Is miRNA Regulation the Key to Controlling Non-Melanoma Skin Cancer Evolution?
Abstract
:1. Introduction
2. Materials and Methods
3. miRNA Function in Oncogenesis, Evolution and Therapy
3.1. The Role of miRNAs in Basal Cell Carcinoma (BCC)
3.2. The Implication of miRNAs in Squamous Cell Carcinoma (SCC)
3.3. Modified miRNAs in Merkel Cell Carcinoma (MCC)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kashyap, M.P.; Sinha, R.; Mukhtar, M.S.; Athar, M. Epigenetic regulation in the pathogenesis of non-melanoma skin cancer. Semin. Cancer Biol. 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Di Meglio, P.; Perera, G.K.; Nestle, F.O. The multitasking organ: Recent insights into skin immune function. Immunity 2011, 23, 857–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrawal, R.; Woodfolk, J.A. Skin barrier defects in atopic dermatitis. Curr. Allergy Asthma Rep. 2014, 14, 433. [Google Scholar] [CrossRef]
- Li, C.; Athar, M. Ionizing Radiation Exposure and Basal Cell Carcinoma Pathogenesis. Radiat. Res. 2016, 185, 217–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Errico, M.; Calcagnile, A.; Canzona, F.; Didona, B.; Posteraro, P.; Cavalieri, R.; Corona, R.; Vorechovsky, I.; Nardo, T.; Stefanini, M.; et al. UV mutation signature in tumor suppressor genes involved in skin carcinogenesis in xeroderma pigmentosum patients. Oncogene 2000, 19, 463–467. [Google Scholar] [CrossRef] [Green Version]
- Moodycliffe, A.M.; Nghiem, D.; Clydesdale, G.; Ullrich, S.E. Immune suppression and skin cancer development: Regulation by NKT cells. Nat. Immunol. 2000, 1, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Gordon, R. Skin Cancer: An Overview of Epidemiology and Risk Factors. Semin. Oncol. Nurs. 2013, 29, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Glass, A.G.; Hoover, R.N. The emerging epidemic of melanoma and squamous cell skin cancer. JAMA 1989, 262, 2097–2100. [Google Scholar] [CrossRef]
- Dubas, L.E.; Ingraffea, A. Nonmelanoma skin cancer. Facial. Plast. Surg. Clin. N. Am. 2013, 21, 43–53. [Google Scholar] [CrossRef]
- Parekh, V.; Seykora, J.T. Cutaneous Squamous Cell Carcinoma. Clin. Lab. Med. 2017, 37, 503–525. [Google Scholar] [CrossRef]
- Green, A.C.; Olsen, C.M. Cutaneous squamous cell carcinoma: An epidemiological review. Br. J. Dermatol. 2017, 177, 373–381. [Google Scholar] [CrossRef]
- Sand, M.; Skrygan, M.; Sand, D.; Georgas, D.; Hahn, S.A.; Gambichler, T.; Altmeyer, P.; Bechara, F.G. Expression of microRNAs in basal cell carcinoma. Br. J. Dermatol. 2012, 167, 847–855. [Google Scholar] [CrossRef]
- Sand, M.; Sand, D.; Altmeyer, P.; Bechara, F.G. MicroRNA in non-melanoma skin cancer. Cancer Biomark. 2012, 11, 253–257. [Google Scholar] [CrossRef]
- Sand, M.; Gambichler, T.; Sand, D.; Skrygan, M.; Altmeyer, P.; Bechara, F.G. MicroRNAs and the skin: Tiny players in the body’s largest organ. J. Dermatol. Sci. 2009, 53, 169–175. [Google Scholar] [CrossRef]
- Tian, J.; Shen, R.; Yan, Y.; Deng, L. miR-186 promotes tumor growth in cutaneous squamous cell carcinoma by inhibiting apoptotic protease activating factor-1. Exp. Ther. Med. 2018, 16, 4010–4018. [Google Scholar] [CrossRef]
- Han, C.; Seebacher, N.A.; Hornicek, F.J.; Kan, Q.; Duan, Z. Regulation of microRNAs function by circular RNAs in human cancer. Oncotarget 2017, 8, 64622–64637. [Google Scholar] [CrossRef] [Green Version]
- Self-Fordham, J.B.; Naqvi, A.R.; Uttamani, J.R.; Kulkarni, V.; Nares, S. MicroRNA: Dynamic Regulators of Macrophage Polarization and Plasticity. Front. Immunol. 2017, 8, 1062. [Google Scholar] [CrossRef] [Green Version]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Broughton, J.P.; Lovci, M.T.; Huang, J.L.; Yeo, G.W.; Pasquinelli, A.E. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol. Cell 2016, 64, 320–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasudevan, S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip. Rev. RNA 2012, 3, 311–330. [Google Scholar] [CrossRef] [PubMed]
