miR-143 Targeting CUX1 to Regulate Proliferation of Dermal Papilla Cells in Hu Sheep
Abstract
:1. Introduction
2. Materials and Methods
2.1. Skin Tissue Collection, RNA and Protein Extraction
2.2. Cell Culture and Transfection
2.3. Cell Proliferation and Cell Cycle
2.4. Target Gene Prediction for miR-143
2.5. Quantitative RT-PCR and Western Blot
2.6. Luciferase Reporter Assay
2.7. Statistical Analysis
3. Results
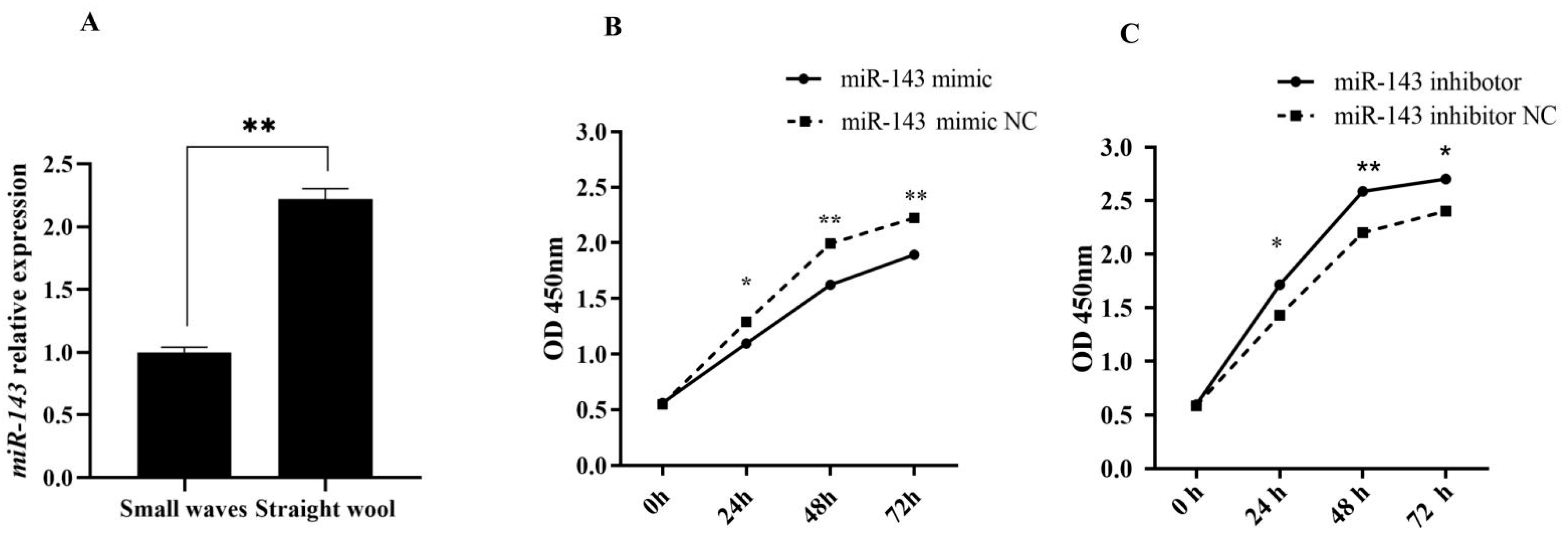
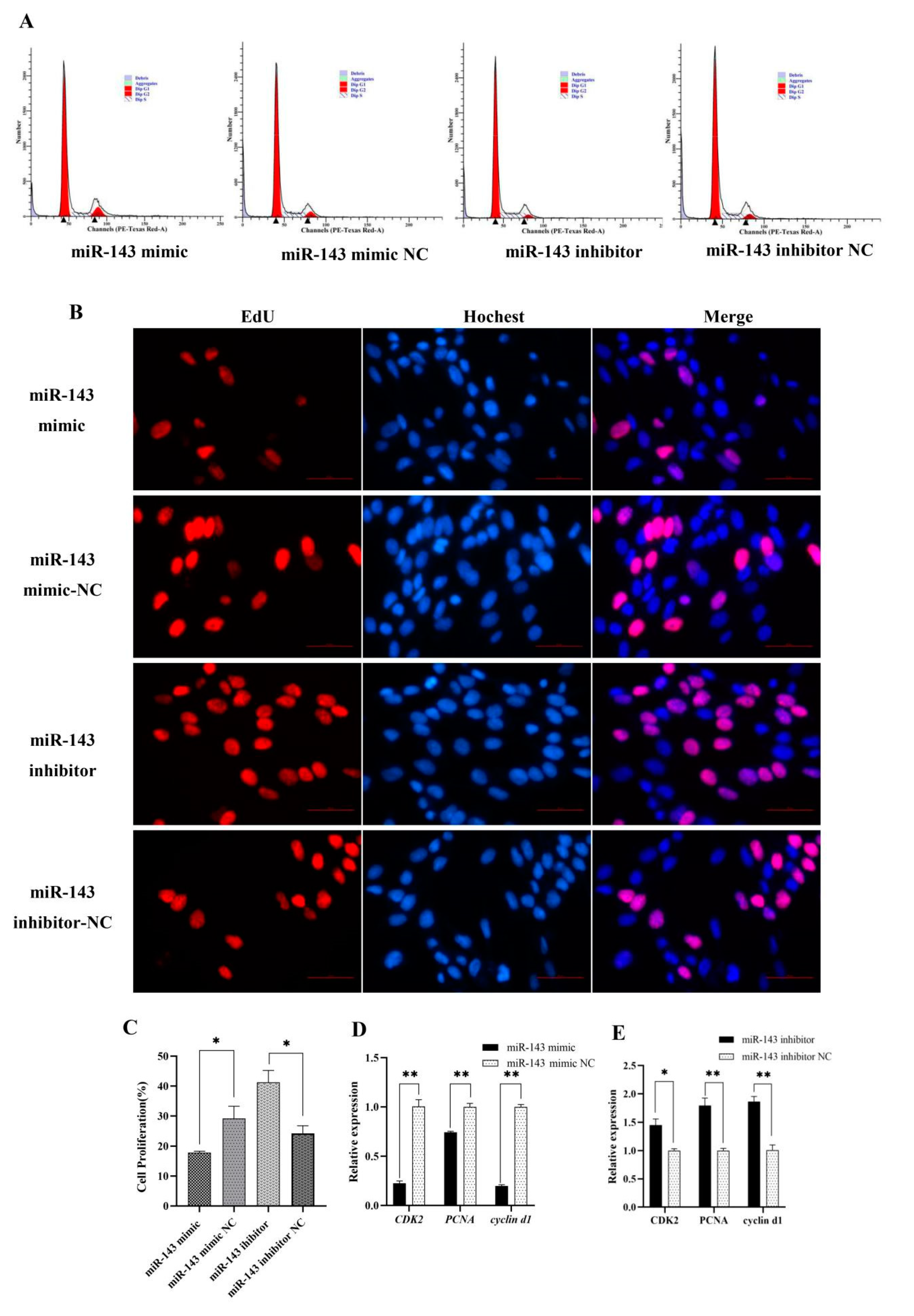
3.1. miR-143 Promoting Dermal Papilla Cells Proliferation
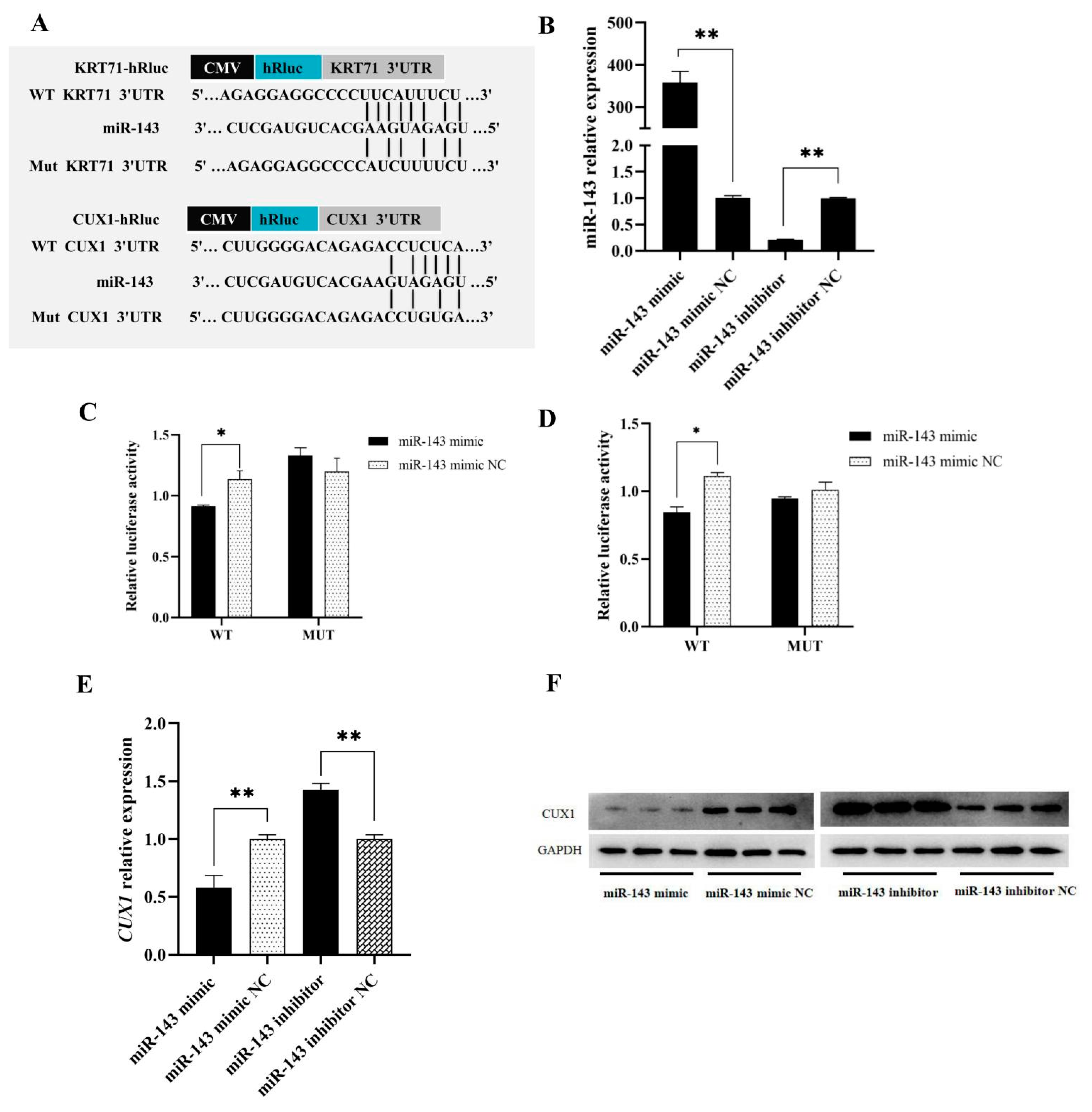
3.2. Target Genes Prediction of miR-143
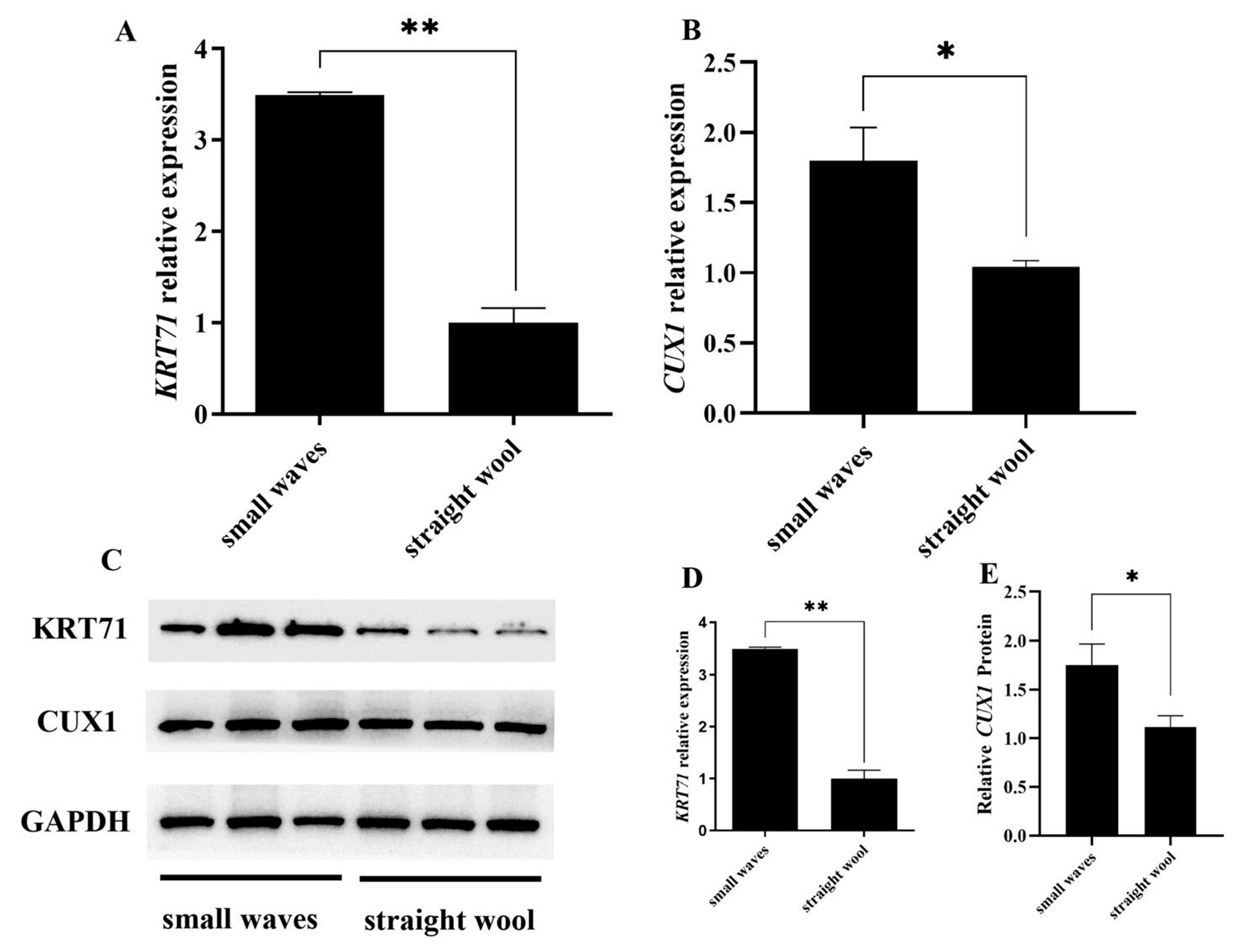
3.3. CUX1 and KRT71 Highly Expressed in Small Waves Group
3.4. CUX1 and KRT71 Targeting to miR-143
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andl, T.; Reddy, S.T.; Gaddapara, T.; Millar, S.E. WNT signals are required for the initiation of hair follicle development. Dev. Cell. 2002, 2, 643–653. [Google Scholar] [CrossRef]
- Lyubimova, A.; Garber, J.J.; Upadhyay, G.; Sharov, A.; Anastasoaie, F.; Yajnik, V.; Cotsarelis, G.; Dotto, G.P.; Botchkarev, V.; Snapper, S.B. Neural Wiskott-Aldrich syndrome protein modulates Wnt signaling and is required for hair follicle cycling in mice. J. Clin. Investig. 2010, 120, 446–456. [Google Scholar] [CrossRef] [Green Version]
- Kulessa, H.; Turk, G.; Hogan, B.L. Inhibition of Bmp signaling affects growth and differentiation in the anagen hair follicle. EMBO J. 2000, 19, 6664–6674. [Google Scholar] [CrossRef] [Green Version]
- Aubin-Houzelstein, G. Notch signaling and the developing hair follicle. Adv. Exp. Med. Biol. 2012, 727, 142–160. [Google Scholar] [CrossRef] [Green Version]
- Driskell, R.R.; Giangreco, A.; Jensen, K.B.; Mulder, K.W.; Watt, F.M. Sox2-positive dermal papilla cells specify hair follicle type in mammalian epidermis. Development 2009, 136, 2815–2823. [Google Scholar] [CrossRef] [Green Version]
