 | 0.313 | *** |
| MHC-S2 a | 227–233 | 181 | 3 | 0.000 | 0.033 | 0.032 | 1.000 | 0.443 | nd |
| MHC-S3 a | 205–208 | 176 | 2 | 0.216 | 0.326 | 0.272 | 0.043 | 0.202 | *** |
| MHC-S4 a | 214–223 | 186 | 4 | 0.081 | 0.094 | 0.092 | −0.197 | 0.093 | nd |
| MHC-S5 a | 255–270 | 185 | 5 | 0.276 | 0.315 | 0.277 | −0.027 | 0.064 | ns |
| MHC-T b | 232–240 | 768 | 5 | 0.362 | 0.517 | 0.451 | 0.087 | 0.160 | *** |
| MCW0312 b | 210–218 | 867 | 4 | 0.379 | 0.494 | 0.406 | −0.055 | 0.134 | *** |
| MHC-D b | 307–319 | 870 | 5 | 0.632 | 0.742 | 0.694 | −0.046 | 0.079 | *** |
| LEI0258 b | 193–539 | 879 | 38 | 0.782 | 0.918 | 0.912 | −0.034 | 0.080 | *** |
| MCW0371 b | 200–210 | 877 | 10 | 0.429 | 0.808 | 0.784 | 0.371 | 0.313 | *** |
| MCW0370 b | 168–182 | 816 | 12 | 0.308 | 0.837 | 0.818 | 0.525 | 0.465 | *** |
| Pop Code 1 | Na | Ne | AR | Ho | He | F |
|---|---|---|---|---|---|---|
| KNC | 1.8 | 1.23 | 1.32 | 0.104 | 0.165 | 0.316 |
| YO | 1.8 | 1.39 | 1.72 | 0.246 | 0.207 | −0.175 |
| YCC | 1.8 | 1.16 | 1.56 | 0.107 | 0.115 | 0.036 |
| CC1 | 1.6 | 1.22 | 1.49 | 0.125 | 0.142 | 0.077 |
| CC2 | 1.4 | 1.24 | 1.39 | 0.100 | 0.150 | 0.166 |
| CC3 | 1.8 | 1.45 | 1.67 | 0.171 | 0.204 | 0.220 |
| CC4 | 1.4 | 1.34 | 1.38 | 0.075 | 0.127 | 0.135 |
| KA | 1.8 | 1.34 | 1.66 | 0.175 | 0.211 | 0.123 |
| TA | 2.0 | 1.47 | 1.82 | 0.159 | 0.238 | 0.284 |
| ND | 2.0 | 1.63 | 1.83 | 0.300 | 0.305 | −0.006 |
| NN | 1.4 | 1.29 | 1.39 | 0.104 | 0.166 | 0.407 |
| AS | 2.0 | 1.56 | 1.91 | 0.425 | 0.345 | −0.103 |
| HI | 2.0 | 1.43 | 1.76 | 0.232 | 0.240 | 0.009 |
| JF | 1.8 | 1.27 | 1.80 | 0.182 | 0.191 | −0.011 |
| ADOL | 1.4 | 1.07 | 1.76 | 0.062 | 0.057 | −0.084 |
| RIR | 1.6 | 1.36 | 1.59 | 0.233 | 0.225 | −0.037 |
| WL | 1.4 | 1.37 | 1.40 | 0.219 | 0.192 | −0.133 |
| HY | 1.6 | 1.19 | 1.76 | 0.123 | 0.132 | 0.373 |
| IR | 1.8 | 1.52 | 1.53 | 0.000 | 0.278 | 1.000 |
| Ross | 1.2 | 1.09 | 1.18 | 0.025 | 0.061 | 0.590 |
| Ab | 1.4 | 1.16 | 1.36 | 0.086 | 0.116 | 0.364 |
| Mean | 1.67 | 1.34 | 0.154 | 0.184 | 0.147 |
| Pop Code 1 | N | Na | Ne | AR | Ho | He | F | FIS |
|---|---|---|---|---|---|---|---|---|
| NG | 49.67 | 3.67 | 2.30 | 2.98 | 0.390 | 0.485 | 0.204 | 0.0585 |
| NL | 49.67 | 3.67 | 2.38 | 2.87 | 0.523 | 0.546 | 0.020 | −0.0390 |
| NR | 43.17 | 4.50 | 3.10 | 3.75 | 0.508 | 0.587 | 0.162 | 0.0307 |
| NW | 47.17 | 5.00 | 2.56 | 3.73 | 0.463 | 0.560 | 0.146 | 0.1348 |
| NY | 50.00 | 4.33 | 3.31 | 3.82 | 0.574 | 0.639 | 0.054 | 0.1193 |
