Evaluation of Reference Genes in Glenea cantor (Fabricius) by Using qRT-PCR
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. Sample Collection
2.3. Total RNA Extraction and cDNA Synthesis
2.4. Primer Design
2.5. qRT-PCR
2.6. Analyses of Candidate Gene Expression
2.7. Expression Validation of the Reference Gene in G. cantor
3. Results
3.1. Evaluation of Amplification Efficiency and Specificity of Primers in G. cantor
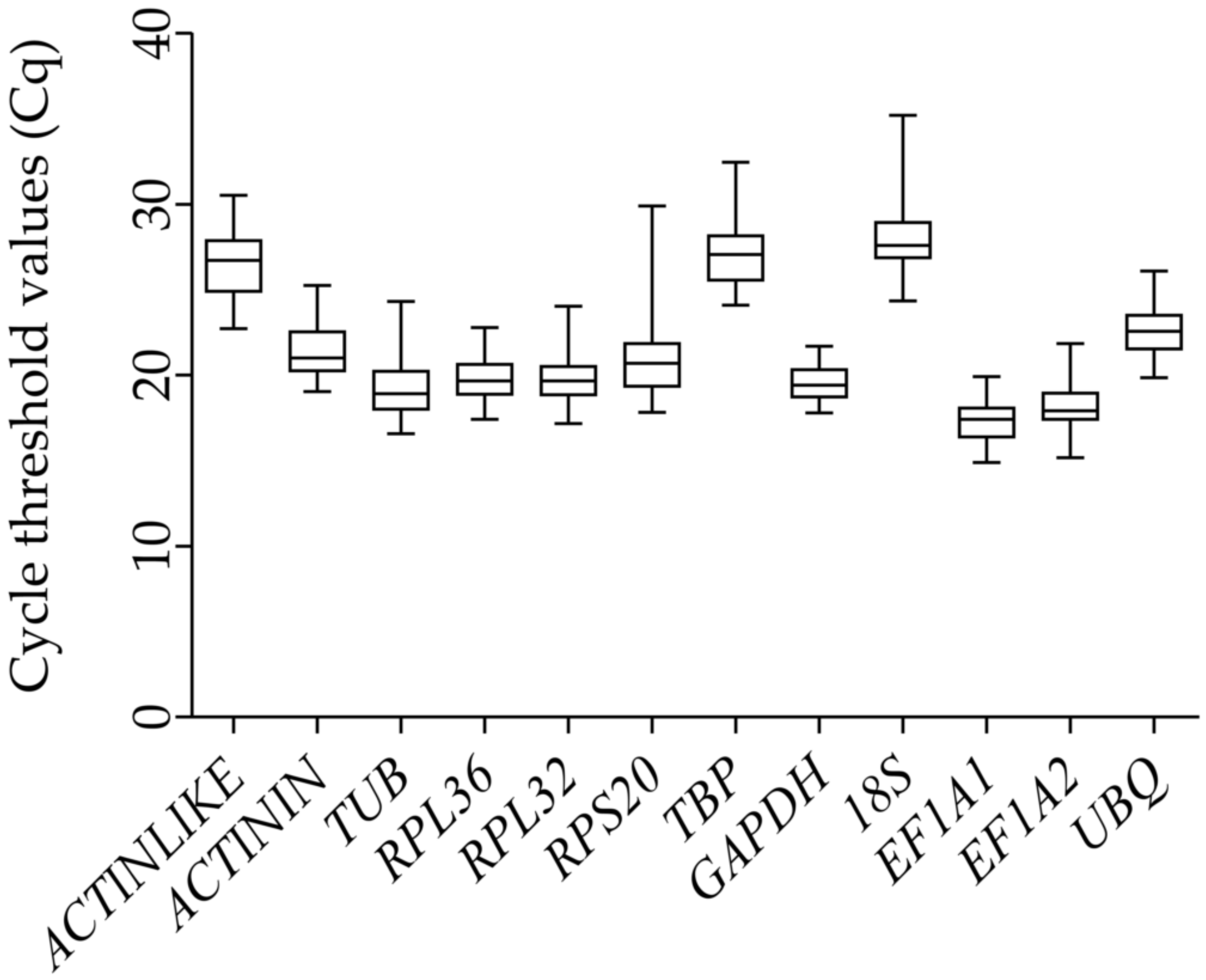
3.2. Expression Range of the Candidate Reference Genes in G. cantor
3.3. Stability Analysis of the Candidate Reference Genes in G. cantor
3.3.1. geNorm
3.3.2. NormFinder
3.3.3. BestKeeper
3.3.4. RefFinder
3.4. Validation of the Reference Gene Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, W.; Tian, M.Y.; Lai, K.P.; Wei, S.G.; Qin, A.Z. Occurrence and control of Glenea cantor. Guangxi Plant Prot. 2006, 19, 1–4. (In Chinese) [Google Scholar] [CrossRef]
- Lu, W.; Wang, Q.; Tian, M.Y.; He, X.Z.; Zeng, X.L.; Zhong, Y.X. Mate location and recognition in Glenea cantor (Fabr.) (Coleoptera: Cerambycidae: Lamiinae): Roles of host plant health, female sex pheromone, and vision. Environ. Entomol. 2007, 36, 864–870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Z.S.; Yang, Y.B.; Dou, F.B.; Zhang, Y.J.; Huang, H.X.; Zheng, X.L.; Wang, X.Y.; Lu, W. Observations on the Ultrastructure of Antennal Sensilla of Adult Glenea cantor (Cerambycidae: Lamiinae). J. Insect Sci. 2020, 20, 1–9. [Google Scholar] [CrossRef]
- Dong, Z.S.; Zhang, Y.J.; Lai, K.P.; Zheng, X.L.; Wang, Q.; Lu, W. Biological characteristics of Glenea cantor Fabricius (Coleoptera: Cerambycidae). J. Environ. Entomol. 2017, 39, 1313–1318. (In Chinese) [Google Scholar]
- Lei, Y.M. Glenea cantor’s damage to kapok trees and its comprehensive control. China Trop. Agric. 2015, 52–54. (In Chinese) [Google Scholar] [CrossRef]
- Lai, K.; Lu, W.; Liu, D.; Luo, Z.; Gao, P. The larval instars and stadia of the longhorn beetle Glenea cantor. Chin. J. Appl Entomol. 2008, 45, 138–140. (In Chinese) [Google Scholar] [CrossRef]
- Lu, W.; Wang, Q.; Tian, M.Y.; Xu, J.; Lv, J.; Qin, A. Mating behavior and sexual selection in a polygamous beetle. Curr. Zool. 2013, 59, 257–264. [Google Scholar] [CrossRef] [Green Version]
- Lu, W.; Wang, Q.; Tian, M.Y.; Xu, J.; Lv, J.; Wei, S.G.; Qin, A.Z. Reproductive traits of Glenea cantor (Coleoptera: Cerambycidae: Lamiinae). J. Econ. Entom. 2013, 106, 215–220. [Google Scholar] [CrossRef]