- Chi, W.; Wu, E.; Morgan, B.A. Dermal papilla cell number specifies hair size, shape and cycling and its reduction causes follicular decline. Development 2013, 140, 1676–1683. [Google Scholar] [CrossRef] [Green Version]
- Preedy, V.R. Handbook of Hair in Health and Disease. Human Health Handbooks No. 1. 2012, Volume 1, pp. 332–351. Available online: https://books.google.co.uk/books?hl=zh-TW&lr=&id=7N3nEX6eL_MC&oi=fnd&pg=PA4&dq=Preedy,+Victor,+R.+Handb.+Hair+Health+Dis&ots=GcKMCrWxsm&sig=rariLE8rfXv9eQjS07LtDlwTTf8#v=onepage&q=Preedy%2C%20Victor%2C%20R.%20Handb.%20Hair%20Health%20Dis&f=false (accessed on 20 June 2021). [CrossRef]
- Cadieu, E.; Neff, M.W.; Quignon, P.; Walsh, K.; Chase, K.; Parker, H.G.; Vonholdt, B.M.; Rhue, A.; Boyko, A.; Byers, A.; et al. Coat variation in the domestic dog is governed by variants in three genes. Science 2009, 326, 150–153. [Google Scholar] [CrossRef] [Green Version]
- Kang, X.; Liu, Y.; Zhang, J.; Xu, Q.; Liu, C.; Fang, M. Characteristics and expression profile of KRT71 screened by suppression subtractive hybridization cDNA library in curly fleece chinese Tan sheep. DNA Cell Biol. 2017, 36, 552–564. [Google Scholar] [CrossRef]
- Tufarelli, C.; Fujiwara, Y.; Zappulla, D.C.; Neufeld, E.J. Hair defects and pup loss in mice with targeted deletion of the first cut repeat domain of the Cux/CDP homeoprotein gene. Dev. Biol. 1998, 200, 69–81. [Google Scholar] [CrossRef]
- Nissimov, J.N.; Das, C.A. Hair curvature: A natural dialectic and review. Biol. Rev. Camb. Philos. Soc. 2014, 89, 723–766. [Google Scholar] [CrossRef]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef]
- Mardaryev, A.N.; Ahmed, M.I.; Vlahov, N.V.; Fessing, M.Y.; Gill, J.H.; Sharov, A.A.; Botchkareva, N.V. Micro-RNA-31 controls hair cycle-associated changes in gene expression programs of the skin and hair follicle. FASEB J. 2010, 24, 3869–3881. [Google Scholar] [CrossRef] [Green Version]
