Defining the Influence of the A12.2 Subunit on Transcription Elongation and Termination by RNA Polymerase I In Vivo
Abstract
:1. Introduction
2. Materials and Methods
2.1. Native Elongating Transcript Sequencing (NET-seq) Library Preparation and Sequencing
2.2. NET-seq Data Formatting and Analysis
3. Results
3.1. WT and rpa12Δ NET-seq Libraries Are Highly Reproducible
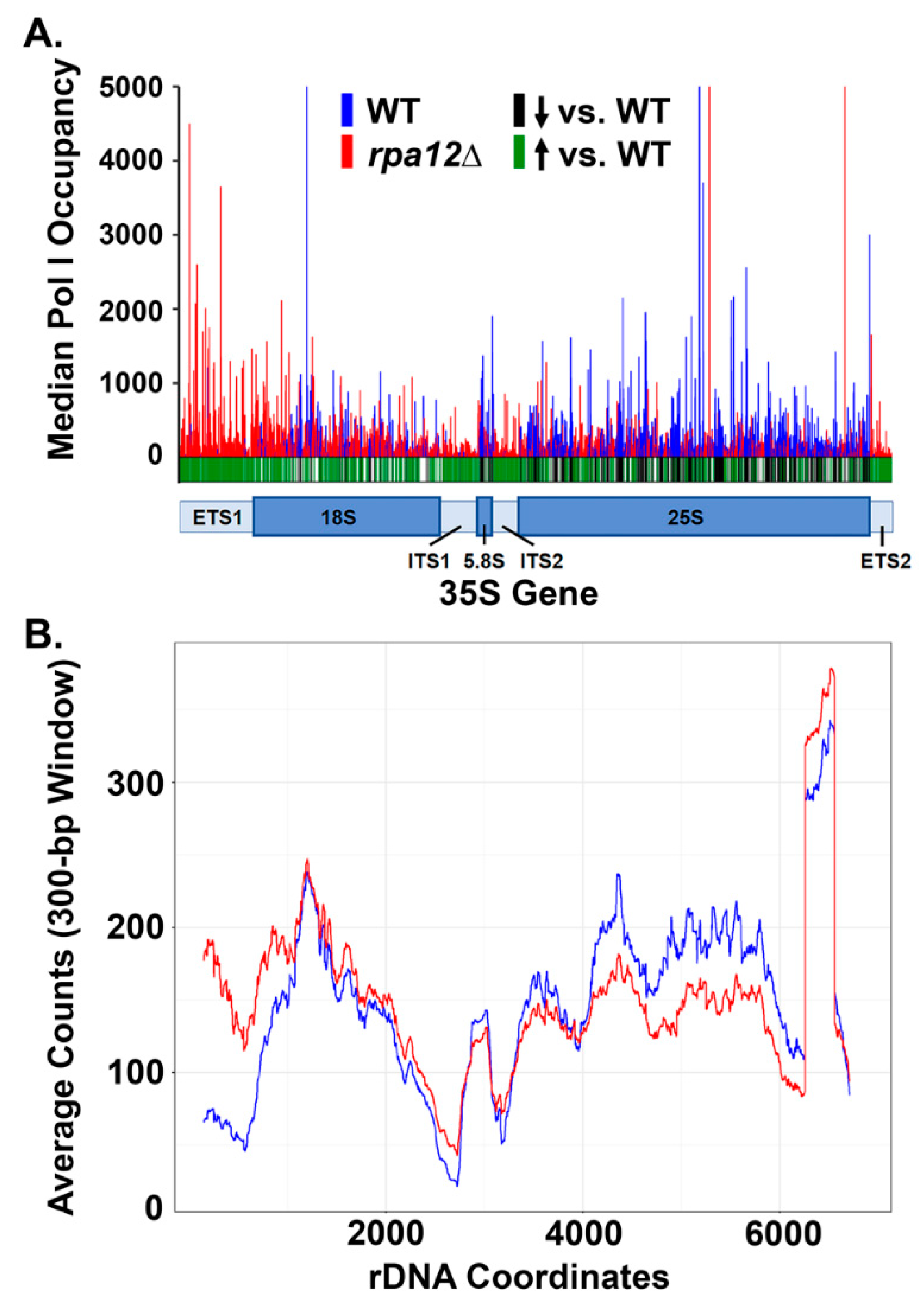
3.2. RPA12 Deletion Alters Pol I Occupancy throughout the 35S Gene
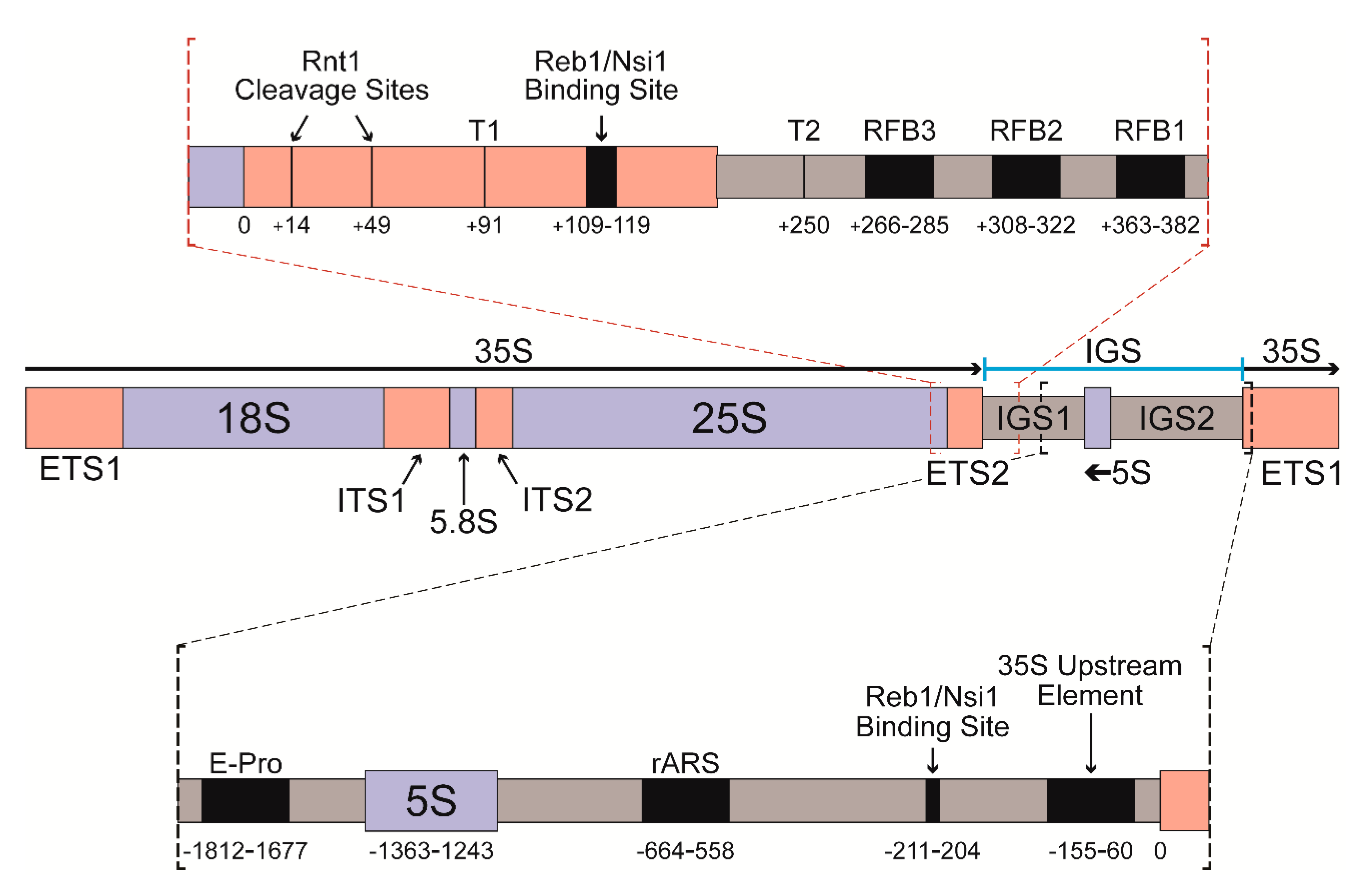
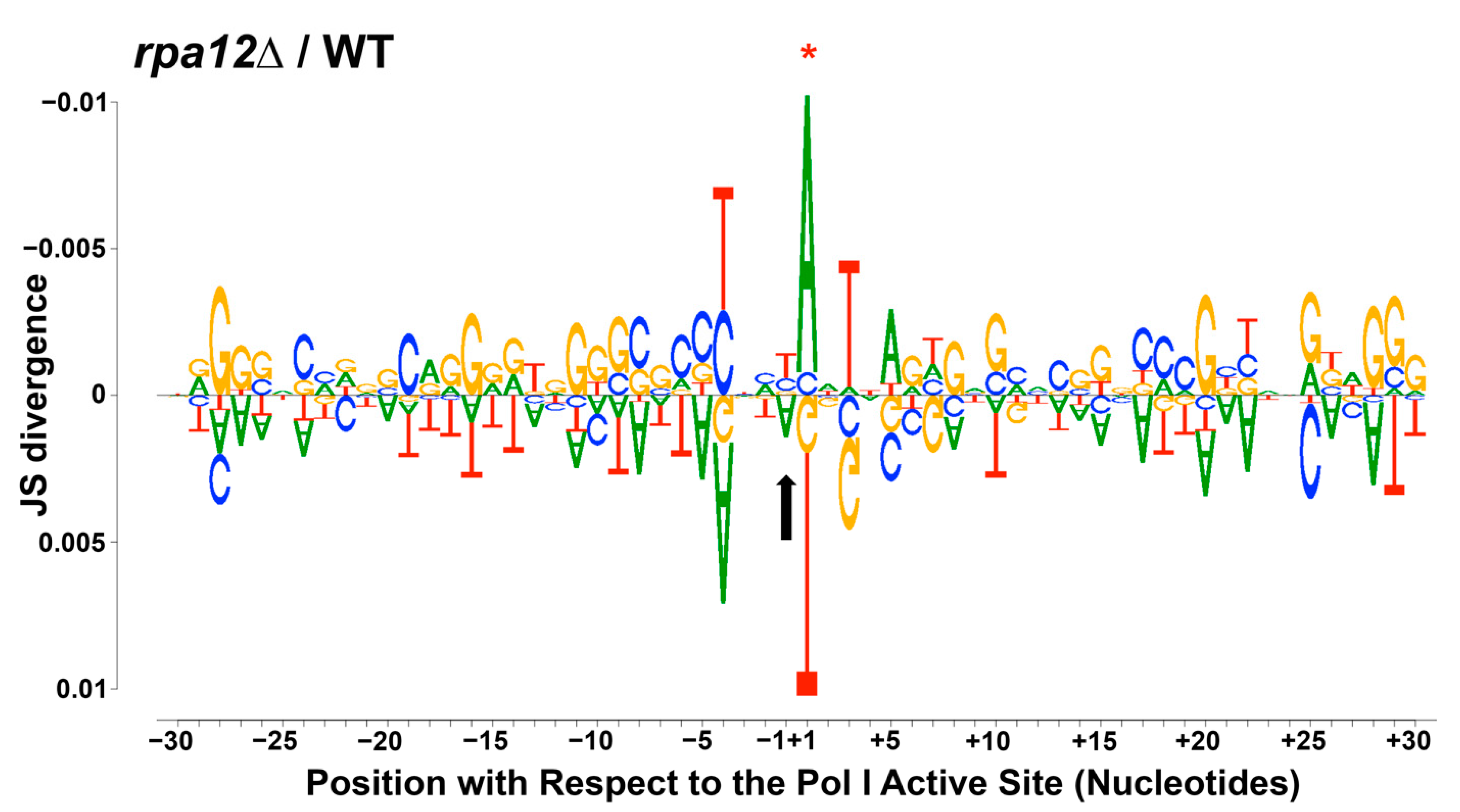
3.3. Changes in Pol I Occupancy Are Template Sequence Dependent
3.4. Deletion of RPA12 Results in Pol I Occupancy Downstream of T1 and T2
4. Discussion
4.1. Pol I Occupancy Patterns within the 35S Gene Change in Response to RPA12 Deletion
4.2. S. cerevisiae Contains a Putative Third Pol I Terminator Region
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Woolford, J.L.; Baserga, S.J. Ribosome Biogenesis in the Yeast Saccharomyces cerevisiae. Genetics 2013, 195, 643–681. [Google Scholar] [CrossRef] [Green Version]
- French, S.L.; Osheim, Y.N.; Cioci, F.; Nomura, M.; Beyer, A.L. In Exponentially Growing Saccharomyces cerevisiae Cells, rRNA Synthesis Is Determined by the Summed RNA Polymerase I Loading Rate Rather than by the Number of Active Genes. Mol. Cell. Biol. 2003, 23, 1558–1568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lang, W.H.; Reeder, R.H. The REB1 Site is an Essential Component of a Terminator for RNA Polymerase I in Saccharo-myces cerevisiae. Mol. Cell. Biol. 1993, 13, 649–658. [Google Scholar] [PubMed] [Green Version]