- Lu, W.; Wang, Q.; Tian, M.Y.; Xu, J.; Qin, A.Z.; He, L.; Jia, B.; Cai, J.J. Host selection and colonization strategies with evidence for a female-produced oviposition attractant in a longhorn beetle. Environ. Entomol. 2011, 40, 1487–1493. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Lu, W.; Nong, C.; Nong, Y.Y. A preliminary study on artificial diet of Glenea cantor. Guangxi Plant Prot. 2007, 20, 9–11. [Google Scholar] [CrossRef]
- Wang, X.Y.; Zheng, X.L.; Lu, W. The complete mitochondrial genome of an Asian longicorn beetle Glenea cantor (Coleoptera: Cerambycidae: Lamiinae). Mitochondrial DNA Part B 2019, 4, 2906–2907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 4, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapman, J.R.; Waldenström, J. With reference to reference genes: A systematic review of endogenous controls in gene expression studies. PLoS ONE 2015, 10, e0141853. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Li, J.L.; Huang, J.X.; Wu, J. Identification of suitable reference genes for miRNA quantitation in bumblebee (Hymenoptera: Apidae) response to reproduction. Apidologie 2019, 50, 40–50. [Google Scholar] [CrossRef] [Green Version]
- Jeon, J.H.; Moon, K.; Kim, Y.; Kim, Y.H. Reference gene selection for qRT-PCR analysis of season- and tissue-specific gene expression profiles in the honey bee Apis mellifera. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Wieczorek, P.; Frąckowiak, P.; Obrępalska-Stęplowska, A. Evaluation of the expression stability of reference genes in Apis mellifera under pyrethroid treatment. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef]
- Chen, L.; Tian, Z.; Wang, X.Y.; Chen, X.M.; Lu, W.; Wang, X.P.; Zheng, X.L. Screening of reference genes for quantitative real-time PCR in Phauda flammans Walker (Lepidoptera: Phaudidae). J. Environ. Entomol. 2020, 3, 1–20. (In Chinese) [Google Scholar]
- Fu, W.; Xie, W.; Zhang, Z.; Wang, S.L.; Wu, Q.J.; Liu, Y.; Zhou, X.M.; Zhou, X.G.; Zhang, Y.J. Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: Plutellidae). Int. J. Biol. Sci. 2013, 9, 792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, J.J.; Sun, L.L.; Zhang, Q.H.; Cao, C.W. Screening and evaluation of the stability of expression of reference genes in Lymantria dispar (Lepidoptera: Erebidae) using qRT-PCR. Gene 2020, 749, 144712. [Google Scholar] [CrossRef]