- Dong, C.; Wang, H.; Xue, L.; Dong, Y.; Yang, L.; Fan, R.; Yu, X.; Tian, X.; Ma, S.; Smith, G.W. Coat color determination by miR-137 mediated down-regulation of microphthalmia-associated transcription factor in a mouse model. RNA 2012, 18, 1679–1686. [Google Scholar] [CrossRef] [Green Version]
- Sonkoly, E.; Wei, T.; Janson, P.C.; Saaf, A.; Lundeberg, L.; Tengvall-Linder, M.; Norstedt, G.; Alenius, H.; Homey, B.; Scheynius, A.; et al. MicroRNAs: Novel regulators involved in the pathogenesis of psoriasis? PLoS ONE 2007, 2, e0000610. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Zhang, Z.; O’Loughlin, E.; Wang, L.; Fan, X.; Lai, E.C.; Yi, R. MicroRNA-205 controls neonatal expansion of skin stem cells by modulating the PI(3)K pathway. Nat. Cell Biol. 2013, 15, 1153–1163. [Google Scholar] [CrossRef]
- Yuan, S.; Li, F.; Meng, Q.; Zhao, Y.; Chen, L.; Zhang, H.; Xue, L.; Zhang, X.; Lengner, C.; Yu, Z. Post-transcriptional regulation of keratinocyte progenitor cell expansion, differentiation and hair follicle regression by miR-22. PLoS Genet. 2015, 11, e1005253. [Google Scholar] [CrossRef]
- Zhang, L.; Stokes, N.; Polak, L.; Fuchs, E. Specific microRNAs are preferentially expressed by skin stem cells to balance self-renewal and early lineage commitment. Cell Stem Cell 2011, 8, 294–308. [Google Scholar] [CrossRef] [Green Version]
- Yi, R.; Poy, M.N.; Stoffel, M.; Fuchs, E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature 2008, 452, 225–229. [Google Scholar] [CrossRef] [Green Version]
- Wanbao Yang, Q.L.B.S. MicroRNA-148b Promotes Proliferation of Hair Follicle Cells by Targeting NFAT5. Frontier of Agricultural Science and Engineering (English Version); Higher Education Press: Wuhan, China, 2016. [Google Scholar] [CrossRef] [Green Version]
- Gao, W.; Sun, W.; Yin, J.; Lv, X.; Bao, J.; Yu, J.; Wang, L.; Jin, C.; Hu, L. Screening candidate microRNAs (miRNAs) in different lambskin hair follicles in Hu sheep. PLoS ONE 2017, 12, e0176532. [Google Scholar] [CrossRef]