- Makarova, J.A.; Shkurnikov, M.U.; Wicklein, D.; Lange, T.; Samatov, T.R.; Turchinovich, A.A.; Tonevitsky, A.G. Intracellular and extracellular microRNA: An update on localization and biological role. Prog. Histochem. Cytochem. 2016, 51, 33–49. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 3, 402. [Google Scholar] [CrossRef] [Green Version]
- Creemers, E.E.; Tijsen, A.J.; Pinto, Y.M. Circulating microRNAs: Novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012, 110, 483–495. [Google Scholar] [CrossRef]
- Nikolouzakis, T.K.; Falzone, L.; Lasithiotakis, K.; Krüger-Krasagakis, S.; Kalogeraki, A.; Sifaki, M.; Spandidos, D.A.; Chrysos, E.; Tsatsakis, A.; Tsiaoussis, J. Current and Future Trends in Molecular Biomarkers for Diagnostic, Prognostic, and Predictive Purposes in Non-Melanoma Skin Cancer. J. Clin. Med. 2020, 9, 2868. [Google Scholar] [CrossRef]
- Rodriguez, A.; Griffiths-Jones, S.; Ashurst, J.L.; Bradley, A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004, 14, 1902–1910. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Jeon, K.; Lee, J.T.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef] [Green Version]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Horsham, J.L.; Ganda, C.; Kalinowski, F.C.; Brown, R.A.; Epis, M.R.; Leedman, P.J. MicroRNA-7: A miRNA with expanding roles in development and disease. Int. J. Biochem. Cell Biol. 2015, 69, 215–224. [Google Scholar] [CrossRef]
- Gerloff, D.; Sunderkötter, C.; Wohlrab, J. Importance of microRNAs in Skin Oncogenesis and Their Suitability as Agents and Targets for Topical Therapy. Skin Pharmacol. Physiol. 2020, 33, 270–279. [Google Scholar] [CrossRef]
- Neagu, M.; Constantin, C.; Cretoiu, S.M.; Zurac, S. miRNAs in the Diagnosis and Prognosis of Skin Cancer. Front. Cell Dev. Biol. 2020, 8, 71. [Google Scholar] [CrossRef] [Green Version]
- Walling, H.W.; Fosko, S.W.; Geraminejad, P.A.; Whitaker, D.C.; Arpey, C.J. Aggressive basal cell carcinoma: Presentation, pathogenesis, and management. Cancer Metastasis Rev. 2004, 23, 389–402. [Google Scholar] [CrossRef]
- Heffelfinger, C.; Ouyang, Z.; Engberg, A.; Leffell, D.J.; Hanlon, A.M.; Gordon, P.B.; Zheng, W.; Zhao, H.; Snyder, M.P.; Bale, A.E. Correlation of Global MicroRNA Expression with Basal Cell Carcinoma Subtype. G3 (Bethesda) 2012, 2, 279–286. [Google Scholar] [CrossRef] [Green Version]
- Sonkoly, E.; Lov, J.; Xu, N.; Meisgen, F.; Wei, T.; Brodin, P.; Jacks, V.; Kasper, M.; Shimokawa, T.; Harada, M.; et al. MicroRNA-203 functions as a tumor suppressor in basal cell carcinoma. Oncogenesis 2012, 1, e3. [Google Scholar] [CrossRef] [Green Version]
- Sand, M.; Bechara, F.G.; Gambichler, T.; Sand, D.; Friedländer, M.R.; Bromba, M.; Schnabel, R.; Hessam, S. Next-generation sequencing of the basal cell carcinoma miRNome and a description of novel microRNA candidates under neoadjuvant vismodegib therapy: An integrative molecular and surgical case study. Ann. Oncol. 2016, 27, 332–338. [Google Scholar] [CrossRef]
- Al-Eryani, L.; Jenkins, S.F.; States, V.A.; Pan, J.; Malone, J.C.; Rai, S.N.; Galandiuk, S.; Giri, A.K.; States, J.C. miRNA expression profiles of premalignant and malignant arsenic-induced skin lesions. PLoS ONE 2018, 13, e0202579. [Google Scholar]
- Hu, P.; Ma, L.; Wu, Z.; Zheng, G.; Li, J. Expression of miR-34a in basal cell carcinoma patients and its relationship with prognosis. J. Buon. 2019, 24, 1283–1288. [Google Scholar]
- Sun, H.; Jiang, P. MicroRNA-451a acts as tumor suppressor in cutaneous basal cell carcinoma. Mol. Genet. Genom. Med. 2018, 6, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Tran, D.C.; Zhu, G.A.; Li, R.; Whitson, R.; Kim, Y.H.; Gupta, A.; Afshari, A.; Antes, T.; Spitale, R.C.; et al. Initial in vitro functional characterization of serum exosomal microRNAs from patients with metastatic basal cell carcinoma. Br. J. Dermatol. 2017, 177, e187–e190. [Google Scholar] [CrossRef] [PubMed]
