Dynamic Molecular Epidemiology Reveals Lineage-Associated Single-Nucleotide Variants That Alter RNA Structure in Chikungunya Virus
Abstract
1. Introduction
1.1. Geographical Spread of Chikungunya Virus
1.2. RNA Structure Conservation in Chikungunya Virus Genomes
1.3. Molecular Epidemiology Reveals RNA Structure-Affecting SNVs
2. Materials and Methods
2.1. Taxon Selection
2.2. Genetic Distance
2.3. CHIKV Nextstrain
2.4. RNA sTructure Modulation via Lineage-Associated SNVs
2.5. Data Availability
3. Results
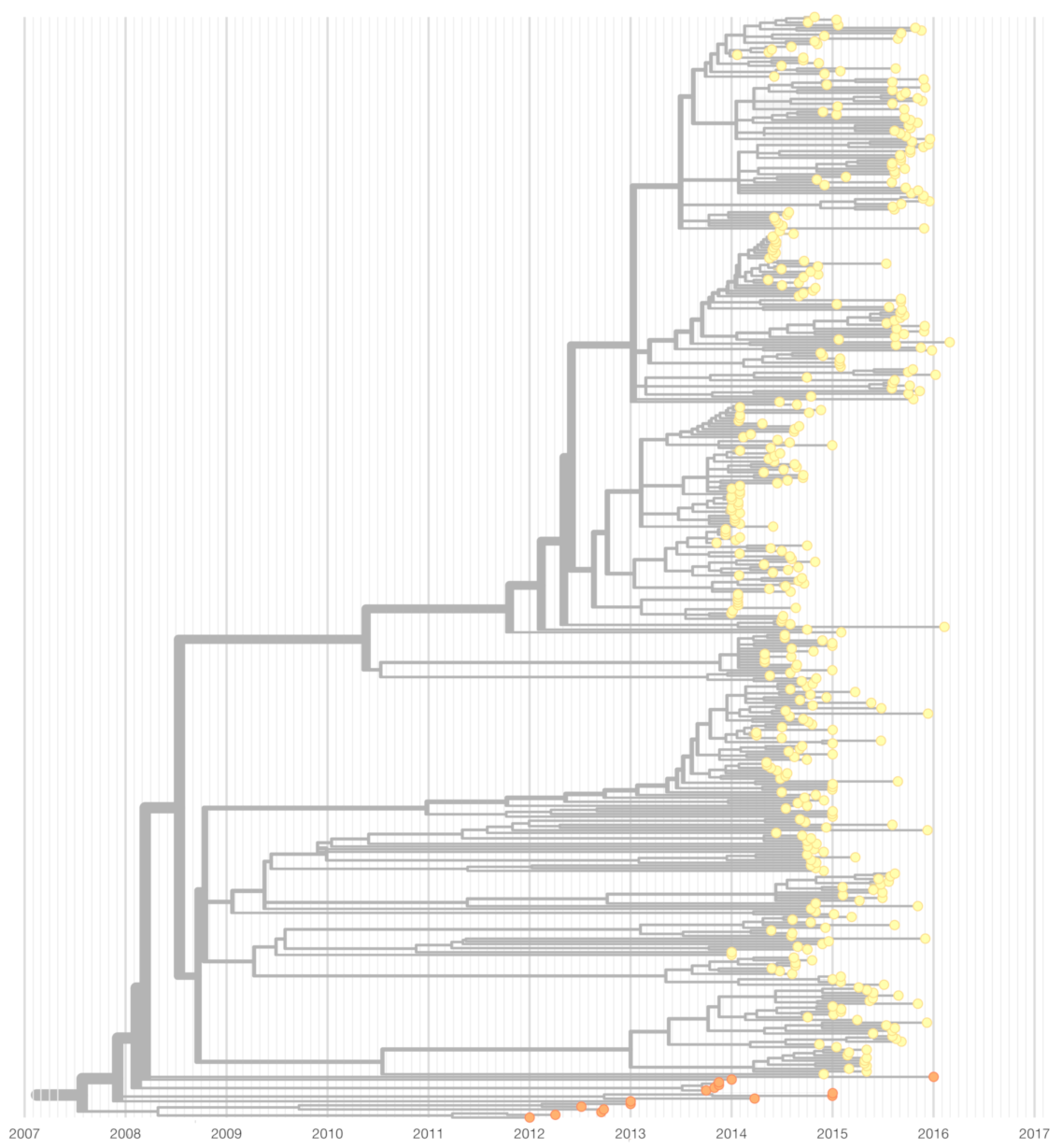
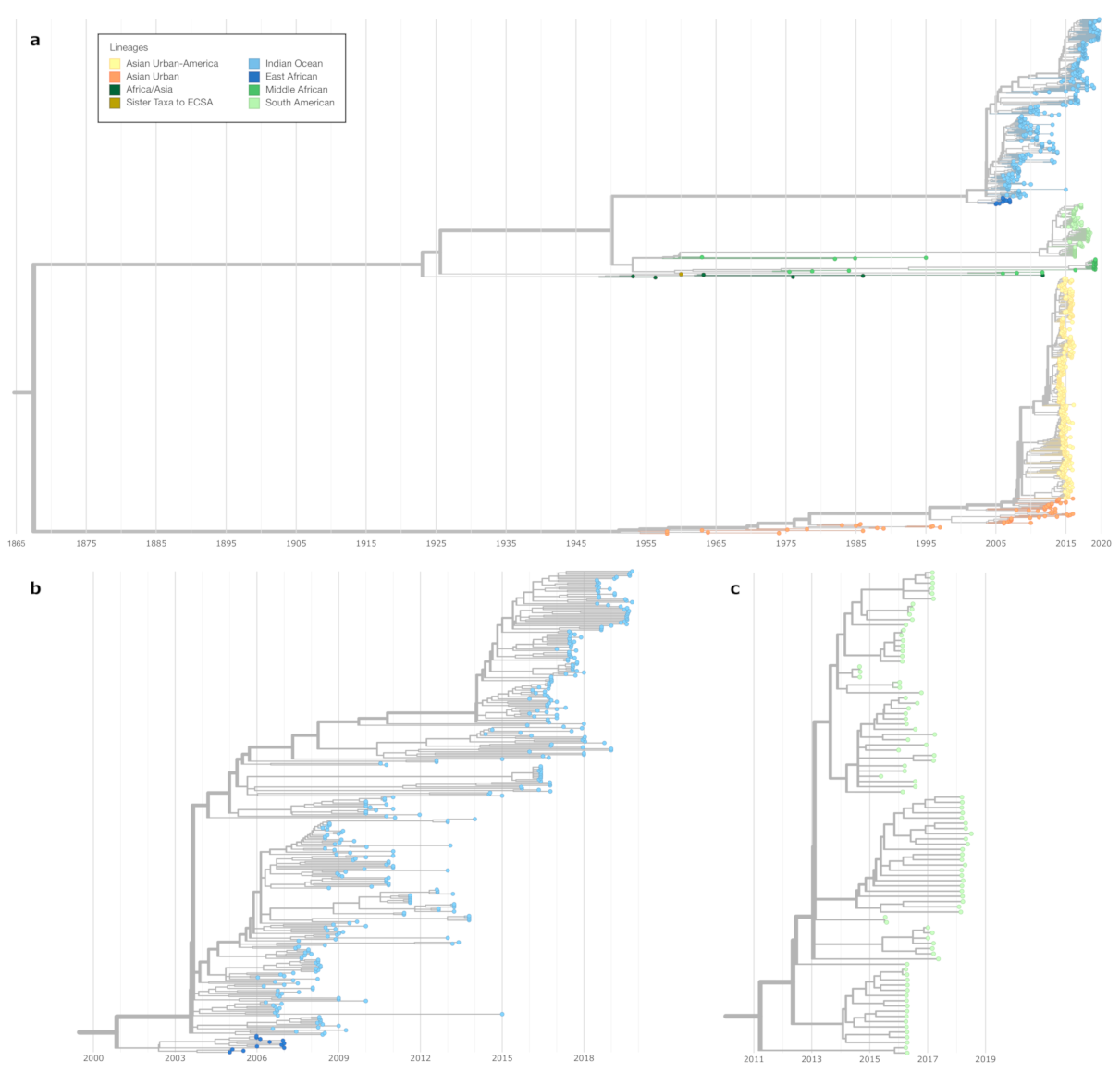
3.1. Genetic Distance between Chikungunya Virus Lineages
3.2. A Nextstrain Build for Chikungunya Virus
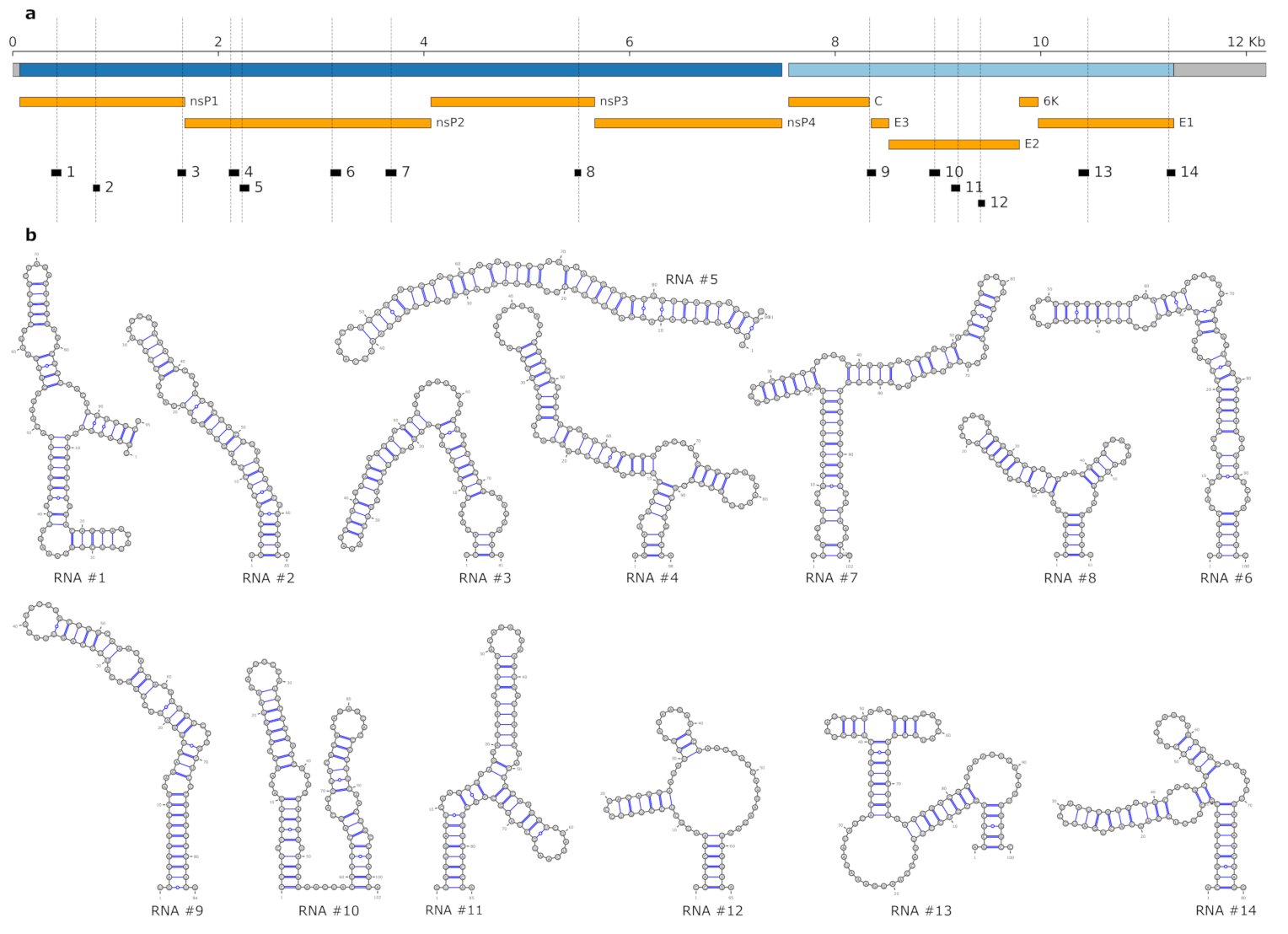
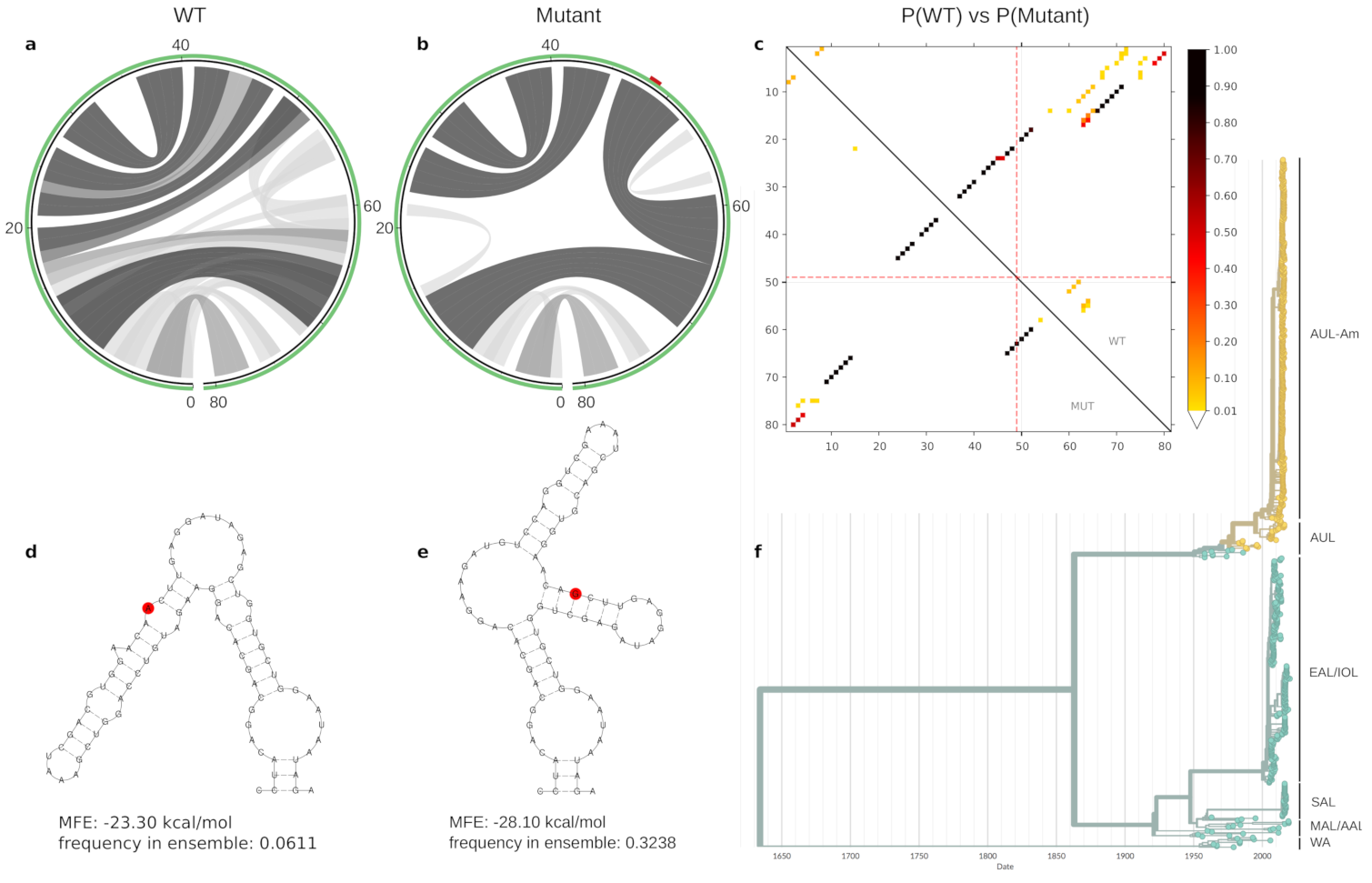
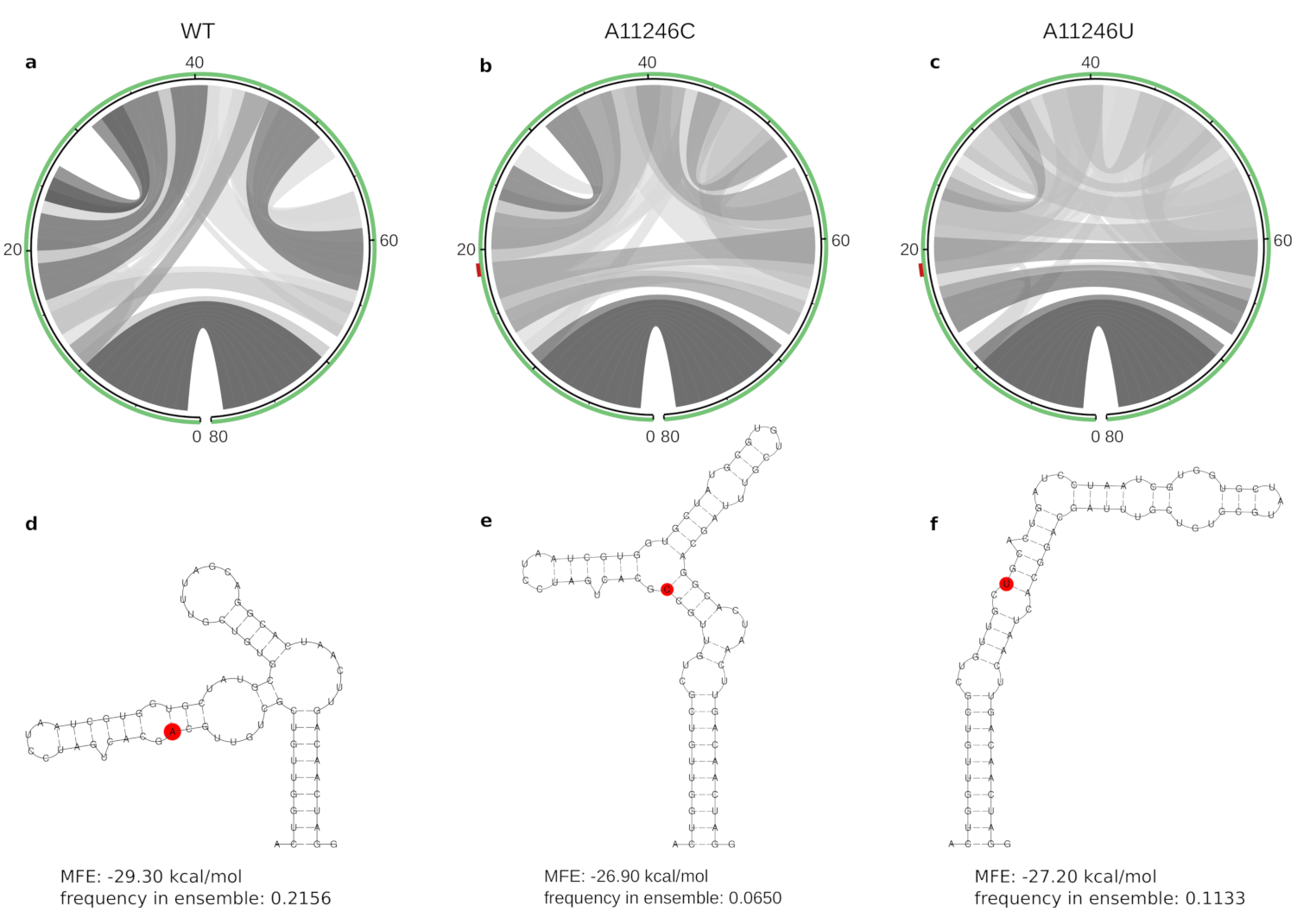
3.3. Lineage-Specific RNA Structures