- Lv, X.; Sun, W.; Yin, J.; Ni, R.; Su, R.; Wang, Q.; Gao, W.; Bao, J.; Yu, J.; Wang, L.; et al. An integrated analysis of microRNA and mRNA expression profiles to identify RNA expression signatures in lambskin hair follicles in Hu Sheep. PLoS ONE 2016, 11, e0157463. [Google Scholar] [CrossRef]
- Sun, W.; Ni, R.; Yin, J.F.; Musa, H.H.; Ding, T.; Chen, L. Genome array of hair follicle genes in lambskin with different patterns. PLoS ONE 2013, 8, e0068840. [Google Scholar] [CrossRef] [Green Version]
- Lv, X.; Gao, W.; Jin, C.; Wang, L.; Wang, Y.; Chen, W.; Zou, S.; Huang, S.; Li, Z.; Wang, J.; et al. Preliminary study on microR-148a and microR-10a in dermal papilla cells of Hu sheep. BMC Genet. 2019, 20, 70. [Google Scholar] [CrossRef] [Green Version]
- Rehmsmeier, M.; Steffen, P.; Hochsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [Green Version]
- Griffiths-Jones, S. miRBase: The microRNA sequence database. Methods Mol. Biol. 2006, 342, 129–138. [Google Scholar] [CrossRef]
- Huang, J.C.; Morris, Q.D.; Frey, B.J. Bayesian inference of microRNA targets from sequence and expression data. J. Comput. Biol. 2007, 14, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Trick, A.Y.; Chen, F.E.; Schares, J.A.; Freml, B.E.; Lor, P.; Yun, Y.; Wang, T.H. High resolution estimates of relative gene abundance with quantitative ratiometric regression PCR (qRR-PCR). Analyst 2021, 146, 6463–6469. [Google Scholar] [CrossRef]
- Park, E.; Cho, M.; Ki, C.S. Correct use of repeated measures analysis of variance. Korean J. Lab. Med. 2009, 29, 1–9. [Google Scholar] [CrossRef]
- Cloete, E.; Khumalo, N.P.; Ngoepe, M.N. The what, why and how of curly hair: A review. Proc. Math. Phys. Eng. Sci. 2019, 475, 20190516. [Google Scholar] [CrossRef] [PubMed]
- Westgate, G.E.; Ginger, R.S.; Green, M.R. The biology and genetics of curly hair. Exp. Dermatol. 2017, 26, 483–490. [Google Scholar] [CrossRef]