- Wan, C.; Li, Y. Integrative analysis of mRNA-miRNA-TFs reveals the key regulatory connections involved in basal cell carcinoma. Arch. Dermatol. Res. 2020, 312, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Farzan, S.F.; Karagas, M.R.; Christensen, B.C.; Li, Z.; Kuriger, J.K.; Nelson, H.H. New Hampshire Skin Cancer Study. RNASEL and MIR146A SNP-SNP interaction as a susceptibility factor for non-melanoma skin cancer. PLoS ONE 2014, 9, e93602. [Google Scholar]
- Alam, M.; Ratner, D. Cutaneous squamous-cell carcinoma. N. Engl. J. Med. 2001, 344, 975–983. [Google Scholar] [CrossRef]
- Inman, G.J.; Wang, J.; Nagano, A.; Alexandrov, L.B.; Purdie, K.J.; Taylor, R.G.; Sherwood, V.; Thomson, J.; Hogan, S.; Spender, L.C.; et al. The genomic landscape of cutaneous SCC reveals drivers and a novel azathioprine associated mutational signature. Nat. Commun. 2018, 9, 3667. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, B.; Wen, X.; Hao, D.; Du, D.; He, G.; Jiang, X. The Roles of lncRNA in Cutaneous Squamous Cell Carcinoma. Front. Oncol. 2020, 10, 158. [Google Scholar] [CrossRef] [Green Version]
- Konicke, K.; López-Luna, A.; Muñoz-Carrillo, J.L.; Servín-González, L.S.; Flores-de la Torre, A.; Olasz, E.; Lazarova, Z. The microRNA landscape of cutaneous squamous cell carcinoma. Drug Discov. Today 2018, 23, 864–870. [Google Scholar] [CrossRef]
- Sand, M.; Gambichler, T.; Skrygan, M.; Sand, D.; Scola, N.; Altmeyer, P.; Bechara, F.G. Expression levels of the microRNA processing enzymes Drosha and dicer in epithelial skin cancer. Cancer Investig. 2010, 28, 649–653. [Google Scholar] [CrossRef]
- Sand, M.; Skrygan, M.; Georgas, D.; Arenz, C.; Gambichler, T.; Sand, D.; Altmeyer, P.; Bechara, F.G. Expression levels of the microRNA maturing microprocessor complex component DGCR8 and the RNA-induced silencing complex (RISC) components argonaute-1, argonaute-2, PACT, TARBP1, and TARBP2 in epithelial skin cancer. Mol. Carcinog. 2012, 51, 916–922. [Google Scholar] [CrossRef]
- Sand, M.; Skrygan, M.; Georgas, D.; Sand, D.; Hahn, S.A.; Gambichler, T.; Altmeyer, P.; Bechara, F.G. Microarray analysis of microRNA expression in cutaneous squamous cell carcinoma. J. Dermatol. Sci. 2012, 68, 119–126. [Google Scholar] [CrossRef]
- Dziunycz, P.; Iotzova-Weiss, G.; Eloranta, J.J.; Lauchli, S.; Hafner, J.; French, L.E.; Altmeyer, P.; Bechara, F.G. Squamous cell carcinoma of the skin shows a distinct microRNA profile modulated by UV radiation. J. Investig. Dermatol. 2010, 130, 2686–2689. [Google Scholar] [CrossRef] [Green Version]
- Yamane, K.; Jinnin, M.; Etoh, T.; Kobayashi, Y.; Shimozono, N.; Fukushima, S.; Masuguchi, S.; Maruo, K.; Inoue, Y.; Ishihara, T.; et al. Down-regulation of miR-124/-214 in cutaneous squamous cell carcinoma mediates abnormal cell proliferation via the induction of ERK. J. Mol. Med. 2013, 91, 69–81. [Google Scholar] [CrossRef]
- Cañueto, J.; Cardeñoso-Álvarez, E.; García-Hernández, J.L.; Galindo-Villardón, P.; Vicente-Galindo, P.; Vicente-Villardón, J.L.; Alonso-López, D.; De Las Rivas, J.; Valero, J.; Moyano-Sanz, E.; et al. MicroRNA (miR)-203 and miR-205 expression patterns identify subgroups of prognosis in cutaneous squamous cell carcinoma. Br. J. Dermatol. 2017, 177, 168–178. [Google Scholar] [CrossRef] [Green Version]
- Stojadinovic, O.; Ramirez, H.; Pastar, I.; Gordon, K.A.; Stone, R.; Choudhary, S.; Badiavas, E.; Nouri, K.; Tomic-Canic, M. MiR-21 and miR-205 are induced in invasive cutaneous squamous cell carcinomas. Arch. Dermatol. Res. 2017, 309, 133–139. [Google Scholar] [CrossRef]
- Zhang, L.; Xiang, P.; Han, X.; Wu, L.; Li, X.; Xiong, Z. Decreased expression of microRNA-20a promotes tumor progression and predicts poor prognosis of cutaneous squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 11446–11451. [Google Scholar]
- Gong, Z.H.; Zhou, F.; Shi, C.; Xiang, T.; Zhou, C.K.; Wang, Q.Q.; Jiang, Y.S.; Gao, S.F. miRNA-221 promotes cutaneous squamous cell carcinoma progression by targeting PTEN. Cell Mol. Biol. Lett. 2019, 24, 9. [Google Scholar] [CrossRef]
- Gal-Yam, E.N.; Saito, Y.; Egger, G.; Jones, P.A. Cancer epigenetics: Modifications, screening, and therapy. Annu. Rev. Med. 2008, 59, 267–280. [Google Scholar] [PubMed]