- Reeder, R.H.; Guevara, P.; Roan, J.G. Saccharomyces cerevisiae RNA Polymerase I Terminates Transcription at the Reb1 Terminator in Vivo. Mol. Cell. Biol. 1999, 19, 7369–7376. [Google Scholar] [CrossRef] [Green Version]
- Lang, W.H.; Reeder, R.H. Transcription Termination of RNA Polymerase I Due to a T-rich Element Interacting with Reb1p. Proc. Natl. Acad. Sci. USA 1995, 92, 9781–9785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lang, W.H.; Morrow, B.; Ju, Q.; Warner, J.R.; Reeder, R.H. A model for transcription termination by RNA polymerase I. Cell 1994, 79, 527–534. [Google Scholar] [CrossRef]
- Kuhn, A.; Bartsch, I.; Grummt, I. Specific interaction of the murine transcription termination factor TTF I with class-I RNA polymerases. Nature 1990, 344, 559–562. [Google Scholar] [CrossRef] [PubMed]
- Evers, R.; Smid, A.; Rudloff, U.; Lottspeich, F.; Grummt, I. Different Domains of the Murine RNA Polymerase I-Specific Termination Factor mTTF-I Serve Distinct Functions in Transcription Termination. EMBO J. 1995, 14, 1248–1256. [Google Scholar] [CrossRef] [PubMed]
- Kawauchi, J.; Mischo, H.; Braglia, P.; Rondon, A.; Proudfoot, N.J. Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes Dev. 2008, 22, 1082–1092. [Google Scholar] [CrossRef] [Green Version]
- Reiter, A.; Hamperl, S.; Seitz, H.; Merkl, P.; Perez-Fernandez, J.; Williams, L.; Gerber, J.; Nemeth, A.; Léger, I.; Gadal, O.; et al. The Reb1-homologue Ydr026c/Nsi1 is Required for Efficient RNA Polymerase I Termina-tion in Yeast. EMBO J. 2012, 31, 3480–3493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Hage, A.; Koper, M.; Kufel, J.; Tollervey, D. Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes Dev. 2008, 22, 1069–1081. [Google Scholar] [CrossRef] [Green Version]