- Zhang, L.L.; Jing, X.D.; Chen, W.; Bai, J.L.; Vasseur, L.; He, W.Y.; You, M.S. Selection of reference genes for expression analysis of plant-derived microRNAs in Plutella xylostella using qRT-PCR and ddPCR. PLoS ONE 2019, 14, e0220475. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.H.; Deng, W.H.; Zhu, F. Reference gene selection for quantitative gene expression analysis in black soldier fly (Hermetia illucens). PLoS ONE 2019, 14, e0221420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, C.F.; Pan, H.P.; Zhang, L.H.; Ou, D.; Lu, Z.T.; Khan, M.M.; Qiu, B.L. Comprehensive Assessment of Candidate Reference Genes for Gene Expression Studies Using RT-qPCR in Tamarixia radiata, a Predominant Parasitoid of Diaphorina citri. Genes 2020, 11, 1178. [Google Scholar] [CrossRef]
- Wang, X.Y.; Xiong, M.; Wang, J.L.; Lei, C.L.; Zhu, F. Reference Gene Stability of a synanthropic fly, Chrysomya megacephala. Parasit. Vector. 2015, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.Y.; Li, Y.; Shang, Y.J.; Ren, L.P.; Chen, W.; Wang, S.W.; Guo, Y.D. Development of Sarcophaga dux (diptera: Sarcophagidae) at constant temperatures and differential gene expression for age estimation of the pupae. J. Therm. Biol. 2020, 93, 102735. [Google Scholar] [CrossRef]
- Li, X.N.; Gong, P.P.; Wang, B.T.; Wang, C.; Li, M.Y.; Zhang, Y.H.; Li, X.R.; Gao, H.F.; Ju, J.S.; Zhu, X.X. Selection and validation of experimental condition-specific reference genes for qRT-PCR in Metopolophium dirhodum (Walker) (Hemiptera: Aphididae). Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Liu, H.Q.; Dong, Y.; Zhang, Y.; Wong, S.M.; Wang, C.C.; Zhou, Y.J.; Xu, Q.F. Determination of suitable RT-qPCR reference genes for studies of gene functions in Laodelphax striatellus (Fallén). Genes 2019, 10, 887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.J.; Zheng, H.L.; Liu, Y.Y.; Li, H.W.; Jiang, Y.H.; Lin, L.B.; Deng, X.Y.; Zhang, Q.L. Selection of reference genes for quantitative real-time PCR in Aquatica leii (Coleoptera: Lampyridae) under five different experimental conditions. Front. Physiol. 2020, 11, 1247. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, D.H.; Siegfried, B.D. Selection of reference genes for normalization of RT-qPCR data in gene expression studies in Anthonomus eugenii Cano (Coleoptera: Curculionidae). Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Li, J.W.; Cheng, B.; Li, W.; Sun, W.X.; Gao, H.Y.; Ju, Q.; Jiang, X.J.; Du, L.; Qu, C.J.; et al. Screening of reference genes for quantitative real-time PCR in Sympiezomias velatus (Coleoptera: Curculionidae). Acta Entomol. Sin. 2018, 61, 1284–1294. (In Chinese) [Google Scholar] [CrossRef]
- Feng, B.; Guo, Q.S.; Mao, B.P.; Du, Y.J. Identification and selection of valid reference genes for assaying gene expression in the chemosensory tissues of Monochamus alternatus (Coleoptera: Cerambycidae) by RT-qPCR. Acta Entomol. Sin. 2016, 59, 427–437. (In Chinese) [Google Scholar] [CrossRef]