- Lodygin, D.; Tarasov, V.; Epanchintsev, A.; Berking, C.; Knyazeva, T.; Körner, H.; Knyazev, P.; Diebold, J.; Hermeking, H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 2008, 7, 2591–2600. [Google Scholar] [CrossRef] [Green Version]
- Vousden, K.H.; Lu, X. Live or let die: The cell’s response to p53. Nat. Rev. Cancer 2002, 2, 594–604. [Google Scholar] [CrossRef] [Green Version]
- Rivlin, N.; Brosh, R.; Oren, M.; Rotter, V. Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes Cancer 2011, 2, 466–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodust, P.M.; Stockfleth, E.; Ulrich, C.; Leverkus, M.; Eberle, J. UV-induced squamous cell carcinoma—A role for antiapoptotic signalling pathways. Br. J. Dermatol. 2009, 161 (Suppl. 3), 107–115. [Google Scholar] [CrossRef] [PubMed]
- Lefort, K.; Brooks, Y.; Ostano, P.; Cario-André, M.; Calpini, V.; Guinea-Viniegra, J.; Albinger-Hegyi, A.; Hoetzenecker, W.; Kolfschoten, I.; Wagner, E.F.; et al. A miR-34a-SIRT6 axis in the squamous cell differentiation network. EMBO J. 2013, 32, 2248–2263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanitz, A.; Imig, J.; Dziunycz, P.J.; Primorac, A.; Galgano, A.; Hofbauer, G.F.L.; Gerber, A.P.; Detmar, M. The expression levels of microRNA-361-5p and its target VEGFA are inversely correlated in human cutaneous squamous cell carcinoma. PLoS ONE 2012, 7, e49568. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Pan, W.; Lin, X.; Hu, Z.; Jin, Y.; Chen, H.; Ma, G.; Qiu, Y.; Chang, L.; Hua, C.; et al. MicroRNA-346 functions as an oncogene in cutaneous squamous cell carcinoma. Tumour Biol. 2016, 37, 2765–2771. [Google Scholar] [CrossRef]
- Xu, N.; Zhang, L.; Meisgen, F.; Harada, M.; Heilborn, J.; Homey, B.; Grandér, D.; Ståhle, M.; Sonkoly, E.; Pivarcsi, A. MicroRNA-125b down-regulates matrix metallopeptidase 13 and inhibits cutaneous squamous cell carcinoma cell proliferation, migration, and invasion. J. Biol. Chem. 2012, 287, 29899–29908. [Google Scholar] [CrossRef] [Green Version]
- Olasz, E.B.; Seline, L.N.; Schock, A.M.; Duncan, N.E.; Lopez, A.; Lazar, J.; Flister, M.J.; Lu, Y.; Liu, P.; Sokumbi, O.; et al. MicroRNA-135b Regulates Leucine Zipper Tumor Suppressor 1 in Cutaneous Squamous Cell Carcinoma. PLoS ONE 2015, 10, e0125412. [Google Scholar] [CrossRef] [Green Version]
- Zhou, M.; Liu, W.; Ma, S.; Cao, H.; Peng, X.; Guo, L.; Zhou, X.; Zheng, L.; Guo, L.; Wan, M.; et al. A novel onco-miR-365 induces cutaneous squamous cell carcinoma. Carcinogenesis 2013, 34, 1653–1659. [Google Scholar] [CrossRef] [Green Version]
- Calder, K.B.; Smoller, B.R. New insights into merkel cell carcinoma. Adv. Anat. Pathol. 2010, 17, 155–161. [Google Scholar] [CrossRef]
- Albores-Saavedra, J.; Batich, K.; Chable-Montero, F.; Sagy, N.; Schwartz, A.M.; Henson, D.E. Merkel cell carcinoma demographics, morphology, and survival based on 3870 cases: A population based study. J. Cutan. Pathol. 2010, 37, 20–27. [Google Scholar] [CrossRef]
- Hughes, M.P.; Hardee, M.E.; Cornelius, L.A.; Hutchins, L.F.; Becker, J.C.; Gao, L. Merkel Cell Carcinoma: Epidemiology, Target, and Therapy. Curr. Dermatol. Rep. 2014, 3, 46–53. [Google Scholar] [CrossRef] [Green Version]
- Feng, H.; Shuda, M.; Chang, Y.; Moore, P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 2008, 319, 1096–1100. [Google Scholar] [CrossRef] [Green Version]
- Ning, M.S.; Kim, A.S.; Prasad, N.; Levy, S.E.; Zhang, H.; Andl, T. Characterization of the Merkel Cell Carcinoma miRNome. J. Skin Cancer 2014, 2014, 289548. [Google Scholar] [CrossRef] [Green Version]
- Renwick, N.; Cekan, P.; Masry, P.A.; McGeary, S.E.; Miller, J.B.; Hafner, M.; Li, Z.; Mihailovic, A.; Morozov, P.; Brown, M.; et al. Multicolor microRNA FISH effectively differentiates tumor types. J. Clin. Investig. 2013, 123, 2694–2702. [Google Scholar] [CrossRef]
- Fan, K.; Gravemeyer, J.; Ritter, C.; Rasheed, K.; Gambichler, T.; Moens, U.; Shuda, M.; Schrama, D.; Becker, J.C. MCPyV Large T Antigen-Induced Atonal Homolog 1 Is a Lineage-Dependency Oncogene in Merkel Cell Carcinoma. J. Investig. Dermatol. 2020, 140, 56–65.e3. [Google Scholar] [CrossRef] [Green Version]
