Long Noncoding RNA HOXA11-AS and Transcription Factor HOXB13 Modulate the Expression of Bone Metastasis-Related Genes in Prostate Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Reagents
2.2. RNA Interference
2.3. Invasion and Proliferation Assays
2.4. Microarray Analysis
2.5. Plasmid Construction
2.6. qPCR Analysis
2.7. Luciferase Assays
2.8. ChIP Assay
2.9. Western Blotting
2.10. Gene Expression Omnibus Dataset Analysis
2.11. Isolation of Extracellular Vesicles (EVs)
2.12. Statistical Analysis
3. Results
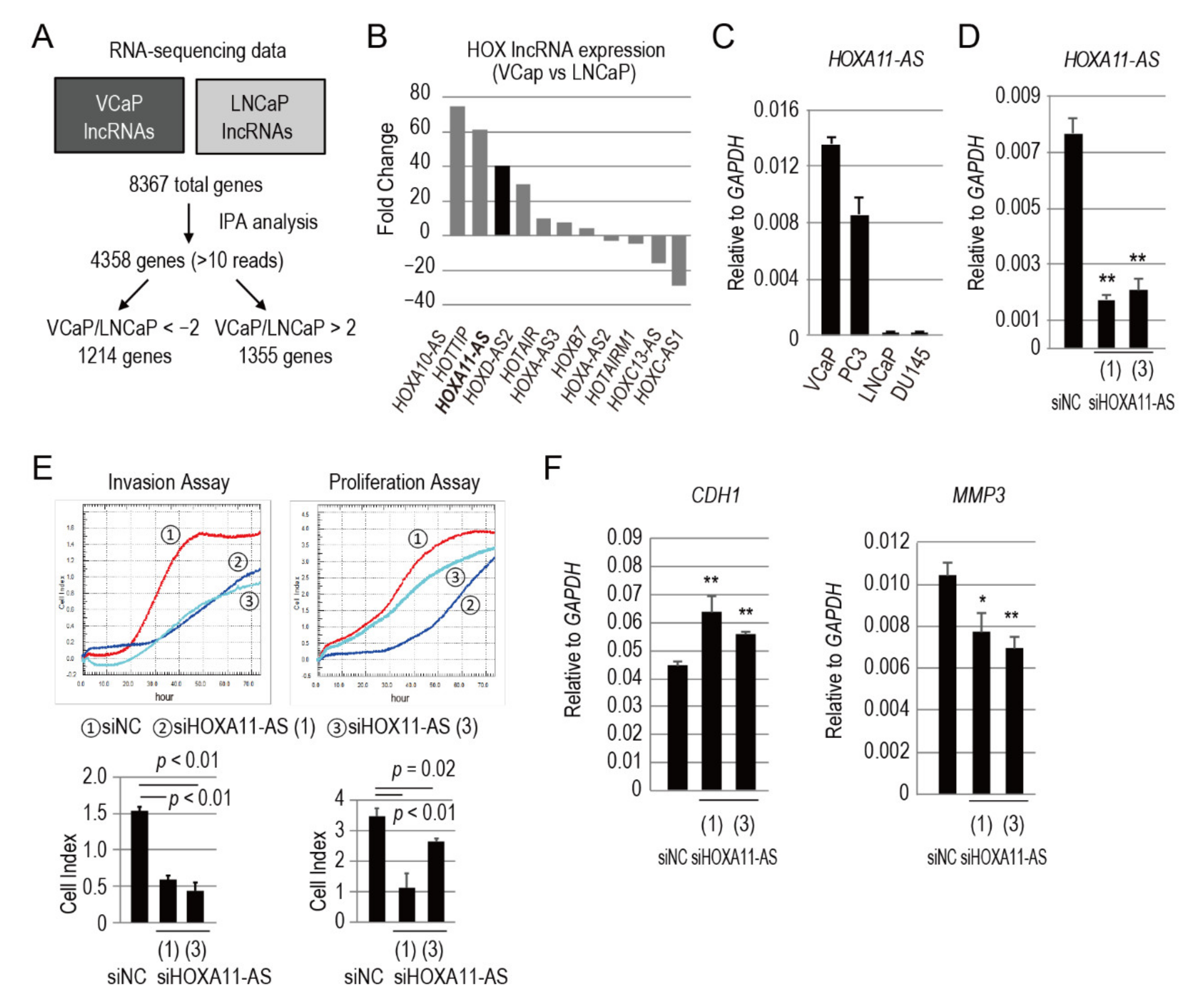
3.1. HOXA11-AS Is Highly Expressed in Cell Lines Derived from Bone Metastases
3.2. HOXA11-AS Promotes the Invasion and Proliferation of Prostate Cancer Cells
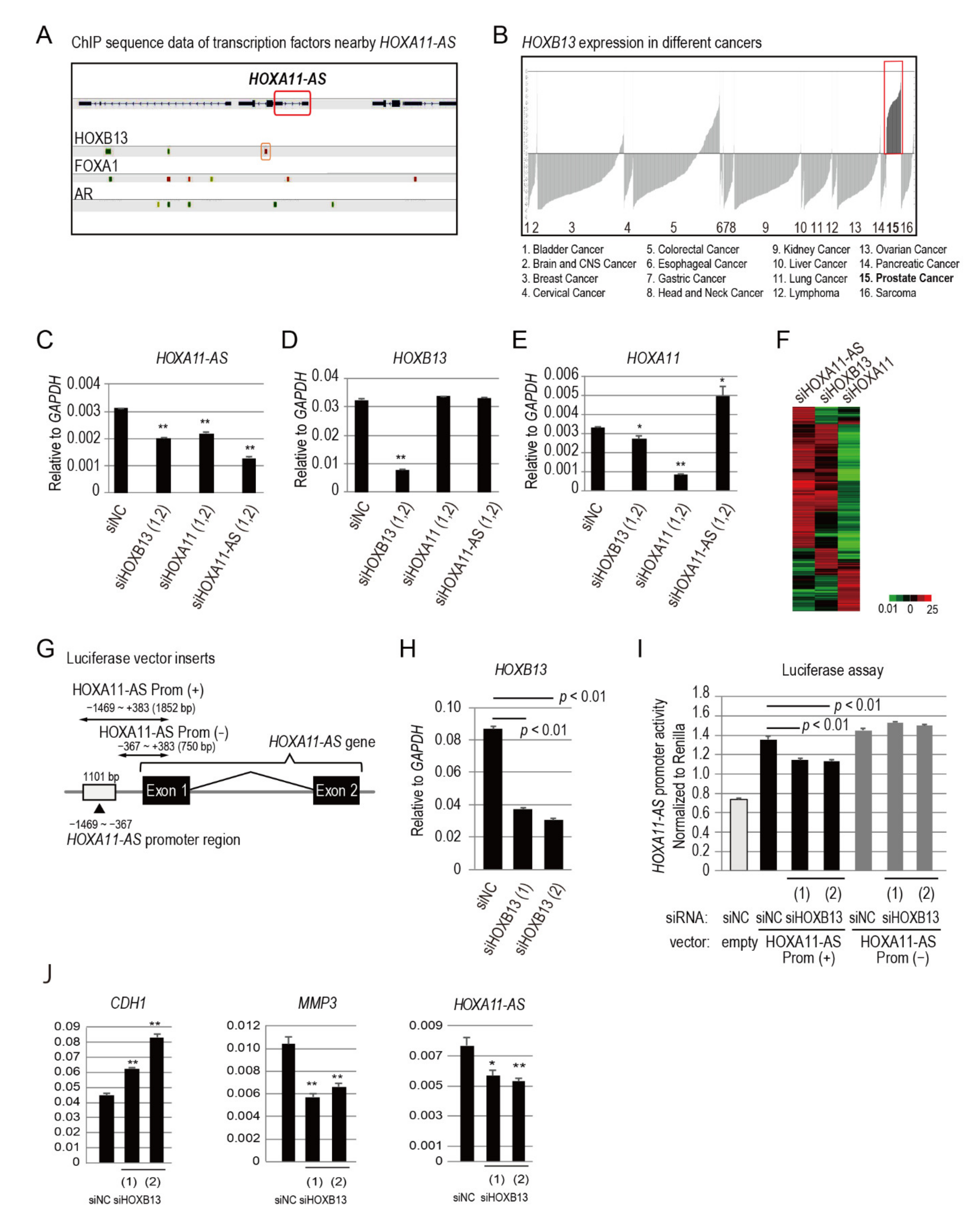
3.3. HOXB13 Is an Upstream Regulator of HOXA11-AS
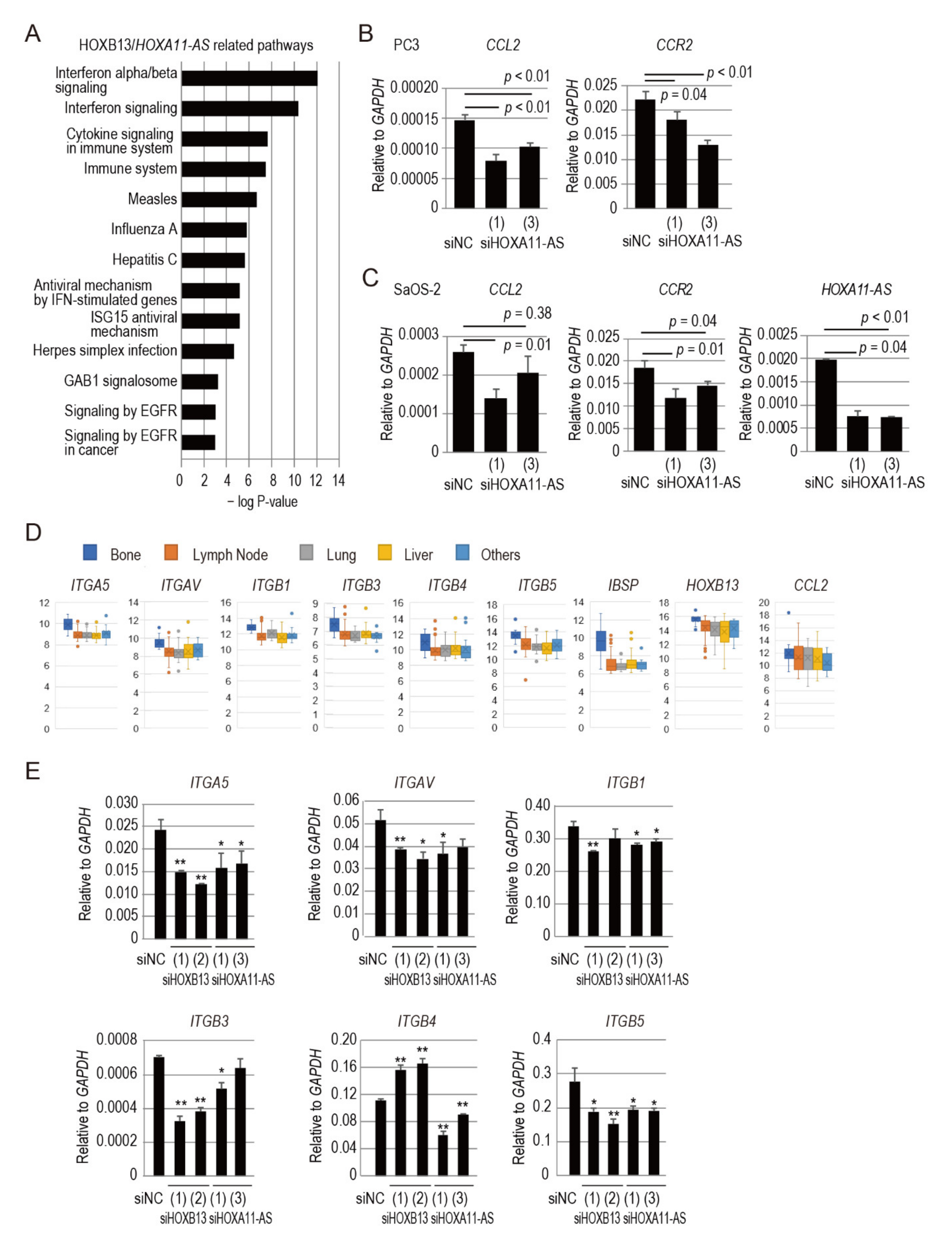
3.4. HOXA11-AS Regulates C-C Motif Chemokine Ligand 2/C-C Chemokine Receptor Type 2 Signaling Associated with Prostate Cancer Bone Metastasis
3.5. HOXB13/HOXA11-AS Axis Regulates Integrin Subunits Specific to Prostate Cancer Bone Metastasis
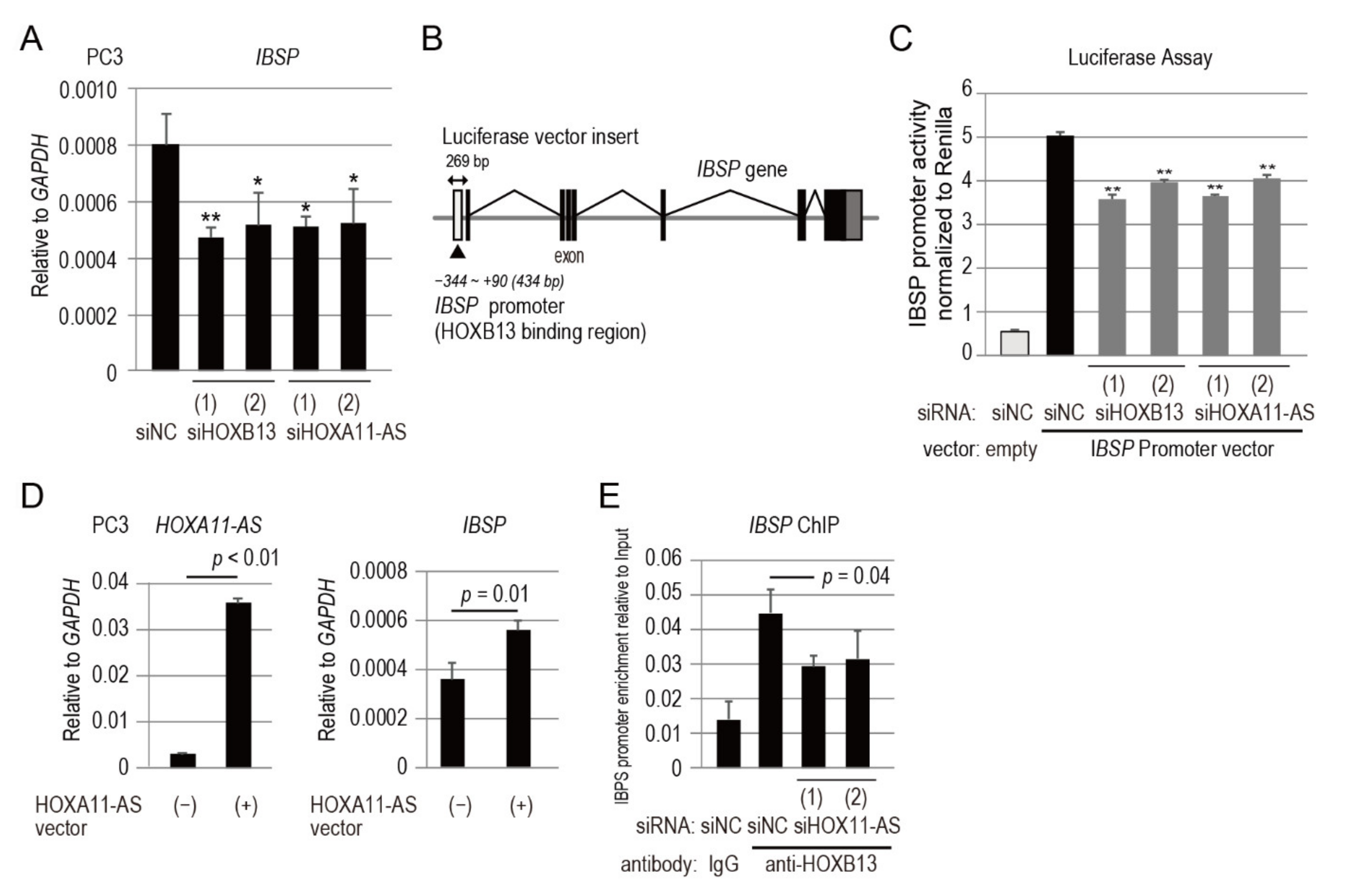
3.6. IBSP Promoter Is Directly Regulated by HOXB13 in Combination with HOXA11-AS
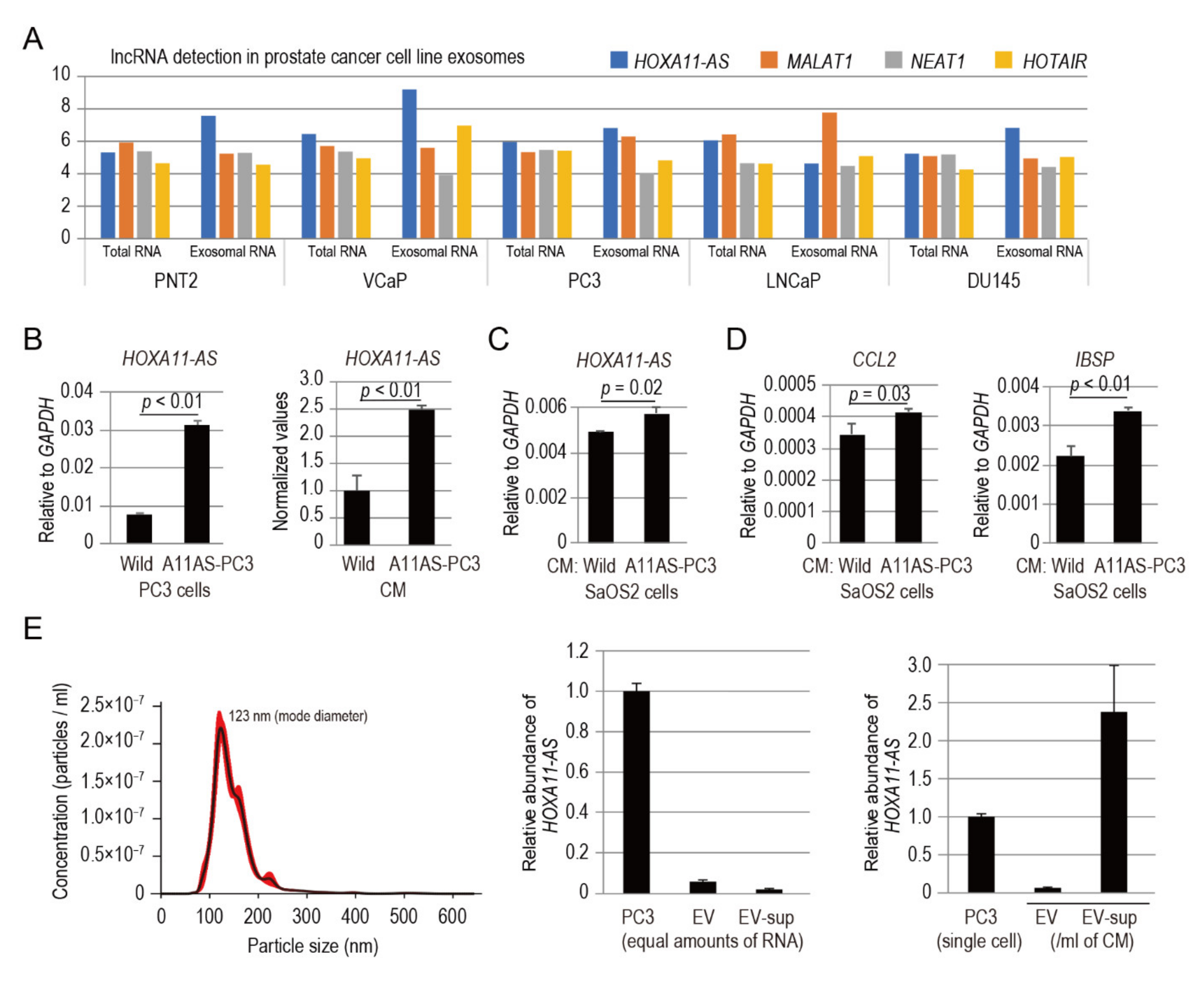
3.7. HOXA11-AS Secreted from Prostate Cancer Cells Modulates the Expression of CCL2 and IBSP in Osteoblastic Cells
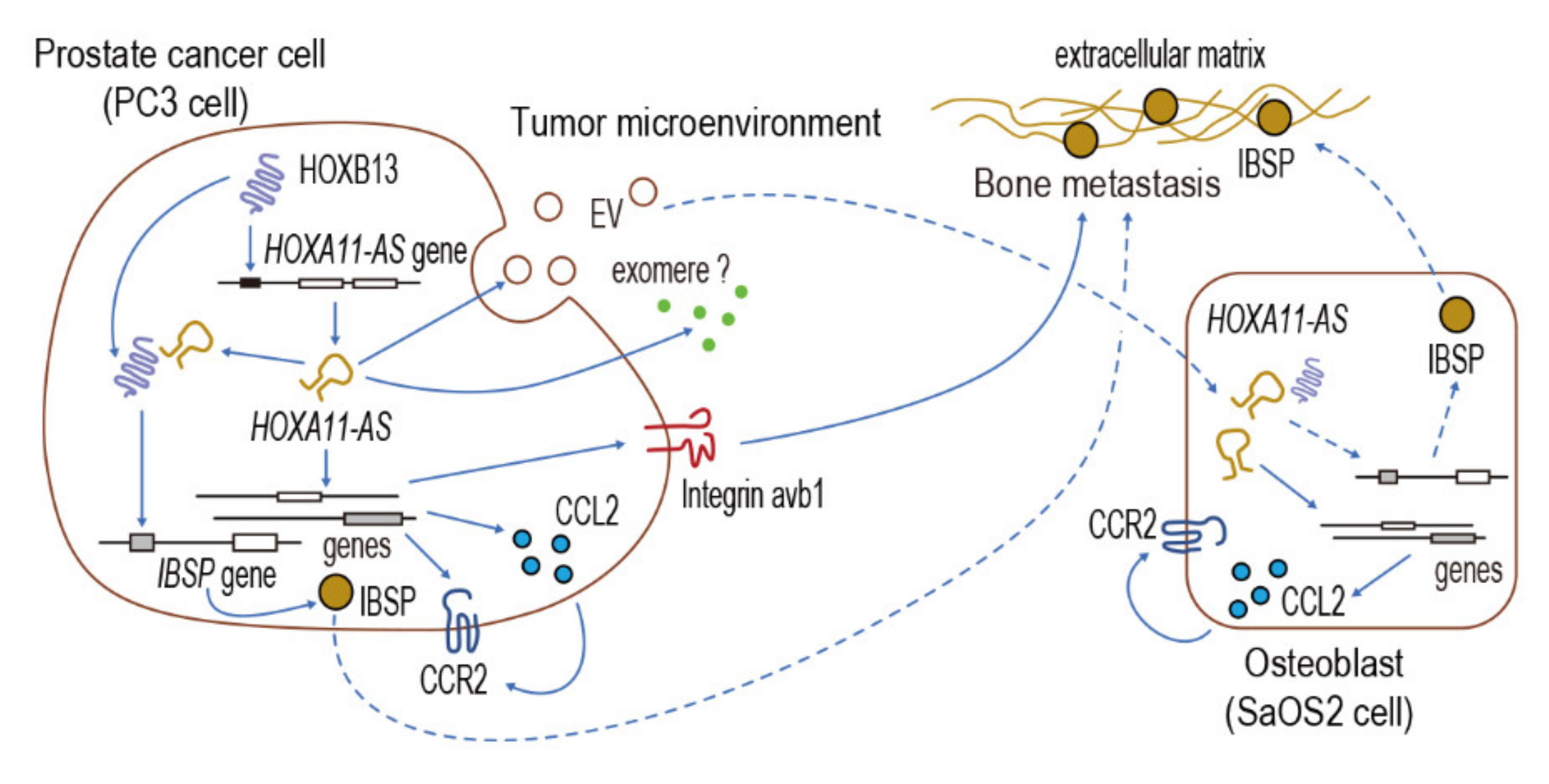
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jinnah, A.H.; Zacks, B.C.; Gwam, C.U.; Kerr, B.A. Emerging and Established Models of Bone Metastasis. Cancers (Basel) 2018, 10, 176. [Google Scholar] [CrossRef]
- Holland, P.W. Evolution of homeobox genes. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Cillo, C.; Faiella, A.; Cantile, M.; Boncinelli, E. Homeobox genes and cancer. Exp. Cell Res. 1999, 248, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bhatlekar, S.; Fields, J.Z.; Boman, B.M. HOX genes and their role in the development of human cancers. J. Mol. Med. (Berlin) 2014, 92, 811–823. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, H.L.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.R.; Feng, F.Y.; Chinnaiyan, A.M. The bright side of dark matter: lncRNAs in cancer. J. Clin. Investig. 2016, 126, 2775–2782. [Google Scholar] [CrossRef]