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Lineage | Divergence |
|---|---|
| AUL-Am | 0.0012 |
| AUL | 0.0128 |
| SAL | 0.003 |
| MAL | 0.0107 |
| IOL | 0.0062 |
| EAL | 0.0011 |
| AAL | 0.0066 |
| WA | 0.0102 |
References
- Chandak, N.; Kashyap, R.; Kabra, D.; Karandikar, P.; Saha, S.; Morey, S.; Purohit, H.; Taori, G.; Daginawala, H. Neurological complications of Chikungunya virus infection. (Original Article) (Clinical report). Neurol. India 2009, 57, 177. [Google Scholar] [PubMed]
- Forrester, N.; Palacios, G.; Tesh, R.; Savji, N.; Guzman, H.; Sherman, M.; Weaver, S.; Lipkin, W. Genome-scale phylogeny of the alphavirus genus suggests a marine origin. J. Virol. 2011, 86, 2729–2738. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.C.; Forrester, N.L. Chikungunya: Evolutionary history and recent epidemic spread. Antivir. Res. 2015, 120, 32–39. [Google Scholar] [CrossRef]
- Her, Z.; Kam, Y.W.; Lin, R.T.; Ng, L.F. Chikungunya: A bending reality. Microbes Infect. 2009, 11, 1165–1176. [Google Scholar] [CrossRef]
- Robinson, M.C. An Epidemic Of Virus Disease In Southern Province, Tanganyika Territory. Trans. R. Soc. Trop. Med. Hyg. 1955, 49, 28–32. [Google Scholar] [CrossRef]
- Ross, R. The Newala epidemic: III. The virus: Isolation, pathogenic properties and relationship to the epidemic. Epidemiol. Infect. 1956, 54, 177–191. [Google Scholar] [CrossRef]
- Pialoux, G.; Gaüzère, B.A.; Jauréguiberry, S.; Strobel, M. Chikungunya, an epidemic arbovirosis. Lancet Infect. Dis. 2007, 7, 319–327. [Google Scholar] [CrossRef]
- Renault, P.; Solet, J.L.; Sissoko, D.; Balleydier, E.; Larrieu, S.; Filleul, L.; Lassalle, C.; Thiria, J.; Rachou, E.; De Valk, H.; et al. A major epidemic of chikungunya virus infection on Réunion Island, France, 2005–2006. Am. J. Trop. Med. Hyg. 2007, 77, 727–731. [Google Scholar] [CrossRef]
- Tsetsarkin, K.A.; Chen, R.; Weaver, S.C. Interspecies transmission and chikungunya virus emergence. Curr. Opin. Virol. 2016, 16, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Schuffenecker, I.; Iteman, I.; Michault, A.; Murri, S.; Frangeul, L.; Vaney, M.C.; Lavenir, R.; Pardigon, N.; Reynes, J.M.; Pettinelli, F.; et al. Genome Microevolution of Chikungunya Viruses Causing the Indian Ocean Outbreak (Chikungunya Virus Genome Microevolution). PLoS Med. 2006, 3, e263. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Countries and Territories Where Chikungunya Cases Have Been Reported (as of 17 September 2019). 2019. Available online: https://www.cdc.gov/chikungunya/geo (accessed on 8 June 2020).