| NO | 45.00 | 3.83 | 2.05 | 2.94 | 0.338 | 0.468 | 0.378 | 0.0682 |
| YO | 59.33 | 5.17 | 2.81 | 3.79 | 0.409 | 0.582 | 0.293 | 0.1242 |
| YCC | 47.83 | 5.83 | 3.10 | 4.00 | 0.553 | 0.612 | 0.081 | 0.0114 |
| CC1 | 19.83 | 3.83 | 2.79 | 3.42 | 0.689 | 0.615 | −0.114 | −0.1023 |
| CC2 | 20.00 | 4.33 | 2.76 | 3.63 | 0.475 | 0.596 | 0.190 | 0.1849 |
| CC3 | 19.00 | 4.00 | 3.17 | 3.71 | 0.569 | 0.644 | 0.102 | 0.0415 |
| CC4 | 20.00 | 4.33 | 2.99 | 3.86 | 0.525 | 0.561 | 0.018 | −0.0689 |
| KA | 18.83 | 6.50 | 5.10 | 5.43 | 0.652 | 0.735 | 0.067 | 0.0992 |
| NU | 17.83 | 6.33 | 4.04 | 5.02 | 0.447 | 0.650 | 0.306 | 0.1983 |
| TA | 34.17 | 8.17 | 4.19 | 4.96 | 0.495 | 0.657 | 0.259 | 0.1559 |
| ND | 36.00 | 5.50 | 3.22 | 4.05 | 0.469 | 0.650 | 0.264 | 0.1396 |
| NN | 45.17 | 5.17 | 2.26 | 3.29 | 0.364 | 0.531 | 0.309 | 0.2137 |
| AS | 16.83 | 5.83 | 3.79 | 4.90 | 0.511 | 0.704 | 0.277 | 0.1979 |
| HI | 38.17 | 6.33 | 3.99 | 4.86 | 0.594 | 0.679 | 0.148 | 0.0866 |
| JF | 18.33 | 3.67 | 2.56 | 3.32 | 0.299 | 0.530 | 0.408 | 0.3778 |
| NIG | 18.50 | 6.00 | 3.73 | 4.77 | 0.507 | 0.645 | 0.206 | 0.0145 |
| RIR | 14.00 | 2.33 | 1.86 | 2.30 | 0.452 | 0.412 | −0.109 | −0.1513 |
| WL | 15.50 | 2.33 | 1.68 | 2.31 | 0.265 | 0.342 | 0.345 | 0.1266 |
| HY | 18.67 | 3.50 | 2.02 | 2.91 | 0.339 | 0.421 | 0.081 | 0.0623 |
| LO | 19.17 | 3.00 | 2.31 | 2.80 | 0.429 | 0.495 | 0.060 | −0.0913 |
| IR | 15.67 | 4.50 | 3.12 | 3.97 | 0.631 | 0.614 | −0.056 | −0.0126 |
| Ross | 15.67 | 4.83 | 2.57 | 3.74 | 0.551 | 0.533 | −0.024 | −0.0334 |
| Ab | 16.67 | 4.00 | 2.25 | 3.27 | 0.468 | 0.531 | 0.101 | 0.12057 |
| Cobb | 16.33 | 4.50 | 3.14 | 3.92 | 0.581 | 0.620 | 0.021 | 0.11709 |
| Source of Variation 1 | d.f. | Variance Component | % Variance | Fixation Index | Significance (p-Value) |
|---|---|---|---|---|---|
| Among groups | 6 | 219.22 | 3.26 | FST = 0.115 | 0.000 |
| Among populations within groups | 22 | 557.33 | 8.28 | FSC = 0.086 | 0.000 |
| Within populations | 1473 | 5955.95 | 88.47 | FCT = 0.033 | 0.081 |
| Genetic Diversity | |||||
|---|---|---|---|---|---|
| Production System 1 | Marker | N 2 | Number of Alleles | Allele Richness 3 | uHe |
| Backyard/Semi-intensive system | MHC-T | 291 | 4 | 2.79 | 0.521 |
| MHC0312 | 314 | 4 | 2.68 | 0.459 | |
| LEI0258 | 316 | 33 | 7.88 | 0.926 | |
| MHC-D | 315 | 5 | 3.71 | 0.741 | |
| MCW0371 | 315 | 10 | 5.05 | 0.766 | |
| MCW0370 | 308 | 11 | 4.62 | 0.817 | |
| 4 Mean (SE) | 309.83 | 11.17 (4.54) | 4.46 (0.79) | 0.705 | |