- Rinn, J.L. lncRNAs: Linking RNA to chromatin. Cold Spring Harb. Perspect. Biol. 2014, 6, a018614. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, Z.; Fan, R.; Jiang, B.; Chen, X.; Chen, Q.; Nie, F.; Lu, K.; Sun, M. Over-expressed long noncoding RNA HOXA11-AS promotes cell cycle progression and metastasis in gastric cancer. Mol. Cancer 2017, 16, 82. [Google Scholar] [CrossRef]
- Sun, M.; Nie, F.; Wang, Y.; Zhang, Z.; Hou, J.; He, D.; Xie, M.; Xu, L.; De, W.; Wang, Z.; et al. LncRNA HOXA11-AS Promotes Proliferation and Invasion of Gastric Cancer by Scaffolding the Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res. 2016, 76, 6299–6310. [Google Scholar] [CrossRef]
- Xue, J.Y.; Huang, C.; Wang, W.; Li, H.B.; Sun, M.; Xie, M. HOXA11-AS: A novel regulator in human cancer proliferation and metastasis. Onco Targets Ther. 2018, 11, 4387–4393. [Google Scholar] [CrossRef]
- Cui, M.; Wang, J.; Li, Q.; Zhang, J.; Jia, J.; Zhan, X. Long non-coding RNA HOXA11-AS functions as a competing endogenous RNA to regulate ROCK1 expression by sponging miR-124-3p in osteosarcoma. Biomed. Pharmacother. 2017, 92, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 3, 22. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, Y.; Kosaka, N.; Konishi, Y.; Ohta, H.; Okamoto, H.; Sonoda, H.; Nonaka, R.; Yamamoto, H.; Ishii, H.; Mori, M.; et al. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat. Commun. 2014, 5, 3591. [Google Scholar] [CrossRef] [PubMed]