- Raisner, R.M.; Hartley, P.D.; Meneghini, M.D.; Bao, M.Z.; Liu, C.L.; Schreiber, S.L.; Rando, O.J.; Madhani, H.D. Histone Variant H2A.Z Marks the 5′ Ends of Both Active and Inactive Genes in Euchromatin. Cell 2005, 123, 233–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartley, P.D.; Madhani, H.D. Mechanisms that Specify Promoter NucleosomeLocationand Identity. Cell 2009, 137, 445–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colin, J.; Candelli, T.; Porrua, O.; Boulay, J.; Zhu, C.; Lacroute, F.; Steinmetz, L.M.; Libri, D. Libri Roadblock Termination by Reb1p Restricts Cryptic and Readthrough Transcription. Mol. Cell 2014, 56, 667–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kufel, J.; Dichtl, B.; Tollervey, D. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA 1999, 5, 909–917. [Google Scholar] [CrossRef] [Green Version]
- Stevens, A.; Poole, T.L. 5′Exonuclease-2 of Saccharomyces cerevisiae Purification and Features of Ribonuclease Activity with Comparison to 5′-Exonuclease-1. J. Biol. Chem. 1995, 270, 16063–16069. [Google Scholar] [CrossRef] [Green Version]
- Van der Sande, C.; Kulkens, T.; Kramer, A.; De Wijs, I.; Van Heerikhuizen, H.; Klootwijk, J.; Planta, R. Termina-tion of Transcription by Yeast RNA Polymerase I. Nucleic Acids Res. 1989, 17, 9127–9146. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, C.