- Mou, C.; Thomason, H.A.; Willan, P.M.; Clowes, C.; Harris, W.E.; Drew, C.F.; Dixon, J.; Dixon, M.J.; Headon, D.J. Enhanced ectodysplasin-A receptor (EDAR) signaling alters multiple fiber characteristics to produce the East Asian hair form. Hum. Mutat. 2008, 29, 1405–1411. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Tan, J.; Yang, Y.; Peng, Q.; Zhang, M.; Li, J.; Lu, D.; Liu, Y.; Lou, H.; Feng, Q.; et al. Genome-wide scans reveal variants at EDAR predominantly affecting hair straightness in Han Chinese and Uyghur populations. Hum. Genet. 2016, 135, 1279–1286. [Google Scholar] [CrossRef]
- Pospiech, E.; Lee, S.D.; Kukla-Bartoszek, M.; Karlowska-Pik, J.; Wozniak, A.; Boron, M.; Zubanska, M.; Bronikowska, A.; Hong, S.R.; Lee, J.H.; et al. Variation in the RPTN gene may facilitate straight hair formation in Europeans and East Asians. J. Dermatol. Sci. 2018, 91, 331–334. [Google Scholar] [CrossRef]
- Demay, M.B. The hair cycle and Vitamin D receptor. Arch. Biochem. Biophys. 2012, 523, 19–21. [Google Scholar] [CrossRef]
- Liu, F.; Chen, Y.; Zhu, G.; Hysi, P.G.; Wu, S.; Adhikari, K.; Breslin, K.; Pospiech, E.; Hamer, M.A.; Peng, F.; et al. Meta-analysis of genome-wide association studies identifies 8 novel loci involved in shape variation of human head hair. Hum. Mol. Genet. 2018, 27, 559–575. [Google Scholar] [CrossRef]
- Rong, E.G.; Yang, H.; Zhang, Z.W.; Wang, Z.P.; Yan, X.H.; Li, H.; Wang, N. Association of methionine synthase gene polymorphisms with wool production and quality traits in Chinese Merino population. J. Anim. Sci. 2015, 93, 4601–4609. [Google Scholar] [CrossRef]
- Salmela, E.; Niskanen, J.; Arumilli, M.; Donner, J.; Lohi, H.; Hytonen, M.K. A novel KRT71 variant in curly-coated dogs. Anim. Genet. 2019, 50, 101–104. [Google Scholar] [CrossRef] [Green Version]
- Bauer, A.; Hadji, R.S.; Brunner, M.T.; Jagannathan, V.; Bucher, I.; Bannoehr, J.; Varjonen, K.; Bond, R.; Bergvall, K.; Welle, M.M.; et al. A second KRT71 allele in curly coated dogs. Anim. Genet. 2019, 50, 97–100. [Google Scholar] [CrossRef] [Green Version]