- Xing, Z.; Li, S.; Liu, Z.; Zhang, C.; Bai, Z. CTCF-induced upregulation of HOXA11-AS facilitates cell proliferation and migration by targeting miR-518b/ACTN4 axis in prostate cancer. Prostate 2020, 80, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Stelloo, S.; Nevedomskaya, E.; Kim, Y.; Hoekman, L.; Bleijerveld, O.B.; Mirza, T.; Wessel, L.F.A.; van Weerden, W.M.; Alteraal, A.F.M.; Bergamn, A.M.; et al. Endogenous androgen receptor proteomic profiling reveals genomic subcomplex involved in prostate tumorigenesis. Oncogene 2018, 37, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Pomerantz, M.M.; Li, F.; Takeda, D.Y.; Lenci, R.; Chonkar, A.; Chabot, M.; Cejas, P.; Vasquez, F.; Cook, J.; Shivdasani, R.A.; et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat. Genet. 2015, 47, 1346–1351. [Google Scholar] [CrossRef]
- Wang, L.; Cui, Y.; Sheng, J.; Yang, Y.; Kuang, G.; Fan, Y.; Jin, J.; Zhang, Q. Epigenetic inactivation of HOXA11, a novel functional tumor suppressor for renal cell carcinoma, is associated with RCC TNM classification. Oncotarget 2017, 8, 21861–21870. [Google Scholar] [CrossRef]
- Viola, A.; Luster, A.D. Chemokines and their receptors: Drug targets in immunity and inflammation. Annu. Rev. Pharmacol. Toxicol. 2008, 48, 171–197. [Google Scholar] [CrossRef]
- Zhang, J.; Lu, Y.; Pienta, K.J. Multiple roles of chemokine (C-C motif) ligand 2 in promoting prostate cancer growth. J. Natl. Cancer Inst. 2010, 102, 522–528. [Google Scholar] [CrossRef]
- Kinashi, T. Intracellular signalling controlling integrin activation in lymphocytes. Nat. Rev. Immunol. 2005, 5, 546–559. [Google Scholar] [CrossRef]
- Ganss, B.; Kim, R.H.; Sodek, J. Bone sialoprotein. Crit. Rev. Oral Biol. Med. 1999, 10, 79–98. [Google Scholar] [CrossRef] [PubMed]
- Tu, Q.; Zhang, J.; Fix, A.; Brewer, E.; Li, Y.P.; Zhang, Z.Y.; Chen, J. Targeted overexpression of BSP in osteoclasts promotes bone metastasis of breast cancer cells. J. Cell Physiol. 2009, 218, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Ruivo, C.F.; Adem, B.; Silva, M.; Melo, S.A. The Biology of Cancer Exosomes: Insights and New Perspectives. Cancer Res. 2017, 77, 6480–6488. [Google Scholar] [CrossRef] [PubMed]