- Veija, T.; Sahi, H.; Koljonen, V.; Bohling, T.; Knuutila, S.; Mosakhani, N. miRNA-34a underexpressed in Merkel cell polyomavirus-negative Merkel cell carcinoma. Virchows Arch. 2015, 466, 289–295. [Google Scholar] [CrossRef]
- Konstatinell, A.; Coucheron, D.H.; Sveinbjørnsson, B.; Moens, U. MicroRNAs as Potential Biomarkers in Merkel Cell Carcinoma. Int. J. Mol. Sci. 2018, 19, 1873. [Google Scholar] [CrossRef] [Green Version]
- Tuaeva, N.O.; Falzone, L.; Porozov, Y.B.; Nosyrev, A.E.; Trukhan, V.M.; Kovatsi, L.; Spandidos, D.A.; Drakoulis, N.; Kalogeraki, A.; Mamoulakis, C.; et al. Translational Application of Circulating DNA in Oncology: Review of the Last Decades Achievements. Cells 2019, 8, 1251. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Zhang, F.; Li, X.; Liu, Q.; Liu, W.; Song, P.; Qiu, Z.; Dong, Y.; Xiang, H. Prognostic role of miR-17-92 family in human cancers: Evaluation of multiple prognostic outcomes. Oncotarget 2017, 8, 69125–69138. [Google Scholar] [CrossRef] [Green Version]
- Bobbili, M.R.; Mader, R.M.; Grillari, J.; Dellago, H. OncomiR-17-5p: Alarm signal in cancer? Oncotarget 2017, 8, 71206–71222. [Google Scholar] [CrossRef] [Green Version]
- Volpicella, M.; Leoni, C.; Costanza, A.; Fanizza, I.; Placido, A.; Ceci, L.R. Genome walking by next generation sequencing approaches. Biology 2012, 1, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liang, Z.; Wang, S.; Lu, S.; Song, Y.; Cheng, Y.; Ying, J.; Liu, W.; Hou, W.; Liu, Y.; et al. Application of next-generation sequencing technology to precision medicine in cancer: Joint consensus of the Tumor Biomarker Committee of the Chinese Society of Clinical Oncology. Cancer Biol. Med. 2019, 16, 189–204. [Google Scholar] [PubMed] [Green Version]
- Martinez-Gutierrez, A.D.; Catalan, O.M.; Vázquez-Romo, R.; Porras Reyes, F.I.; Alvarado-Miranda, A.; Lara Medina, F.; Bargallo-Rocha, J.E.; Orozco Moreno, L.T.; De León, D.C.; Herrera, L.A.; et al. miRNA profile obtained by next generation sequencing in metastatic breast cancer patients is able to predict the response to systemic treatments. Int. J. Mol. Med. 2019, 44, 1267–1280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garofoli, M.; Volpicella, M.; Guida, M.; Porcelli, L.; Azzariti, A. The Role of Non-Coding RNAs as Prognostic Factor, Predictor of Drug Response or Resistance and Pharmacological Targets, in the Cutaneous Squamous Cell Carcinoma. Cancers 2020, 12, 2552. [Google Scholar] [CrossRef]
- Corsten, M.F.; Miranda, R.; Kasmieh, R.; Krichevsky, A.M.; Weissleder, R.; Shah, K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007, 67, 8994–9000. [Google Scholar] [CrossRef] [Green Version]
- Tivnan, A.; Tracey, L.; Buckley, P.G.; Alcock, L.C.; Davidoff, A.M.; Stallings, R.L. MicroRNA-34a is a potent tumor suppressor molecule in vivo in neuroblastoma. BMC Cancer 2011, 11, 33. [Google Scholar] [CrossRef] [Green Version]
- Babar, I.A.; Cheng, C.J.; Booth, C.J.; Liang, X.; Weidhaas, J.B.; Saltzman, W.M. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc. Natl. Acad. Sci. USA 2012, 109, E1695–E1704. [Google Scholar] [CrossRef] [Green Version]
- Skourti, E.; Logotheti, S.; Kontos, C.K.; Pavlopoulou, A.; Dimoragka, P.T.; Trougakos, I.P.; Gorgoulis, V.; Scorilas, A.; Michalopoulos, I.; Zoumpourlis, V. Progression of mouse skin carcinogenesis is associated with the orchestrated deregulation of mir-200 family members, mir-205 and their common targets. Mol. Carcinog. 2016, 55, 1229–1242. [Google Scholar] [CrossRef]
- Li, P.; He, Q.Y.; Luo, C.Q.; Qian, L.Y. Circulating miR-221 expression level and prognosis of cutaneous malignant melanoma. Med. Sci. Monit. 2014, 20, 2472–2477. [Google Scholar]
- Liu, K.; Jin, J.; Rong, K.; Zhuo, L.; Li, P. MicroRNA 675 inhibits cell proliferation and invasion in melanoma by directly targeting metadherin. Mol. Med. Rep. 2018, 17, 3372–3379. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Wang, Q.; Zhu, X.H. MiR-135b is a novel oncogenic factor in cutaneous melanoma by targeting LATS2. Melanoma Res. 2019, 29, 119–125. [Google Scholar] [CrossRef]