- Pierard-Franchimont, C.; Paquet, P.; Quatresooz, P.; Pierard, G.E. Mechanobiology and cell tensegrity: The root of ethnic hair curling? J. Cosmet. Dermatol. 2011, 10, 163–167. [Google Scholar] [CrossRef]
- Ji, K.; Zhang, P.; Zhang, J.; Fan, R.; Liu, Y.; Yang, S.; Hu, S.; Liu, X.; Dong, C. MicroRNA 143-5p regulates alpaca melanocyte migration, proliferation and melanogenesis. Exp. Dermatol. 2018, 27, 166–171. [Google Scholar] [CrossRef]
- Qi, S.; Liu, B.; Zhang, J.; Liu, X.; Dong, C.; Fan, R. Knockdown of microRNA1435p by STTM technology affects eumelanin and pheomelanin production in melanocytes. Mol. Med. Rep. 2019, 20, 2649–2656. [Google Scholar] [CrossRef]
- Han, W.; Yang, F.; Wu, Z.; Guo, F.; Zhang, J.; Hai, E.; Shang, F.; Su, R.; Wang, R.; Wang, Z.; et al. Inner mongolian cashmere goat secondary follicle development regulation research based on mRNA-miRNA co-analysis. Sci. Rep. 2020, 10, 4519. [Google Scholar] [CrossRef]
- Guoping, M.; Ran, L.; Yanru, Q. miR-143 inhibits cell proliferation of gastric cancer cells through targeting GATA6. Oncol. Res. 2018, 26, 1023–1029. [Google Scholar] [CrossRef]
- Sun, X.; Dai, G.; Yu, L.; Hu, Q.; Chen, J.; Guo, W. miR-143-3p inhibits the proliferation, migration and invasion in osteosarcoma by targeting FOSL2. Sci. Rep. 2018, 8, 606. [Google Scholar] [CrossRef] [Green Version]
- Cordes, K.R.; Sheehy, N.T.; White, M.P.; Berry, E.C.; Morton, S.U.; Muth, A.N.; Lee, T.H.; Miano, J.M.; Ivey, K.N.; Srivastava, D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 2009, 460, 705–710. [Google Scholar] [CrossRef] [Green Version]
- Papanota, A.M.; Tsiakanikas, P.; Kontos, C.K.; Malandrakis, P.; Liacos, C.I.; Ntanasis-Stathopoulos, I.; Kanellias, N.; Gavriatopoulou, M.; Kastritis, E.; Avgeris, M.; et al. A molecular signature of circulating microRNA can predict osteolytic bone disease in multiple myeloma. Cancers 2021, 13, 3877. [Google Scholar] [CrossRef]