- Ahadi, A.; Khoury, S.; Losseva, M.; Tran, N. A comparative analysis of lncRNAs in prostate cancer exosomes and their parental cell lines. Genom. Data 2016, 9, 7–9. [Google Scholar] [CrossRef]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef]
- Chen, Y.G.; Satpathy, A.T.; Chang, H.Y. Gene regulation in the immune system by long noncoding RNAs. Nat. Immunol. 2017, 18, 962–972. [Google Scholar] [CrossRef]
- Hur, K.; Kim, S.H.; Kim, J.M. Potential Implications of Long Noncoding RNAs in Autoimmune Diseases. Immune Netw. 2019, 19, e4. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, T.; Zhou, W.; Li, J.; Li, X.; Wang, Q.; Jin, X.; Yin, J.; Chen, L.; Zhang, Y.; et al. Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nat. Commun. 2020, 11, 1000. [Google Scholar] [CrossRef]
- Yu, W.; Peng, W.; Jiang, H.; Sha, H.; Li, J. LncRNA HOXA11-AS promotes proliferation and invasion by targeting miR-124 in human non-small cell lung cancer cells. Tumour Biol. 2017, 39, 1010428317721440. [Google Scholar] [CrossRef]
- Zhao, X.; Li, X.; Zhou, L.; Ni, J.; Yan, W.; Ma, R.; Wu, J.; Feng, J.; Chen, P. LncRNA HOXA11-AS drives cisplatin resistance of human LUAD cells via modulating miR-454-3p/Stat3. Cancer Sci. 2018, 109, 3068–3079. [Google Scholar] [CrossRef]
- Li, N.; Yang, M.; Shi, K.; Li, W. Long non-coding RNA HOXA11-AS in human cancer: A meta-analysis. Clin. Chim. Acta 2017, 474, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Sun, Q.; Zhang, L.; Zhou, X.; Cheng, X.; Zhou, D.; Ye, F.; Lin, J.; Wang, W. The lncRNA HOXA11-AS functions as a competing endogenous RNA to regulate PADI2 expression by sponging miR-125a-5p in liver metastasis of colorectal cancer. Oncotarget 2017, 8, 70642–70652. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Jia, G.; Qu, Y.; Du, Q.; Liu, B.; Liu, B. Long Non-Coding RNA (LncRNA) HOXA11-AS Promotes Breast Cancer Invasion and Metastasis by Regulating Epithelial-Mesenchymal Transition. Med. Sci. Monit. 2017, 23, 3393–3403. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.L.; Zhao, Y.N.; Shi, Z.Z.; Gu, G.Y.; Cong, D.; Wei, C.; Bai, Y.S. HOXA11-AS promotes the migration and invasion of hepatocellular carcinoma cells by inhibiting miR-124 expression by binding to EZH2. Hum. Cell 2019, 32, 504–514. [Google Scholar] [CrossRef]