- Delatte, H.; Desvars, A.; Bouetard, A.; Bord, S.; Gimonneau, G.; Vourc, G.; Fontenille, D. Blood-feeding behavior of Aedes albopictus, a Vector of Chikungunya on la Reunion. (Report). Vector-Borne Zoonotic Dis. 2010, 10, 249. [Google Scholar] [CrossRef]
- Rochlin, I.; Ninivaggi, D.V.; Hutchinson, M.L.; Farajollahi, A. Climate change and range expansion of the Asian tiger mosquito (Aedes albopictus) in Northeastern USA: Implications for public health practitioners. PLoS ONE 2013, 8, e60874. [Google Scholar] [CrossRef]
- Powers, A.M.; Brault, A.C.; Tesh, R.B.; Weaver, S.C. Re-emergence of Chikungunya and O’nyong-nyong viruses: Evidence for distinct geographical lineages and distant evolutionary relationships. J. Gen. Virol. 2000, 81, 471–479. [Google Scholar] [CrossRef]
- Volk, S.M.; Chen, R.; Tsetsarkin, K.A.; Adams, A.P.; Garcia, T.I.; Sall, A.A.; Nasar, F.; Schuh, A.J.; Holmes, E.C.; Higgs, S.; et al. Genome-Scale Phylogenetic Analyses of Chikungunya Virus Reveal Independent Emergences of Recent Epidemics and Various Evolutionary Rates. J. Virol. 2010, 84, 6497–6504. [Google Scholar] [CrossRef]
- De Bernardi Schneider, A.; Ochsenreiter, R.; Hostager, R.; Hofacker, I.L.; Janies, D.; Wolfinger, M.T. Updated Phylogeny of Chikungunya Virus Suggests Lineage-Specific RNA Architecture. Viruses 2019, 11, 798. [Google Scholar] [CrossRef]
- Nunes, M.R.T.; Faria, N.R.; de Vasconcelos, J.M.; Golding, N.; Kraemer, M.U.; de Oliveira, L.F.; da Silva Azevedo, R.d.S.; da Silva, D.E.A.; da Silva, E.V.P.; da Silva, S.P.; et al. Emergence and potential for spread of Chikungunya virus in Brazil. BMC Med. 2015, 13, 102. [Google Scholar] [CrossRef]
- Teixeira, M.G.; Andrade, A.M.; Maria da Conceição, N.C.; Castro, J.S.; Oliveira, F.L.; Goes, C.S.; Maia, M.; Santana, E.B.; Nunes, B.T.; Vasconcelos, P.F. East/Central/South African genotype Chikungunya virus, Brazil, 2014. Emerg. Infect. Dis. 2015, 21, 906. [Google Scholar] [CrossRef]
- White, S.K.; Mavian, C.; Salemi, M.; Morris, J.G., Jr.; Elbadry, M.A.; Okech, B.A.; Lednicky, J.A.; Dunford, J.C. A new “American” subgroup of African-lineage Chikungunya virus detected in and isolated from mosquitoes collected in Haiti, 2016. PLoS ONE 2018, 13, e0196857. [Google Scholar] [CrossRef]
- Rezza, G.; Nicoletti, L.; Angelini, R.; Romi, R.; Finarelli, A.; Panning, M.; Cordioli, P.; Fortuna, C.; Boros, S.; Magurano, F.; et al. Infection with Chikungunya virus in Italy: An outbreak in a temperate region. Lancet 2007, 370, 1840–1846. [Google Scholar] [CrossRef]
- Lanciotti, R.S.; Kosoy, O.L.; Laven, J.J.; Panella, A.J.; Velez, J.O.; Lambert, A.J.; Campbell, G.L. Chikungunya virus in US travelers returning from India, 2006. Emerg. Infect. Dis. 2007, 13, 764. [Google Scholar] [CrossRef]
- De Lamballerie, X.; Leroy, E.; Charrel, R.; Ttsetsarkin, K.; Higgs, S.; Gould, E. Chikungunya virus adapts to tiger mosquito via evolutionary convergence: A sign of things to come? Virol. J. 2008, 5, 33. [Google Scholar] [CrossRef]
- Zeller, H.; Van Bortel, W.; Sudre, B. Chikungunya: Its history in Africa and Asia and its spread to new regions in 2013–2014. J. Infect. Dis. 2016, 214, S436–S440. [Google Scholar] [CrossRef]
- Yactayo, S.; Staples, J.E.; Millot, V.; Cibrelus, L.; Ramon-Pardo, P. Epidemiology of Chikungunya in the Americas. J. Infect. Dis. 2016, 214, S441–S445. [Google Scholar] [CrossRef]
- Halstead, S.B. Reappearance of chikungunya, formerly called dengue, in the Americas. Emerg. Infect. Dis. 2015, 21, 557. [Google Scholar] [CrossRef]
- Strauss, J.H.; Strauss, E.G. The Alphaviruses: Gene Expression, Replication, and Evolution. Microbiol. Mol. Biol. R. 1994, 58, 491–562. [Google Scholar] [CrossRef]
- Li, X.F.; Jiang, T.; Deng, Y.Q.; Zhao, H.; Yu, X.D.; Ye, Q.; Wang, H.J.; Zhu, S.Y.; Zhang, F.C.; Qin, E.D.; et al. Complete genome sequence of a Chikungunya virus isolated in Guangdong, China. J. Virol. 2012, 86, 8904–8905. [Google Scholar] [CrossRef]