| Marker | N | Number of alleles | Allele richness | uHe | |
| Intensive system for native chicken | MHC-T | 376 | 5 | 2.05 | 0.512 |
| MHC0312 | 453 | 4 | 2.04 | 0.527 | |
| LEI0258 | 459 | 26 | 4.94 | 0.884 | |
| MHC-D | 454 | 5 | 3.41 | 0.736 | |
| MCW0371 | 458 | 10 | 3.76 | 0.814 | |
| MCW0370 | 412 | 11 | 3.79 | 0.828 | |
| 4 Mean (SE) | 435.3 | 10.17 (3.38) | 3.33 (0.46) | 0.717 | |
| Marker | N | Number of alleles | Allele richness | uHe | |
| Intensive system for commercial chicken | MHC-T | 101 | 3 | 1.90 | 0.468 |
| MHC0312 | 100 | 2 | 1.97 | 0.249 | |
| LEI0258 | 104 | 18 | 5.46 | 0.861 | |
| MHC-D | 101 | 4 | 3.24 | 0.674 | |
| MCW0371 | 104 | 8 | 3.83 | 0.774 | |
| MCW0370 | 96 | 10 | 4.23 | 0.790 | |
| 4 Mean (SE) | 101.00 | 7.5 (2.45) | 3.44(0.56) | 0.636 | |
| Pop Code 1 | Total N. ha 2 | N ha 3 (F > 5%) | Unique Haplotypes 4 |
|---|---|---|---|
| RIR | 4 | 4 | 0 |
| WL | 13 | 9 | 11 |
| NG | 40 | 4 | 29 |
| NL | 22 | 4 | 13 |
| NR | 17 | 9 | 5 |
| NW | 17 | 5 | 7 |
| NY | 22 | 7 | 4 |
| NO | 13 | 5 | 4 |
| YOG | 34 | 5 | 20 |
| HI | 32 | 3 | 21 |
| ND | 36 | 3 | 24 |
| NN | 18 | 4 | 11 |
| AS | 26 | 3 | 22 |
| JF | 14 | 7 | 13 |
| KA | 31 | 4 | 19 |
| NU | 24 | 8 | 19 |
| TA | 46 | 1 | 30 |
| NIG | 20 | 7 | 14 |
| YCC | 27 | 5 | 7 |
| CC1 | 14 | 6 | 3 |
| CC2 | 15 | 7 | 5 |
| CC3 | 16 | 8 | 3 |
| CC4 | 19 | 8 | 7 |
| IN | 14 | 6 | 4 |
| Ab | 13 | 8 | 4 |
| Cobb | 15 | 6 | 7 |
| Ross | 19 | 4 | 6 |
| HL | 13 | 6 | 6 |
| LO | 12 | 6 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manjula, P.; Kim, M.; Cho, S.; Seo, D.; Lee, J.H. High Levels of Genetic Variation in MHC-Linked Microsatellite Markers from Native Chicken Breeds. Genes 2021, 12, 240. https://doi.org/10.3390/genes12020240
Manjula P, Kim M, Cho S, Seo D, Lee JH. High Levels of Genetic Variation in MHC-Linked Microsatellite Markers from Native Chicken Breeds. Genes. 2021; 12(2):240. https://doi.org/10.3390/genes12020240
Chicago/Turabian StyleManjula, Prabuddha, Minjun Kim, Sunghyun Cho, Dongwon Seo, and Jun Heon Lee. 2021. "High Levels of Genetic Variation in MHC-Linked Microsatellite Markers from Native Chicken Breeds" Genes 12, no. 2: 240. https://doi.org/10.3390/genes12020240
APA StyleManjula, P., Kim, M., Cho, S., Seo, D., & Lee, J. H. (2021). High Levels of Genetic Variation in MHC-Linked Microsatellite Markers from Native Chicken Breeds. Genes, 12(2), 240. https://doi.org/10.3390/genes12020240