- Zhao, Y.; Yamashita, T.; Ishikawa, M. Regulation of tumor invasion by HOXB13 gene overexpressed in human endometrial cancer. Oncol. Rep. 2005, 13, 721–726. [Google Scholar] [CrossRef]
- Miao, J.; Wang, Z.; Provencher, H.; Muir, B.; Dahiya, S.; Carney, E.; Leong, C.O.; Sgroi, D.C.; Orsulic, S. HOXB13 promotes ovarian cancer progression. Proc. Natl. Acad. Sci. USA 2007, 104, 17093–17098. [Google Scholar] [CrossRef]
- Lopez, R.; Garrido, E.; Pina, P.; Hidalgo, A.; Lazos, M.; Ochoa, R.; Salcedo, M. HOXB homeobox gene expression in cervical carcinoma. Int. J. Gynecol. Cancer 2006, 16, 329–335. [Google Scholar] [CrossRef]
- Maeda, K.; Hamada, J.; Takahashi, Y.; Tada, M.; Yamamoto, Y.; Sugihara, T.; Moriuchi, T. Altered expressions of HOX genes in human cutaneous malignant melanoma. Int. J. Cancer 2005, 114, 436–441. [Google Scholar] [CrossRef]
- Ma, X.J.; Wang, Z.; Ryan, P.D.; Isakoff, S.J.; Barmettler, A.; Fuller, A.; Muir, B.; Mohapatra, G.; Salunga, R.; Tuggle, J.T.; et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell 2004, 5, 607–616. [Google Scholar] [CrossRef]
- Okuda, H.; Toyota, M.; Ishida, W.; Furihata, M.; Tsuchiya, M.; Kamada, M.; Tokito, T.; Shuin, T. Epigenetic inactivation of the candidate tumor suppressor gene HOXB13 in human renal cell carcinoma. Oncogene 2006, 25, 1733–1742. [Google Scholar] [CrossRef]
- Xie, B.; Bai, B.; Xu, Y.; Liu, Y.; Lv, Y.; Gao, X.; Wu, F.; Fang, Z.; Lou, Y.; Pan, H.; et al. Tumor-suppressive function and mechanism of HOXB13 in right-sided colon cancer. Signal Transduct. Target Ther. 2019, 4, 51. [Google Scholar] [CrossRef] [PubMed]
- Cantile, M.; Scognamiglio, G.; La Sala, L.; La Mantia, E.; Scaramuzza, V.; Valentino, E.; Tatangelo, F.; Losito, S.; Pezzullo, L.; Chifalo, M.G.; et al. Aberrant expression of posterior HOX genes in well differentiated histotypes of thyroid cancers. Int. J. Mol. Sci. 2013, 14, 21727–21740. [Google Scholar] [CrossRef] [PubMed]