- Tewari, R.S.; Ala, U.; Accornero, P.; Baratta, M.; Miretti, S. Circulating skeletal muscle related microRNAs profile in Piedmontese cattle during different age. Sci. Rep. 2021, 11, 15815. [Google Scholar] [CrossRef]
- Rong, Z.W.; Soucie, E.; Sung, M.N.; Martin-Soudant, N.; Berube, G.; Leduy, L.; Nepveu, A. Exon/intron structure and alternative transcripts of the CUTL1 gene. Gene 2000, 241, 75–85. [Google Scholar] [CrossRef]
- Alcalay, N.I.; Vanden, H.G. Regulation of cell proliferation and differentiation in the kidney. Front. Biosci. (Landmark Ed.) 2009, 14, 4978–4991. [Google Scholar] [CrossRef]
- Luong, M.X.; van der Meijden, C.M.; Xing, D.; Hesselton, R.; Monuki, E.S.; Jones, S.N.; Lian, J.B.; Stein, J.L.; Stein, G.S.; Neufeld, E.J.; et al. Genetic ablation of the CDP/Cux protein C terminus results in hair cycle defects and reduced male fertility. Mol. Cell. Biol. 2002, 22, 1424–1437. [Google Scholar] [CrossRef] [Green Version]
- Ellis, T.; Gambardella, L.; Horcher, M.; Tschanz, S.; Capol, J.; Bertram, P.; Jochum, W.; Barrandon, Y.; Busslinger, M. The transcriptional repressor CDP (Cutl1) is essential for epithelial cell differentiation of the lung and the hair follicle. Genes Dev. 2001, 15, 2307–2319. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, A.; Farooq, M.; Fujikawa, H.; Inoue, A.; Ohyama, M.; Ehama, R.; Nakanishi, J.; Hagihara, M.; Iwabuchi, T.; Aoki, J.; et al. A missense mutation within the helix initiation motif of the keratin K71 gene underlies autosomal dominant woolly hair/hypotrichosis. J. Investig. Dermatol. 2012, 132, 2342–2349. [Google Scholar] [CrossRef] [Green Version]
- Harel, S.; Christiano, A.M. Keratin 71 mutations: From water dogs to woolly hair. J. Investig. Dermatol. 2012, 132, 2315–2317. [Google Scholar] [CrossRef] [Green Version]
| Gene/miRNA Name | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Product Length (bp) |
|---|---|---|---|
| CUX1 | GCACGACATTGAGACGGAG | AGCTATGGTCTCAGCCTGGT | 160 |