-D.; Geiger, S.R.; Baumli, S.; Gartmann, M.; Gerber, J.; Jennebach, S.; Mielke, T.; Tschochner, H.; Beckmann, R.; Cramer, P. Functional Architecture of RNA Polymerase I. Cell 2007, 131, 1260–1272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prescott, E.M.; Osheim, Y.N.; Jones, H.S.; Alen, C.M.; Roan, J.G.; Reeder, R.H.; Beyer, A.L.; Proudfoot, N.J. Transcriptional Termination by RNA Polymerase I Requires the Small Subunit Rpa12P. Proc. Natl. Acad. Sci. USA 2004, 101, 6068–6073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mullem, V.V.; Landrieux, E.; Vandenhaute, J.; Thuriaux, P. Rpa12p, a ConservedRNA Polymerase I Subunit with Two Functional Domains. Mol. Microbiol. 2002, 43, 1105–1113. [Google Scholar] [CrossRef]
- Nogi, Y.; Yano, R.; Dodd, J.; Carles, C.; Nomura, M. Gene RRN4 in Saccharomyces cerevisiae Encodes the A12.2 Subunit of RNA Polymerase I and Is Essential Only at High Tempuratures. Mol. Cell Biol. 1993, 13, 114–122. [Google Scholar]
- Jaiswal, R.; Choudhury, M.; Zaman, S.; Singh, S.; Santosh, V.; Bastia, D.; Escalante, C.R. Functional architecture of the Reb1-Ter complex of Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 2016, 113, E2267–E2276. [Google Scholar] [CrossRef] [Green Version]
- Zhao, A.; Guo, A.; Liu, Z.; Pape, L. Molecular Cloning and Analysis of Schizosaccharomyces pombe Reb1p: Se-quence-specific Recognition of Two Sites in the Far Upstream rDNA Intergenic Spacer. Nucleic Acids Res. 1997, 25, 904–910. [Google Scholar] [CrossRef] [Green Version]
- Appling, F.D.; Scull, C.E.; Lucius, A.L.; Schneider, D.A. The A12.2 Subunit Is an Intrinsic Destabilizer of the RNA Polymerase I Elongation Complex. Biophys. J. 2018, 114, 2507–2515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Appling, F.D.; Schneider, D.A.; Lucius, A.L. Multisubunit RNA Polymerase Cleavage Factors Modulate the Kinetics and Energetics of Nucleotide Incorporation: An RNA Polymerase I Case Study. Biochemistry 2017, 56, 5654–5662. [Google Scholar] [CrossRef]