- Ye, B.H.; Chen, Y.W.; Shu, J.P.; Zhang, W.; Zhang, Y.B.; Li, H.B.; Song, Q.Y. Screening and application of reference genes for qRT-PCR in bamboo wireworm. J. Zhejiang A F Univ. 2021, 38, 1–8. (In Chinese) [Google Scholar] [CrossRef]
- Toutges, M.J.; Hartzer, K.; Lord, J.; Oppert, B. Evaluation of reference genes for quantitative polymerase chain reaction across life cycle stages and tissue types of Tribolium castaneum. J. Agric. Food Chem. 2010, 58, 8948–8951. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Etschmann, B.; Wilcken, B.; Stoevesand, K.; Von Der Schulenburg, A.; Sterner-Kock, A. Selection of reference genes for quantitative real-time PCR analysis in canine mammary tumors using the GeNorm algorithm. Vet. Pathol. 2006, 43, 934–942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hruz, T.; Wyss, M.; Docquier, M.; Pfaffl, M.W.; Masanetz, S.; Borghi, L.; Verbrugghe, P.; Kalaydjieva, L.; Bleuler, S.; Laule, O.; et al. RefGenes: Identification of reliable and condition specific reference genes for RT-qPCR data normalization. BMC Genom. 2011, 12, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozera, B.; Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef] [Green Version]
- Wong, M.L.; Medrano, J.F. Real-time PCR for mRNA quantitation. Biotechniques 2005, 39, 75–85. [Google Scholar] [CrossRef]
- Bustin, S.A. Developments in real-time PCR research and molecular diagnostics. Expert Rev. Mol. Diagn. 2010, 10, 713–715. [Google Scholar] [CrossRef] [PubMed]
- Hellemans, J.; Vandesompele, J. Selection of reliable reference genes for RT-qPCR analysis [M]. In Quantitative Real-Time PCR; Humana Press: New York, NY, USA, 2014; pp. 19–26. [Google Scholar]
- Eisenberg, E.; Levanon, E.Y. Human housekeeping genes, revisited. Trends Genet. 2013, 29, 569–574. [Google Scholar] [CrossRef]
- Fu, X.H.; Meyer-Rochow, V.B. Selection and Validation of Suitable Reference Genes for RT-qPCR Analysis in the Rare Aquatic Firefly Aquatica leii (Coleoptera: Lampyridae). Insects 2021, 12, 359. [Google Scholar] [CrossRef] [PubMed]
- Lü, J.; Chen, S.M.; Guo, M.J.; Ye, C.Y.; Qiu, B.L.; Yang, C.X.; Pan, H.P. Selection of appropriate reference genes for RT-qPCR analysis in Propylea japonica (Coleoptera: Coccinellidae). PLoS ONE 2018, 13, e0208027. [Google Scholar] [CrossRef] [Green Version]
- Lü, J.; Chen, S.M.; Guo, M.J.; Ye, C.Y.; Qiu, B.L.; Wu, J.H.; Yang, C.X.; Pan, H.P. Selection and validation of reference genes for RT-qPCR analysis of the ladybird beetle Henosepilachna vigintioctomaculata. Frontiers in physiology. Front. Physiol. 2018, 9, 1614. [Google Scholar] [CrossRef] [Green Version]