- D’Aguanno, S.; Valentini, E.; Tupone, M.G.; Desideri, M.; Di Martile, M.; Spagnuolo, M.; Buglioni, S.; Ercolani, C.; Falcone, I.; De Dominici, M.; et al. Semaphorin 5A drives melanoma progression: Role of Bcl-2, miR-204 and c-Myb. J. Exp. Clin. Cancer Res. 2018, 37, 278. [Google Scholar] [CrossRef]
- Sánchez-Sendra, B.; Martinez-Ciarpaglini, C.; González-Muñoz, J.F.; Murgui, A.; Terrádez, L.; Monteagudo, C. Downregulation of intratumoral expression of miR-205, miR-200c and miR-125b in primary human cutaneous melanomas predicts shorter survival. Sci. Rep. 2018, 8, 17076. [Google Scholar] [CrossRef]
- Sabarimurugan, S.; Madurantakam Royam, M.; Das, A.; Das, S.; Gothandam, K.M.; Jayaraj, R. Systematic Review and Meta-analysis of the Prognostic Significance of miRNAs in Melanoma Patients. Mol. Diagn. Ther. 2018, 22, 653–669. [Google Scholar] [CrossRef]
- Kozar, I.; Cesi, G.; Margue, C.; Philippidou, D.; Kreis, S. Impact of BRAF kinase inhibitors on the miRNomes and transcriptomes of melanoma cells. Biochim. Biophys. Acta Gen. Subj. 2017, 1861 Pt 11, 2980–2992. [Google Scholar] [CrossRef]
- Fattore, L.; Mancini, R.; Acunzo, M.; Romano, G.; Laganà, A.; Pisanu, M.E. miR-579-3p controls melanoma progression and resistance to target therapy. Proc. Natl. Acad. Sci. USA 2016, 113, E5005–E5013. [Google Scholar] [CrossRef] [Green Version]
- Caporali, S.; Amaro, A.; Levati, L.; Alvino, E.; Lacal, P.M.; Mastroeni, S.; Ruffini, F.; Bonmassar, L.; Antonini Cappellini, G.C.; Felli, N.; et al. miR-126-3p down-regulation contributes to dabrafenib acquired resistance in melanoma by up-regulating ADAM9 and VEGF-A. J. Exp. Clin. Cancer Res. 2019, 38, 272. [Google Scholar] [CrossRef]
- Li, N.; Liu, Y.; Pang, H.; Lee, D.; Zhou, Y.; Xiao, Z. Methylation-Mediated Silencing of MicroRNA-211 Decreases the Sensitivity of Melanoma Cells to Cisplatin. Med. Sci. Monit. 2019, 25, 1590–1599. [Google Scholar] [CrossRef]
- Prabhakar, K.; Rodrίguez, C.I.; Jayanthy, A.S.; Mikheil, D.M.; Bhasker, A.I.; Perera, R.J.; Setaluri, V. Role of miR-214 in regulation of β-catenin and the malignant phenotype of melanoma. Mol. Carcinog. 2019, 58, 1974–1984. [Google Scholar] [CrossRef]
- Huber, V.; Vallacchi, V.; Fleming, V.; Hu, X.; Cova, A.; Dugo, M. Tumor-derived microRNAs induce myeloid suppressor cells and predict immunotherapy resistance in melanoma. J. Clin. Investig. 2018, 128, 5505–5516. [Google Scholar] [CrossRef] [Green Version]
- de Planell-Saguer, M.; Rodicio, M.C. Analytical aspects of microRNA in diagnostics: A review. Anal. Chim. Acta. 2011, 699, 134–152. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, E.; Hershkovitz, L.; Itzhaki, O.; Hajdu, S.; Nemlich, Y.; Ortenberg, R.; Gefen, N.; Edry, L.; Modai, S.; Keisari, Y.; et al. Regulation of cancer aggressive features in melanoma cells by microRNAs. PLoS ONE 2011, 6, e18936. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, Z.W. Expression of miR-203 is decreased and associated with the prognosis of melanoma patients. Int. J. Clin. Exp. Pathol. 2015, 8, 13249–13254. [Google Scholar] [PubMed]
- Hunt, E.A.; Broyles, D.; Head, T.; Deo, S.K. MicroRNA Detection: Current Technology and Research Strategies. Annu. Rev. Anal. Chem. 2015, 8, 217–237. [Google Scholar] [CrossRef]
- Ouyang, T.; Liu, Z.; Han, Z.; Ge, Q. MicroRNA Detection Specificity: Recent Advances and Future Perspective. Anal. Chem. 2019, 91, 3179–3186. [Google Scholar] [CrossRef] [Green Version]
- Kappel, A.; Backes, C.; Huang, Y.; Zafari, S.; Leidinger, P.; Meder, B.; Schwarz, H.; Gumbrecht, W.; Meese, E.; Staehler, C.F.; et al. MicroRNA in vitro diagnostics using immunoassay analyzers. Clin. Chem. 2015, 61, 600–607. [Google Scholar] [CrossRef] [Green Version]
- Ozsolak, F.; Milos, P.M. RNA sequencing: Advances, challenges and opportunities. Nat. Rev. Genet. 2011, 12, 87–98. [Google Scholar] [CrossRef]