- Scull, C.E.; Lucius, A.L.; Schneider, D.A. The N-terminal domain of the A12.2 subunit stimulates RNA polymerase I transcription elongation. Biophys. J. 2021, 120, 1883–1893. [Google Scholar] [CrossRef] [PubMed]
- Clarke, A.M.; Engel, K.L.; Giles, K.E.; Petit, C.M.; Schneider, D.A. NETSeq reveals heterogeneous nucleotide incorporation by RNA polymerase I. Proc. Natl. Acad. Sci. USA 2018, 115, E11633–E11641. [Google Scholar] [CrossRef] [Green Version]
- Huffines, A.; Edwards, Y.; Schneider, D. Spt4 Promotes Pol I Processivity and Transcription Elongation. Genes 2021, 12, 413. [Google Scholar] [CrossRef] [PubMed]
- Scull, C.E.; Clarke, A.M.; Lucius, A.L.; Schneider, D.A. Downstream sequence-dependent RNA cleavage and pausing by RNA polymerase I. J. Biol. Chem. 2020, 295, 1288–1299. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Wickham, H.; Francois, R.; Henry, L.; Muller, K. dplyr: A Grammar of Data Manipulation. 2018. Available online: https://CRAN.R-project.org/package=dplyr (accessed on 10 November 2021).
- Wickham, H. The Split-Apply-Combine Strategy for Data Analysis. J. Stat. Softw. 2011, 40, 1–29. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Wagih, O. ggseqlogo: A ‘ggplot2′ Extension for Drawing Publication-Ready Sequence Logos. 2017. Available online: https://rdrr.io/cran/ggseqlogo/ (accessed on 10 November 2021).
- Kassambara, A. ggpubr: ‘ggplot2′ Based Publication Ready Plots. 2018. Available online: https://rpkgs.datanovia.com/ggpubr/ (accessed on 10 November 2021).