- Hyde, J.L.; Chen, R.; Trobaugh, D.W.; Diamond, M.S.; Weaver, S.C.; Klimstra, W.B.; Wilusz, J. The 5’ and 3’ ends of alphavirus RNAs–non-coding is not non-functional. Virus Res. 2015, 206, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Wang, E.; Tsetsarkin, K.A.; Weaver, S.C. Chikungunya virus 3’ untranslated region: Adaptation to mosquitoes and a population bottleneck as major evolutionary forces. PLoS Pathog. 2013, 9, e1003591. [Google Scholar] [CrossRef]
- Filomatori, C.V.; Bardossy, E.S.; Merwaiss, F.; Suzuki, Y.; Henrion, A.; Saleh, M.C.; Alvarez, D.E. RNA recombination at Chikungunya virus 3’UTR as an evolutionary mechanism that provides adaptability. PLoS Pathog. 2019, 15, e1007706. [Google Scholar] [CrossRef]
- Kiening, M.; Ochsenreiter, R.; Hellinger, H.J.; Rattei, T.; Hofacker, I.; Frishman, D. Conserved secondary structures in viral mRNAs. Viruses 2019, 11, 401. [Google Scholar] [CrossRef]
- Ochsenreiter, R.; Hofacker, I.L.; Wolfinger, M.T. Functional RNA Structures in the 3’UTR of Tick-Borne, Insect-Specific and No-Known-Vector Flaviviruses. Viruses 2019, 11, 298. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Leibowitz, J.L. The structure and functions of coronavirus genomic 3’ and 5’ ends. Virus Res. 2015, 206, 120–133. [Google Scholar] [CrossRef]
- Kutchko, K.M.; Madden, E.A.; Morrison, C.; Plante, K.S.; Sanders, W.; Vincent, H.A.; Cruz Cisneros, M.C.; Long, K.M.; Moorman, N.J.; Heise, M.T.; et al. Structural divergence creates new functional features in alphavirus genomes. Nucleic Acids Res. 2018, 46, 3657–3670. [Google Scholar] [CrossRef]
- Madden, E.A.; Plante, K.S.; Morrison, C.R.; Kutchko, K.M.; Sanders, W.; Long, K.M.; Taft-Benz, S.; Cisneros, M.C.C.; White, A.M.; Sarkar, S.; et al. Using SHAPE-MaP to model RNA secondary structure and identify 3’UTR variation in chikungunya virus. J. Virol. 2020, 94, e00701-20. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, M.; Kinney, R.M.; Kaaden, O.R. The Alphavirus 3’-Nontranslated Region: Size Heterogeneity and Arrangement of Repeated Sequence Elements. Virology 1998, 240, 100–108. [Google Scholar] [CrossRef]
- Halvorsen, M.; Martin, J.S.; Broadaway, S.; Laederach, A. Disease-associated mutations that alter the RNA structural ensemble. PLoS Genet. 2010, 6, e1001074. [Google Scholar] [CrossRef]
- Martin, J.S.; Halvorsen, M.; Davis-Neulander, L.; Ritz, J.; Gopinath, C.; Beauregard, A.; Laederach, A. Structural effects of linkage disequilibrium on the transcriptome. RNA 2012, 18, 77–87. [Google Scholar] [CrossRef]
- Wan, Y.; Qu, K.; Zhang, Q.C.; Flynn, R.A.; Manor, O.; Ouyang, Z.; Zhang, J.; Spitale, R.C.; Snyder, M.P.; Segal, E.; et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature 2014, 505, 706–709. [Google Scholar] [CrossRef]
- Corley, M.; Solem, A.; Qu, K.; Chang, H.Y.; Laederach, A. Detecting riboSNitches with RNA folding algorithms: A genome-wide benchmark. Nucleic Acid Res. 2015, 43, 1859–1868. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Wei, R.; Zhou, Z.; Huang, L.; Wang, Y.; Tang, J.; Zou, Y.; Shi, L.; Gu, X.; Davis, M.J.; et al. Integrative Analysis of Somatic Mutations in Non-coding Regions Altering RNA Secondary Structures in Cancer Genomes. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Lin, J.; Chen, Y.; Zhang, Y.; Ouyang, Z. Identification and analysis of RNA structural disruptions induced by single nucleotide variants using Riprap and RiboSNitchDB. NAR Genom. Bioinform. 2020, 2, lqaa057. [Google Scholar] [CrossRef]
- De Bernardi Schneider, A.; Ford, C.T.; Hostager, R.; Williams, J.; Cioce, M.; Çatalyürek, Ü.V.; Wertheim, J.O.; Janies, D. StrainHub: A phylogenetic tool to construct pathogen transmission networks. Bioinformatics 2020, 36, 945–947. [Google Scholar] [CrossRef]
- Campbell, F.; Didelot, X.; Fitzjohn, R.; Ferguson, N.; Cori, A.; Jombart, T. outbreaker2: A modular platform for outbreak reconstruction. BMB Bioinform. 2018, 19, 1–8. [Google Scholar] [CrossRef]