- Tanase, C.P.; Albulescu, R.; Neagu, M. Application of 3D hydrogel microarrays in molecular diagnostics: Advantages and limitations. Expert Rev. Mol. Diagn. 2011, 11, 461–464. [Google Scholar] [CrossRef]
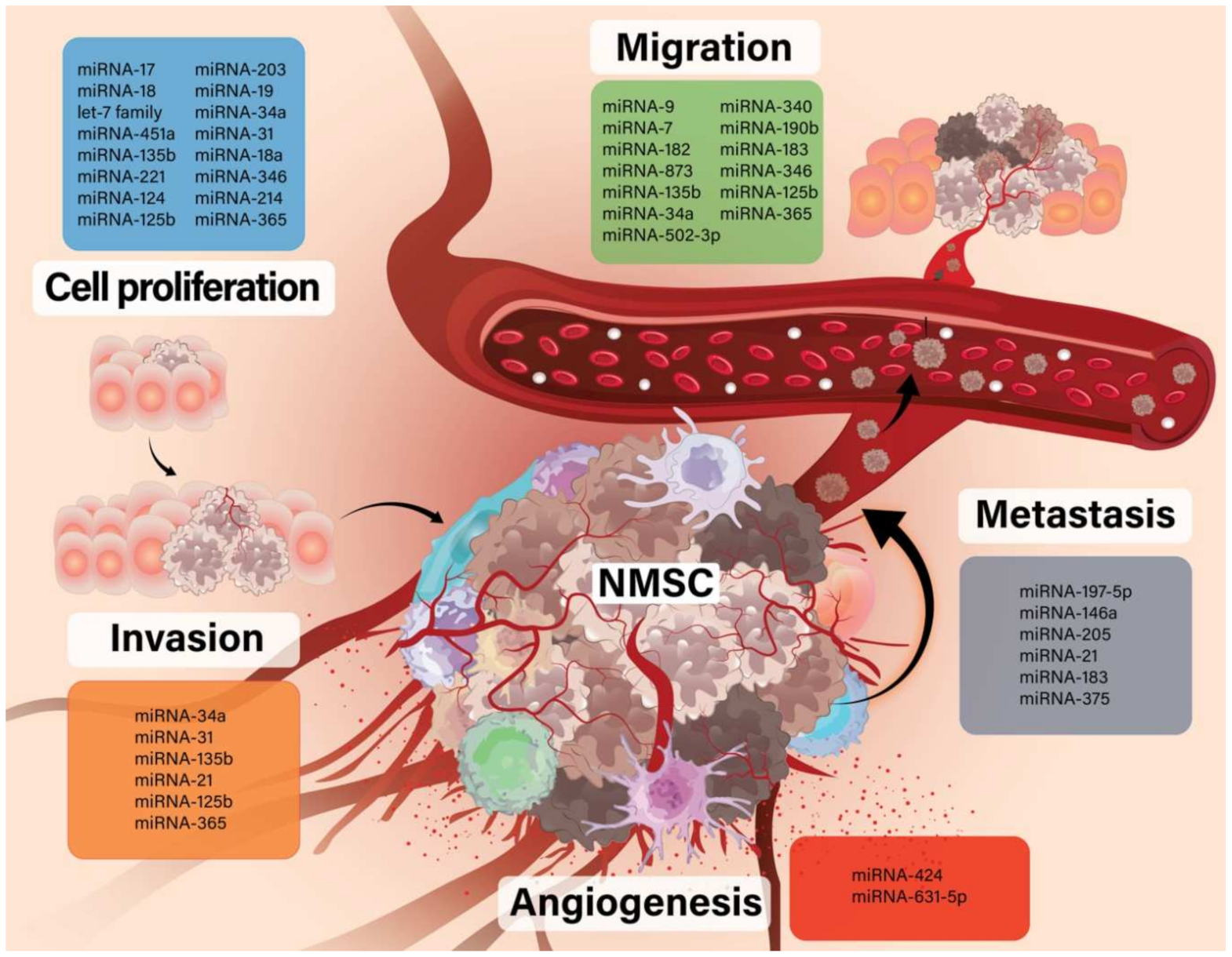
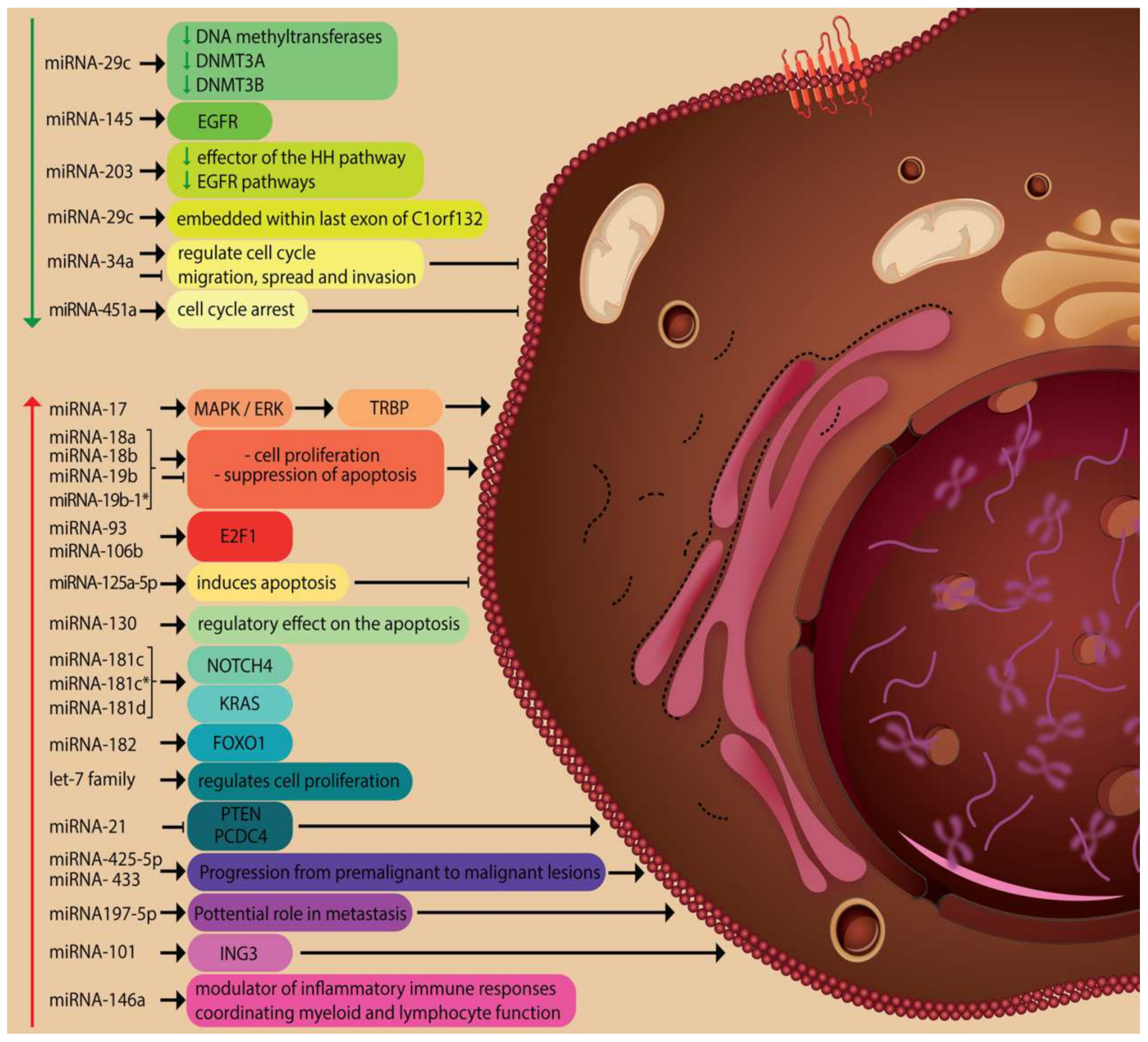
| Nr | Ref | Expression | miRNA | Probe | Species | Role |
|---|---|---|---|---|---|---|
| 1 | Sand M. et al. [13,14] | upregulated | miRNA-17 | tissue | human | pro-growth miRNA regulated in vitro by MAPK (mitogen-activated protein kinase)/ ERK-induced phosphorylation of TRBP (TAR-RNA binding protein) |
| miRNA-18a miRNA-18b | cell proliferation and the suppression of apoptosis | |||||
| miRNA-19b miRNA-19b-1* | responsible for enhanced cell pro- liferation and the suppression of apoptosis | |||||
| miRNA-93 | transcription factor E2F1 (E2 promoter binding factor 1) is a target gene of miRNA-93 | |||||
| miRNA-106b | transcription factor E2F1 is a target gene of miRNA-106b | |||||
| miRNA-125a-5p | induces apoptosis | |||||
| miRNA-130a | regulatory effect on the apoptosis | |||||
| miRNA-181c miRNA-181c* miRNA-181d | targets NOTCH4( neurogenic locus notch homolog-4) and KRAS (Kirsten rat sarcoma virus) | |||||
| miRNA-182 | negatively regulate human Forkheadbox O1 (FOXO1) | |||||
| miRNA-455-3p miRNA-455-5p miRNA-542-5p | not mentioned | |||||
| downregulated | miRNA-29c | downregulates DNA methyltransferases DNMT3A and DNMT3B | ||||
| miRNA-29c* miRNA-139-5p miRNA-140-3p | not mentioned | |||||
| miRNA-145 | targets EGFR | |||||
| miRNA-572 miRNA-638 miRNA-2861 miRNA-3196 | not mentioned | |||||
| 2 | Heffelfinger et al. [36] | upregulated | let-7 family | tissue | human | involved in regulating cell proliferation |
| miRNA-21 | represses a variety of tumor suppressors such as PTEN (Phosphatase And Tensin Homolog) and PCDC4 (Programmed cell death protein 4) | |||||
| miRNA-148a miRNA-143 miRNA-378 | not mentioned | |||||
| 3 | Sonkoly E et al. [37] | downregulated | miRNA-203 | tissue | human | downstream effector of the HH pathway and EGFR pathways. Potential therapeutic target for the treatment of BCC |
| 4 | Al-Eryani L et al. [39] | upregulated | miRNA-425-5p | tissue (arsenic induced lession) | human | Premalignant lesions progression to malignancy |
| miRNA- 433 | ||||||
| downregulated | miRNA-29c | encoded in the last exon of C1orf132 (chromosome 1 open reading frame 132), and the transcript converts an unknown open reading frame | ||||
| miRNA-381 miRNA-452 miRNA487b miRNA-494 miRNA-590-5p | not mentioned | |||||
| 5 | Hu P et al. [40] | downregulated | miRNA-34a | blood | human | can regulate cell cycle and inhibit the migration, spread and invasion of tumor cells |
| 6 | Sun H, Jiang P [41] | downregulated | miRNA-451a | tissue | human & mouse | limits cell proliferation by cell cycle arrest induction, suggesting the potential therapeutic target of miRNA-451a in BCC |
| 7 | Chang J et al. [42] | upregulated | miRNA197-5p | blood | human | Potential role in metastasis process |
| 8 | Wan C, Li Y [43] | miRNA-101 | tissue | human | targets ING3 | |
| miRNA-7b miRNA-141 miRNA-9 miRNA-200a miRNA-203 miRNA-7c miRNA-132 miRNA-203 miRNA-495 miRNA-385 miRNA-220a miRNA-30e miRNA-29b miRNA-103 miRNA-130a miRNA-144 | not mentioned | |||||
| 9 | Farzan SF et al. [44] | upregulated | miRNA-146a | blood | human | modulator of inflammatory immune responses, coordinating myeloid and lymphocyte function to impact aspects of both innate and adaptive immunity |
| Nr | Ref | Expression | miRNA | Probe | Species | Role |
|---|---|---|---|---|---|---|
| 1 | Sand M. et al. [51] | upregulated | miRNA-31 | tissue | human | downregulate the tumor suppressor RhoBTB1 (Rho Related BTB Domain Containing 1) in the cSCC cell line A-431, determing cell proliferation and invasion |
| miRNA-135b | miRNA-135b can regulate cell migration and tumor invasiveness in early stages of SCC progression and can act as an oncogenic miRNA in human keratinocytes | |||||