- Brechka, H.; Bhanvadia, R.R.; VanOpstall, C.; Vander, G.D.J. HOXB13 mutations and binding partners in prostate development and cancer: Function, clinical significance, and future directions. Genes Dis. 2017, 4, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, T.N.; Burger, J.A.; Glodek, A.; Fujii, N.; Burger, M. CXCR4 chemokine receptor and integrin signaling co-operate in mediating adhesion and chemoresistance in small cell lung cancer (SCLC) cells. Oncogene 2005, 24, 4462–4471. [Google Scholar] [CrossRef]
- Hood, J.D.; Cheresh, D.A. Role of integrins in cell invasion and migration. Nat. Rev. Cancer 2002, 2, 91–100. [Google Scholar] [CrossRef]
- Hamidi, H.; Ivaska, J. Every step of the way: Integrins in cancer progression and metastasis. Nat. Rev. Cancer 2018, 18, 533–548. [Google Scholar] [CrossRef]
- Seguin, L.; Desgrosellier, J.S.; Weis, S.M.; Cheresh, D.A. Integrins and cancer: Regulators of cancer stemness, metastasis, and drug resistance. Trends Cell Biol. 2015, 25, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Hamidi, H.; Pietila, M.; Ivaska, J. The complexity of integrins in cancer and new scopes for therapeutic targeting. Br. J. Cancer 2016, 115, 1017–1023. [Google Scholar] [CrossRef] [PubMed]
- Raab-Westphal, S.; Marshall, J.F.; Goodman, S.L. Integrins as Therapeutic Targets: Successes and Cancers. Cancers (Basel) 2017, 9, 110. [Google Scholar] [CrossRef]
- Sottnik, J.L.; Daignault-Newton, S.; Zhang, X.; Morrissey, C.; Hussain, M.H.; Keller, E.T.; Hall, C.L. Integrin alpha2beta 1 (alpha2beta1) promotes prostate cancer skeletal metastasis. Clin. Exp. Metastasis 2013, 30, 569–578. [Google Scholar] [CrossRef] [PubMed]
- De, S.; Chen, J.; Narizhneva, N.V.; Heston, W.; Brainard, J.; Sage, E.H.; Byzova, T.V. Molecular pathway for cancer metastasis to bone. J. Biol. Chem. 2003, 278, 39044–39050. [Google Scholar] [CrossRef] [PubMed]