- De Maio, N.; Worby, C.J.; Wilson, D.J.; Stoesser, N. Bayesian reconstruction of transmission within outbreaks using genomic variants. PLoS Comput. Biol. 2018, 14, e1006117. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef]
- Kinganda-Lusamaki, E.; Black, A.; Mukadi, D.; Hadfield, J.; Mbala-Kingebeni, P.; Pratt, C.; Aziza, A.; Diagne, M.; White, B.; Bisento, N.; et al. Operationalizing genomic epidemiology during the Nord-Kivu Ebola outbreak, Democratic Republic of the Congo. medRxiv 2020. [Google Scholar] [CrossRef]
- Popa, A.; Genger, J.W.; Nicholson, M.D.; Penz, T.; Schmid, D.; Aberle, S.W.; Agerer, B.; Lercher, A.; Endler, L.; Colaco, H.; et al. Genomic epidemiology of superspreading events in Austria reveals mutational dynamics and transmission properties of SARS-CoV-2. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2016, 44, D67–D72. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Köster, J.; Rahmann, S. Snakemake—A scalable bioinformatics workflow engine. Bioinformatics 2012, 28, 2520–2522. [Google Scholar] [CrossRef]
- Lorenz, R.; Stadler, P.F. RNA Secondary Structures with Limited Base Pair Span: Exact Backtracking and an Application. Genes 2021, 12, 14. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Bernhart, S.H.; Zu Siederdissen, C.H.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithm. Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]
- Miladi, M.; Raden, M.; Diederichs, S.; Backofen, R. MutaRNA: Analysis and visualization of mutation-induced changes in RNA structure. Nucleic Acids Res. 2020, 37, 1–5. [Google Scholar] [CrossRef]
- Sagulenko, P.; Puller, V.; Neher, R.A. TreeTime: Maximum-likelihood phylodynamic analysis. Virus Evol. 2018, 4, vex042. [Google Scholar] [CrossRef]
- Lopez-Delisle, L.; Rabbani, L.; Wolff, J.; Bhardwaj, V.; Backofen, R.; Grüning, B.; Ramírez, F.; Manke, T. pyGenomeTracks: Reproducible plots for multivariate genomic data sets. Bioinformatics 2020, btaa692. [Google Scholar] [CrossRef] [PubMed]
- Tsetsarkin, K.A.; Vanlandingham, D.L.; McGee, C.E.; Higgs, S. A single mutation in Chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007, 3, e201. [Google Scholar] [CrossRef]
- McNaughton, A.L.; Revill, P.A.; Littlejohn, M.; Matthews, P.C.; Ansari, M.A. Analysis of genomic-length HBV sequences to determine genotype and subgenotype reference sequences. J. Gen. Virol. 2020, 101, 1–13. [Google Scholar] [CrossRef]
- Bbosa, N.; Kaleebu, P.; Ssemwanga, D. HIV subtype diversity worldwide. Curr. Opin. HIV Aids 2019, 14, 153–160. [Google Scholar] [CrossRef]
- De Bernardi Schneider, A.; Osiowy, C.; Hostager, R.; Krarup, H.; Borresen, M.; Tanaka, Y.; Morriseau, T.; Wertheim, J.O. Analysis of Hepatitis B virus genotype D in Greenland suggests the presence of a novel quasi-subgenotype. Front. Microbiol. 2021. [Google Scholar] [CrossRef]
- Robertson, D.L.; Anderson, J.; Bradac, J.; Carr, J.; Foley, B.; Funkhouser, R.; Gao, F.; Hahn, B.; Kalish, M.; Kuiken, C.; et al. HIV-1 nomenclature proposal. Science 2000, 288, 55–56. [Google Scholar] [CrossRef]
- Souza, T.M.A.; Azeredo, E.L.; Badolato-Corrêa, J.; Damasco, P.V.; Santos, C.; Petitinga-Paiva, F.; Nunes, P.C.G.; Barbosa, L.S.; Cipitelli, M.C.; Chouin-Carneiro, T.; et al. First report of the East-Central South African genotype of Chikungunya virus in Rio de Janeiro, Brazil. PLoS Curr. 2017, 9. [Google Scholar] [CrossRef]
- Schuster, P. Quasispecies on Fitness Landscapes. In Quasispecies: From Theory to Experimental Systems; Domingo, E., Schuster, P., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 61–120. [Google Scholar] [CrossRef]
- Geoghegan, J.L.; Holmes, E.C. Virus Evolution. In Fields Virology; Howley, P.M., Knipe, D.M., Eds.; Wolters Kluwer Health: Philadelphia, PA, USA, 2021. [Google Scholar]
- Faure, G.; Ogurtsov, A.Y.; Shabalina, S.A.; Koonin, E.V. Adaptation of mRNA structure to control protein folding. RNA Biol. 2017, 14, 1649–1654. [Google Scholar] [CrossRef]