- Pan, H.P.; Yang, X.W.; Siegfried, B.D.; Zhou, X.G. A comprehensive selection of reference genes for RT-qPCR analysis in a predatory lady beetle, Hippodamia convergens (Coleoptera: Coccinellidae). PLoS ONE 2015, 10, e0125868. [Google Scholar] [CrossRef]
- Yang, C.X.; Preisser, E.L.; Zhang, H.j.; Liu, Y.; Dai, L.Y.; Pan, H.P.; Zhou, X.G. Selection of reference genes for RT-qPCR analysis in Coccinella septempunctata to assess un-intended effects of RNAi transgenic plants. Front. Plant Sci. 2016, 7, 1672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.W.; Pan, H.P.; Yuan, L.; Zhou, X.G. Reference gene selection for RT-qPCR analysis in Harmonia axyridis, a global invasive lady beetle. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Jing, D.P.; Zhang, T.T.; Bai, S.X.; He, K.L.; Prabu, S.; Luan, J.B.; Wang, Z.Y. Sexual-biased gene expression of olfactory-related genes in the antennae of Conogethes pinicolalis (Lepidoptera: Crambidae). BMC Genom. 2020, 21, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Zhang, Y.B.; Xu, K.K.; Wang, Y.W.; Yang, W.J. Selection and Validation of Reference Genes for Gene Expression Analysis in Tuta absoluta Meyrick (Lepidoptera: Gelechiidae). Insects 2021, 12, 589. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.C.; Tian, J.C.; Lu, Y.H.; Xu, H.X.; Zang, L.S.; Lu, Z.X.; Jin, L.H. Selection of reference genes for RT-qPCR analysis in Trichogramma chilonis (Hymenoptera: Trichogrammatidae). J. Asia-Pac. Entomol 2021, in press. [Google Scholar] [CrossRef]
- Huo, L.X.; Bai, X.P.; Che, W.N.; Ning, S.F.; Lv, L.; Zhang, L.S.; Zhou, J.C.; Dong, H. Selection and evaluation of qPCR reference genes for expression analysis in the tiny egg parasitoid wasp, Trichogramma dendrolimi Matsumura (Hymenoptera: Trichogrammatidae). bioRxiv 2021, 2021–2027. [Google Scholar] [CrossRef]
- Deng, Y.C.; Zhao, H.X.; Yang, S.; Zhang, L.; Zhang, L.N.; Hou, C.S. Screening and validation of reference genes for RT-qPCR under different honey bee viral infections and dsRNA treatment. Front. Microbiol. 2020, 11, 1715. [Google Scholar] [CrossRef] [PubMed]
- Gurusamy, D.; Howell, J.L.; Chereddy, S.C.R.R.; Mogilicherla, K.; Palli, S.R. Improving RNA interference in the southern green stink bug, Nezara viridula. J. Pest Sci. 2021, 94, 1461–1472. [Google Scholar] [CrossRef]