- Fisher, L.W.; Jain, A.; Tayback, M.; Fedarko, N.S. Small integrin binding ligand N-linked glycoprotein gene family expression in different cancers. Clin. Cancer Res. 2004, 10, 8501–8511. [Google Scholar] [CrossRef]
- Waltregny, D.; Bellahcene, A.; van Riet, I.; Fisher, L.W.; Young, M.; Fernandez, P. Prognostic value of bone sialoprotein expression in clinically localized human prostate cancer. J. Natl. Cancer Inst. 1998, 90, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- Bellahcene, A.; Maloujahmoum, N.; Fisher, L.W.; Pastorino, H.; Tagliabue, E.; Menard, S.; Castronovo, V. Expression of bone sialoprotein in human lung cancer. Calcif. Tissue Int. 1997, 61, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Bellahcene, A.; Albert, V.; Pollina, L.; Basolo, F.; Fisher, L.W.; Castronovo, V. Ectopic expression of bone sialoprotein in human thyroid cancer. Thyroid 1998, 8, 637–641. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.H.; Shapiro, H.S.; Li, J.J.; Wrana, J.L.; Sodek, J. Characterization of the human bone sialoprotein (BSP) gene and its promoter sequence. Matrix Biol. 1994, 14, 31–40. [Google Scholar] [CrossRef]
- Nakayama, Y.; Nakajima, Y.; Kato, N.; Takai, H.; Kim, D.S.; Arai, M.; Mezawa, M.; Araki, S.; Sodek, J.; Ogata, Y. Insulin-like growth factor-I increases bone sialoprotein (BSP) expression through fibroblast growth factor-2 response element and homeodomain protein-binding site in the proximal promoter of the BSP gene. J. Cell Physiol. 2006, 208, 326–335. [Google Scholar] [CrossRef]
- Krishnan, R.; Klumpers, D.D.; Park, C.Y.; Rajendran, K.; Trepat, X.; van Bezu, J.; van Hinsbergh, V.W.M.; Carman, V.C.; Brain, J.D.; Fredberg, J.J.; et al. Substrate stiffening promotes endothelial monolayer disruption through enhanced physical forces. Am. J. Physiol. Cell Physiol. 2011, 300, C146–C154. [Google Scholar] [CrossRef]
- Rajski, M.; Vogel, B.; Baty, F.; Rochlitz, C.; Buess, M. Global gene expression analysis of the interaction between cancer cells and osteoblasts to predict bone metastasis in breast cancer. PLoS ONE 2012, 7, e29743. [Google Scholar] [CrossRef]