| KRT71 | GGCTCATCCAGAGAATCCGC | GAGCATTGTCACCCCTCTGT | 102 |
| GAPDH | TCTCAAGGGCATTCTAGGCTAC | GCCGAATTCATTGTCGTACCAG | 151 |
| CDK2 | AGAAGTGGCTGCATCACAAG | TCTCAGAATCTCCAGGGAATAG | 92 |
| PCNA | CGAGGGCTTCGACACTTAC | GTCTTCATTGCCAGCACATT | 97 |
| Cyclin d1 | CCGAGGAGAACAAGCAGATC | GAGGGTGGGTTGGAAATG | 91 |
| miR-143 | CGCGTGAGATGAAGCACTG | AGTGCAGGGTCCGAGGTATT | variable |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT | 95 |
| Gene Name | Primer | Forward Primer (5′-3′) |
|---|---|---|
| CUX1 | CUX1-WT-F | CCctcgagCCATCTCCCTTTAAAGAGGG |
| CUX1-WI-R | TTgcggccgcGACTTAGAGCTCGCTGTCC | |
| KRT71 | KRT71-WT-F | CCctcgagTTGTCCCAGCTCCTGCTT |
| KRT71-WT-R | TTgcggccgcTATTGAGGATGCAG | |
| CUX1 | CUX1-MUT1-F | CTTGGGGACAGAGACCTGTGAGCTGTGACACTG |
| CUX1-MUT1-R | CAGTGTCACAGCTCACAGGTCTCTGTCCCCAAG | |
| KRT71 | KRT71-MUT1-F | GGGAAGAGGAGGCCCCATCTTTTCTCCTTTC |
| KRT71-MUT1-R | GAAAGGAGAAAAGATGGGGCCTCCTCTTCCC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, T.; Huang, S.; Lv, X.; Wang, S.; Getachew, T.; Mwacharo, J.M.; Haile, A.; Sun, W. miR-143 Targeting CUX1 to Regulate Proliferation of Dermal Papilla Cells in Hu Sheep. Genes 2021, 12, 2017. https://doi.org/10.3390/genes12122017
Hu T, Huang S, Lv X, Wang S, Getachew T, Mwacharo JM, Haile A, Sun W. miR-143 Targeting CUX1 to Regulate Proliferation of Dermal Papilla Cells in Hu Sheep. Genes. 2021; 12(12):2017. https://doi.org/10.3390/genes12122017
Chicago/Turabian StyleHu, Tingyan, Sainan Huang, Xiaoyang Lv, Shanhe Wang, Tesfaye Getachew, Joram M. Mwacharo, Aynalem Haile, and Wei Sun. 2021. "miR-143 Targeting CUX1 to Regulate Proliferation of Dermal Papilla Cells in Hu Sheep" Genes 12, no. 12: 2017. https://doi.org/10.3390/genes12122017
APA StyleHu, T., Huang, S., Lv, X., Wang, S., Getachew, T., Mwacharo, J. M., Haile, A., & Sun, W. (2021). miR-143 Targeting CUX1 to Regulate Proliferation of Dermal Papilla Cells in Hu Sheep. Genes, 12(12), 2017. https://doi.org/10.3390/genes12122017