- Bai, Y.; Lv, Y.N.; Zeng, M.; Jia, P.Y.; Lu, H.N.; Zhu, Y.B.; Li, S.; Cui, Y.Y.; Luan, Y.X. Selection of Reference Genes for Normalization of Gene Expression in Thermobia domestica (Insecta: Zygentoma: Lepismatidae). Genes 2021, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Kyre, B.R.; Rodrigues, T.B.; Rieske, L.K. RNA interference and validation of reference genes for gene expression analyses using qPCR in southern pine beetle, Dendroctonus frontalis. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, H.; Chu, X.; Ma, Q.; Liang, G.; Wu, S.; Wang, R.; Tigabu, M.; Zhang, F.; Hu, X. Molecular cloning and expression analysis of the endogenous cellulase gene MaCel1 in Monochamus alternatus. Forests 2020, 11, 1372. [Google Scholar] [CrossRef]






| Species | Condition | Optimal Reference Gene | Reference |
|---|---|---|---|
| Anthonomus eugenii Cano | developmental stages | EF1-α, 18S and RPL12 | [28] |
| sexes | RPS23 and RPL12 | ||
| low temperature | GAPDH and α-TUB | ||
| high temperature | α-TUB and RPS23 | ||
| all temperatures | α-TUB and GAPDH | ||
| starvation | RPL12 and α-TUB | ||
| Aquatica leii | tissues | α-tubulin and β-tubulin | [27] |
| temperatures | β-tubulin, EF1A and GST | ||
| sexes | β-actin and EF1A | ||
| developmental stages | β-tubulin GST and GAPDH and SDHA | ||
| larvae exposed to different concentrations of benzo(a)pyrene | α-tubulin and EF1A | ||
| Sympiezomias velatus | tissues | TUB, TUA, RPS20 and RPL12 | [29] |
| Monochamus alternatus | different chemosensory tissues at different developmental stages and in different genders | GAPDH and TUB | [30] |
| Melanotus cribricollis | infectious conditions of Metarhizium pingshaense | PRS27 and RPS3 | [31] |
| Tribolium castaaneum (Herbst) | developmental stages | RPS6, RPL13a, RPS3 and RPS18 | [32] |
| Gene Name (Abbreviation) | GenBank Accession Number | Primer Sequence (5′–3′) | Amplicon Size (bp) | PCR Efficiency | Regression Coefficient (R2) |
|---|---|---|---|---|---|
| ACTINLIKE | MW462107 | F: TTCAAACTGGCGGAAGGGTT R: GGGCCGGTCTTATATCCACG | 100 | 1.005 | 0.996 |
| ACTININ | MW462108 | F: GTCAACGCAAGACTGCTCCA R: TCAGGGCGATGACGATGAAT | 106 | 0.992 | 0.998 |
| TUB | MW462097 | F: CTACACCATCGGCAAGGAAA R: CTCCGAAAGAGTGGAAGATCAG | 107 | 0.947 | 0.997 |
| RPL36 | MW462098 | F: GAAATTCGTGCGAGACCTCATC R: GGCGGCGCTTAAGGAATTTA | 119 | 0.959 | 0.998 |
| RPL32 | MW462099 | F: TCAAGGGCCAGTTCTTGATG R: ACTTTCCTGAATCCGGTTGG | 85 | 0.960 | 0.999 |
| RPS20 | MW462100 | F: CCAGTACGTATGCCCACAAA R: ACCTGTCCCAGGTCTTAGAA | 82 | 0.910 | 0.999 |
| TBP | MW462101 | F: GAGACTGGTGCTGCTCATATT R: GCGCATCTTTGATGTCTTGTC | 80 | 0.927 | 0.984 |
| GAPDH | MW462102 | F: CAAGGCTGGAATCTCTCTCAA R: GTTGATCAAGTCGATGACCCT | 97 | 0.938 | 0.995 |
| 18S | MW462103 | F: CGGTGGAAAGAGAGGTAGAAG R: CAACGCCGAAATGCTGATAG | 104 | 1.094 | 0.961 |
| EF1A1 | MW462104 | F: TGGCGATGCTGCCATTAT R: GGACAGCGAAACGTCCTAAT | 92 | 1.081 | 0.951 |