| Lineage | AUL-Am | AUL | SAL | MAL | IOL | EAL | AAL | sECSA |
|---|---|---|---|---|---|---|---|---|
| AUL | 0.01108 | |||||||
| SAL | 0.06902 | 0.06622 | ||||||
| MAL | 0.06840 | 0.06548 | 0.02432 | |||||
| IOL | 0.06988 | 0.06734 | 0.02954 | 0.02631 | ||||
| EAL | 0.06763 | 0.06533 | 0.02574 | 0.02259 | 0.00584 | |||
| AAL | 0.06390 | 0.06067 | 0.03145 | 0.02887 | 0.03141 | 0.02807 | ||
| sECSA | 0.06302 | 0.06014 | 0.02957 | 0.02758 | 0.03038 | 0.02690 | 0.01918 | |
| WA | 0.19383 | 0.19228 | 0.17696 | 0.17829 | 0.17917 | 0.17750 | 0.17443 | 0.17505 |
| Lineage | TMRCA | Date Confidence Interval | Year of First Isolation |
|---|---|---|---|
| AAL | 17-04-1948 | (18-10-1946,14-12-1949) | 1953 |
| AUL | 05-02-1951 | (13-09-1949, 04-01-1953) | 1958 |
| AUL-Am | 12-03-2008 | (24-12-2007, 10-11-2008) | 2013 |
| EAL | 24-05-2002 | (15-02-2001, 20-04-2003) | 2005 |
| IOL | 03-08-2003 | (20-10-2002, 14-01-2004) | 2006 |
| MAL | 31-01-1953 | (20-05-1951, 14-01-1955) | 1962 |
| SAL | 22-03-2011 | (24-06-2009, 15-09-2012) | 2014 |
| WA | 16-01-1954 | (13-05-1952, 26-09-1955) | 1964 |
| Variant | Type | Protein | AA Mutation | RNA # | Locus | z-Score | BP Distance | Lineage Association | ||
|---|---|---|---|---|---|---|---|---|---|---|
| G432A | N | nsP1 | E > A | 1 | 378–472 | −30.00 | −2.474 | −27.00 | 29 | G: AUL, AUL-Am |
| U810C | S | nsP1 | – | 2 | 783–847 | −21.60 | −2.103 | −18.70 | 42 | U: WA, AUL, AUL-Am, * |
| A1653G | S | nsP1 | – | 3 | 1605–1685 | −23.30 | −2.640 | −28.10 | 15 | A: AUL , AUL-Am |
| U2122C | S | nsP2 | – | 4 | 2105–2202 | −31.90 | −2.171 | −28.30 | 29 | U: AUL, AUL-Am |
| G2232A | S | nsP2 | – | 5 | 2210–2300 | −31.50 | −2.771 | −27.40 | 36 | G: AUL, AUL-Am |
| C3108U | S | nsP2 | – | 6 | 3093–3192 | −28.20 | −2.141 | −25.60 | 42 | C: WA, AUL, AUL-Am |
| C3682U | S | nsP2 | – | 7 | 3630–3731 | −42.20 | −4.325 | −40.10 | 22 | C: AUL, AUL-Am, * |
| U5508A | N | nsP3 | D > E | 8 | 5467–5527 | −18.10 | −2.370 | −16.20 | 15 | U: , AUL-Am |
| G8336C | S | C | – | 9 | 8312–8395 | −37.10 | −3.023 | −34.40 | 27 | G: , AUL-Am |
| C8358U | S | C | – | 9 | 8312–8395 | −37.10 | −3.023 | −36.40 | 27 | C: AUL, AUL-AM, SAL, MAL , * |
| G8969A | N | E2 | R > K | 10 | 8918–9019 | −38.60 | −2.447 | −36.10 | 40 | A: , AUL-Am |
| C9197U | S | E2 | – | 11 | 9130–9214 | −22.40 | −2.443 | −20.40 | 34 | C: WA, AUL, AUL-AM |
| A9414C | S | E2 | – | 12 | 9392–9456 | −17.60 | −2.568 | −15.80 | 18 | A: , AUL-Am |
| A10460C | N | E1 | I > V | 13 | 10369–10468 | −31.60 | −2.623 | −31.70 | 18 | A: AUL, AUL-AM |
| A11246C | S | E1 | – | 14 | 11,228–11,307 | −29.30 | −3.049 | −26.90 | 25 | A: AUL, AUL-Am, AAL, SAL, MAL; C: EAL, IOL |
| A11246U | S | E1 | – | 14 | 11,228–11,307 | −29.30 | −3.049 | −27.20 | 36 | A: AUL, AUL-Am, AAL, SAL, MAL; U: WA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spicher, T.; Delitz, M.; Schneider, A.d.B.; Wolfinger, M.T. Dynamic Molecular Epidemiology Reveals Lineage-Associated Single-Nucleotide Variants That Alter RNA Structure in Chikungunya Virus. Genes 2021, 12, 239. https://doi.org/10.3390/genes12020239
Spicher T, Delitz M, Schneider AdB, Wolfinger MT. Dynamic Molecular Epidemiology Reveals Lineage-Associated Single-Nucleotide Variants That Alter RNA Structure in Chikungunya Virus. Genes. 2021; 12(2):239. https://doi.org/10.3390/genes12020239
Chicago/Turabian StyleSpicher, Thomas, Markus Delitz, Adriano de Bernardi Schneider, and Michael T. Wolfinger. 2021. "Dynamic Molecular Epidemiology Reveals Lineage-Associated Single-Nucleotide Variants That Alter RNA Structure in Chikungunya Virus" Genes 12, no. 2: 239. https://doi.org/10.3390/genes12020239
APA StyleSpicher, T., Delitz, M., Schneider, A. d. B., & Wolfinger, M. T. (2021). Dynamic Molecular Epidemiology Reveals Lineage-Associated Single-Nucleotide Variants That Alter RNA Structure in Chikungunya Virus. Genes, 12(2), 239. https://doi.org/10.3390/genes12020239