- Kimura, G.; Sugisaki, Y.; Masugi, Y.; Nakazawa, N. Calcification in human osteoblasts cultured in medium conditioned by the prostatic cancer cell line PC-3 and prostatic acid phosphatase. Urol. Int. 1992, 48, 25–30. [Google Scholar] [CrossRef]
- Ma, P.; Pan, Y.; Li, W.; Sun, C.; Liu, J.; Xu, T.; Shu, Y. Extracellular vesicles-mediated noncoding RNAs transfer in cancer. J. Hematol. Oncol. 2017, 10, 57. [Google Scholar] [CrossRef] [PubMed]
- Gezer, U.; Ozgur, E.; Cetinkaya, M.; Isin, M.; Dalay, N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol. Int. 2014, 38, 1076–1079. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Tesic, M.M.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Higginbotham, J.N.; Jeppesen, D.K.; Yang, Y.P.; Li, W.; McKinley, E.T.; Graves-Deal, R.; Ping, J.; Britain, C.M.; Dorsett, K.A.; et al. Transfer of Functional Cargo in Exomeres. Cell Rep. 2019, 27, 940–954.e6. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Misawa, A.; Kondo, Y.; Takei, H.; Takizawa, T. Long Noncoding RNA HOXA11-AS and Transcription Factor HOXB13 Modulate the Expression of Bone Metastasis-Related Genes in Prostate Cancer. Genes 2021, 12, 182. https://doi.org/10.3390/genes12020182
Misawa A, Kondo Y, Takei H, Takizawa T. Long Noncoding RNA HOXA11-AS and Transcription Factor HOXB13 Modulate the Expression of Bone Metastasis-Related Genes in Prostate Cancer. Genes. 2021; 12(2):182. https://doi.org/10.3390/genes12020182
Chicago/Turabian StyleMisawa, Aya, Yukihiro Kondo, Hiroyuki Takei, and Toshihiro Takizawa. 2021. "Long Noncoding RNA HOXA11-AS and Transcription Factor HOXB13 Modulate the Expression of Bone Metastasis-Related Genes in Prostate Cancer" Genes 12, no. 2: 182. https://doi.org/10.3390/genes12020182
APA StyleMisawa, A., Kondo, Y., Takei, H., & Takizawa, T. (2021). Long Noncoding RNA HOXA11-AS and Transcription Factor HOXB13 Modulate the Expression of Bone Metastasis-Related Genes in Prostate Cancer. Genes, 12(2), 182. https://doi.org/10.3390/genes12020182