| EF1A2 | MW462105 | F: GGAGAATTCGAAGCTGGTATCT R: CGCCAACAATGAGTTGTCTTAC | 92 | 0.981 | 0.998 |
| UBQ | MW462106 | F: AGGTGGCATGCAGATCTTT R: CTTGGCCTTGACGTTCTCTAT | 94 | 1.011 | 1.000 |
| cellulase | OL757647 | F: GCGCTTGGGCTGAAAATTTG R: ACTACACTGGGCTGCTGATTAC | 148 | 0.966 | 0.998 |
| Tissues | Developmental Stages | Sexes | All Samples | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rank | Gene Name | SD (CP) | CV (CP, %) | Gene | SD (CP) | CV (CP, %) | Gene | SD (CP) | CV (CP, %) | Gene Name | SD (CP) | CV (CP, %) |
| 1 | GAPDH | 0.87 | 4.47 | RPL36 | 0.76 | 3.88 | GAPDH | 0.67 | 3.45 | GAPDH | 0.83 | 4.26 |
| 2 | EF1A1 | 0.92 | 5.33 | GAPDH | 0.76 | 3.90 | RPL36 | 1.02 | 5.21 | EF1A1 | 0.97 | 5.66 |
| 3 | RPL32 | 0.98 | 4.96 | RPL32 | 1.01 | 5.20 | EF1A1 | 1.14 | 6.74 | RPL32 | 1.02 | 5.20 |
| 4 | EF1A2 | 0.99 | 5.56 | EF1A1 | 1.03 | 6.02 | RPL32 | 1.30 | 6.64 | RPL36 | 1.05 | 5.29 |
| 5 | TUB | 1.01 | 5.30 | TBP | 1.25 | 4.72 | ACTINLIKE | 1.33 | 5.28 | EF1A2 | 1.16 | 6.38 |
| 6 | UBQ | 1.07 | 4.77 | ACTINLIKE | 1.28 | 5.04 | ACTININ | 1.47 | 6.84 | UBQ | 1.31 | 5.79 |
| 7 | RPL36 | 1.15 | 5.81 | EF1A2 | 1.44 | 7.68 | TBP | 1.52 | 5.71 | ACTININ | 1.42 | 6.61 |
| 8 | 18S | 1.24 | 4.43 | RPS20 | 1.51 | 7.39 | EF1A2 | 1.55 | 8.34 | TBP | 1.48 | 5.45 |
| 9 | ACTININ | 1.28 | 6.03 | ACTININ | 1.68 | 7.68 | UBQ | 1.84 | 8.19 | 18S | 1.49 | 5.32 |
| 10 | ACTINLIKE | 1.30 | 4.81 | UBQ | 1.85 | 8.05 | 18S | 1.89 | 6.80 | ACTINLIKE | 1.51 | 5.70 |
| 11 | RPS20 | 1.40 | 6.63 | 18S | 2.05 | 7.28 | RPS20 | 1.94 | 9.35 | RPS20 | 1.51 | 7.24 |
| 12 | TBP | 1.51 | 5.53 | TUB | 2.46 | 12.17 | TUB | 2.52 | 12.89 | TUB | 1.56 | 8.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, R.-R.; Huang, Z.-Y.; Qin, C.-W.; Zheng, X.-L.; Lu, W.; Wang, X.-Y. Evaluation of Reference Genes in Glenea cantor (Fabricius) by Using qRT-PCR. Genes 2021, 12, 1984. https://doi.org/10.3390/genes12121984
Su R-R, Huang Z-Y, Qin C-W, Zheng X-L, Lu W, Wang X-Y. Evaluation of Reference Genes in Glenea cantor (Fabricius) by Using qRT-PCR. Genes. 2021; 12(12):1984. https://doi.org/10.3390/genes12121984
Chicago/Turabian StyleSu, Ran-Ran, Zhong-Yan Huang, Chao-Wei Qin, Xia-Lin Zheng, Wen Lu, and Xiao-Yun Wang. 2021. "Evaluation of Reference Genes in Glenea cantor (Fabricius) by Using qRT-PCR" Genes 12, no. 12: 1984. https://doi.org/10.3390/genes12121984
APA StyleSu, R.-R., Huang, Z.-Y., Qin, C.-W., Zheng, X.-L., Lu, W., & Wang, X.-Y. (2021). Evaluation of Reference Genes in Glenea cantor (Fabricius) by Using qRT-PCR. Genes, 12(12), 1984. https://doi.org/10.3390/genes12121984