- Wilke, C.O. cowplot: Streamlined Plot Theme and Plot Annotations for ‘ggplot2’. 2017. Available online: https://rdrr.io/cran/cowplot/ (accessed on 10 November 2021).
- Bengtsson, H. matrixStats: Functions that Apply to Rows and Columns of Matrices (and to Vectors). 2018. Available online: https://github.com/HenrikBengtsson/matrixStats (accessed on 10 November 2021).
- Wickham, H. Scales: Scale Functions for Visualization. 2017. Available online: https://cran.r-project.org/web/packages/scales/index.html (accessed on 10 November 2021).
- Nettling, M.; Treutler, H.; Grau, J.; Keilwagen, J.; Posch, S.; Grosse, I. DiffLogo: A comparative visualization of sequence motifs. BMC Bioinform. 2015, 16, 387. [Google Scholar] [CrossRef]
- Churchman, L.S.; Weissman, J.S. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 2011, 469, 368–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sigurdsson, S.; Dirac-Svejstrup, A.B.; Svejstrup, J.Q. Evidence that Transcript Cleavage Is Essential for RNA Polymerase II Transcription and Cell Viability. Mol. Cell 2010, 38, 202–210. [Google Scholar] [CrossRef]
- Scull, C.E.; Ingram, Z.M.; Lucius, A.L.; Schneider, D.A. A Novel Assay for RNA Polymerase I Transcription Elongation Sheds Light on the Evolutionary Divergence of Eukaryotic RNA Polymerases. Biochemistry 2019, 58, 2116–2124. [Google Scholar] [CrossRef]
- Saeki, H.; Svejstrup, J.Q. Stability, Flexibility, and Dynamic Interactions of Colliding RNA Polymerase II Elongation Complexes. Mol. Cell 2009, 35, 191–205. [Google Scholar] [CrossRef]
- Németh, A.; Perez-Fernandez, J.; Merkl, P.; Hamperl, S.; Gerber, J.; Griesenbeck, J.; Tschochner, H. RNA Polymerase I Termi-nation: Where is the End? Biochim. Biophys. Acta Gene Regul. Mech. 2013, 1829, 306–317. [Google Scholar] [CrossRef]
- McStay, B.; Reeder, R.H. A Termination Site for Xenous RNA Polymerase I Also Acts as an Element of an Adjacent Promoter. Cell 1986, 47, 913–920. [Google Scholar] [CrossRef]
- Grummt, I.; Rosenbauer, H.; Niedermeyer, I.; Maier, U.; Öhrlein, A. A repeated 18 bp sequence motif in the mouse rDNA spacer mediates binding of a nuclear factor and transcription termination. Cell 1986, 45, 837–846. [Google Scholar] [CrossRef]
- Turowski, T.W.; Petfalski, E.; Goddard, B.D.; French, S.L.; Helwak, A.; Tollervey, D. Nascent Transcript Folding Plays a Major Role in Determining RNA Polymerase Elongation Rates. Mol. Cell 2020, 79, 488–503. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clarke, A.M.; Huffines, A.K.; Edwards, Y.J.K.; Petit, C.M.; Schneider, D.A. Defining the Influence of the A12.2 Subunit on Transcription Elongation and Termination by RNA Polymerase I In Vivo. Genes 2021, 12, 1939. https://doi.org/10.3390/genes12121939
Clarke AM, Huffines AK, Edwards YJK, Petit CM, Schneider DA. Defining the Influence of the A12.2 Subunit on Transcription Elongation and Termination by RNA Polymerase I In Vivo. Genes. 2021; 12(12):1939. https://doi.org/10.3390/genes12121939
Chicago/Turabian StyleClarke, Andrew M., Abigail K. Huffines, Yvonne J. K. Edwards, Chad M. Petit, and David A. Schneider. 2021. "Defining the Influence of the A12.2 Subunit on Transcription Elongation and Termination by RNA Polymerase I In Vivo" Genes 12, no. 12: 1939. https://doi.org/10.3390/genes12121939
APA StyleClarke, A. M., Huffines, A. K., Edwards, Y. J. K., Petit, C. M., & Schneider, D. A. (2021). Defining the Influence of the A12.2 Subunit on Transcription Elongation and Termination by RNA Polymerase I In Vivo. Genes, 12(12), 1939. https://doi.org/10.3390/genes12121939