| miRNA-424 | determines angiogenesis, regulates cell-autonomous angiogenic functions | |||||
| miRNA-21* miRNA-374a miRNA-196a | not mentioned | |||||
| miRNA-18a | associated with the Sonic Hedgehog pathway, correlated with molecular pathogenesis of cSCC | |||||
| miRNA-766 miRNA-128 | not mentioned | |||||
| miRNA-130b | downregulate the tumor suppressor protein 53-induced nuclear protein 1 (TP53INP1) | |||||
| miRNA-455-5p | not mentioned | |||||
| miRNA-21 | targets phosphatase and tensin homolog (PTEN), PDC4 (Programed Cell Death 4) and BTG2 (B-cell translocation gene 2) | |||||
| downregulated | miRNA-30a* miRNA-133b miRNA-101 miRNA-4324 miRNA-136 | not mentioned | ||||
| miRNA-378 | targets insulin-like growth factor 1 receptor (IGF1R) and caspase 3; reduced expression in basal cell carcinoma | |||||
| miRNA-204 miRNA-497 miRNA-29c miRNA-214 | not mentioned | |||||
| miRNA-145 | inhibits actin-binding protein Fascin homolog 1 (FSCN1) in esophageal squamous cell carcinoma; down-regulated in basal cell carcinoma | |||||
| miRNA-199a-5p miRNA-125b | not mentioned | |||||
| miRNA-140-3p | targets CD38; down-regulated in basal cell carcinoma | |||||
| miRNA-26a | downregulation of oncogene Histone-lysine N-methyltransferase (EZH2) | |||||
| 2 | P Dziunycz et al. [52] | upregulated | miRNA-21 | tissue | human | essential role in the development or maintenance of SCC of the skin |
| miRNA-184 miRNA-205 | not mentioned | |||||
| downregulated | miRNA-203 | unleash p63 expression, leading to decreased cell senescence and supporting SCC formation | ||||
| miRNA-378 | not mentioned | |||||
| 3 | Yamane et al. [53] | downregulated | miRNA-124 miRNA-214 | Tissue and serum | human | lead to overexpression of ERK1/2. May lead to the development of useful biomarkers for early detection of this tumor and to new treatments using miRNA |
| 4 | Zhang L et al. [56] | downregulated | miRNA-20a | tissue | human | might play important roles in the tumorigenesis and progression of CSCC patients, may serve as a novel molecular marker to predict the tumor progression and inferior prognosis of CSCC patients |
| 5 | Gong et al. [57] | upregulated | miRNA-221 | blood | human | significantly promotes cell proliferation |
| 6 | Kanitz et al. [64] | downregulated | miRNA-361-5p | tissue | human | regulator of VEGFA expression |
| 7 | Chen et al. [65] | downregulated | miRNA-346 | tissue | human | promotes the cSCC cell proliferation and migration through directly targeting SRCIN1 (SRC Kinase Signaling Inhibitor 1). This study may provide a new therapeutic target for cSCC. |
| 8 | Xu et al. [66] | downregulated | miRNA-125b | tissue | human | potential therapeutic biomarker. Matrix metalloproteinase (MMP)13 was considered a direct target of miRNA-125b |
| 9 | Olasz et al. [67] | upregulated | miRNA-135b | tissue | human | miRNA-135b can regulate cell migration and tumor invasiveness in early stages of SCC progression and can act as an oncogenic miRNA in human keratinocytes |
| 9 | Zhou M et al. [68] | upregulated | miRNA-365 | tissue | human | downregulates NFIB (nuclear Factor I B) and inhibits the expression of cyclin-dependent kinase CDK6 and CDK4 |
| Nr | Ref | Expression | miRNA | Probe | Species | Role |
|---|---|---|---|---|---|---|
| 1 | Ning et al. [73] | upregulated | miRNA-9 miRNA-502-3p miRNA-7 miRNA-340 miRNA-182 miRNA-190b miRNA-873 miRNA-183 | tissue | human | increasing tumor motility and colony formation. Potential diagnostic and therapeutic applications in cases of MCPyV-positive MCC |
| downregulated | miRNA-3170 miRNA-125b miRNA-374c | not mentioned | ||||
| 2 | Renwick et al. [74] | upregulated | miRNA-205 miRNA-375 | tissue | human | microRNAs downregulate the expression of gene targets through interaction with their three prime untranslated region (3′ UTR). miRNA-375 targets MTPN(myotrophin) gene, which encodes the myotrophin protein, further regulating hormone release and exocytosis. Potential diagnostic role by discerning BCC from MCC |
| 3 | Veija et al. [76] | downregulated | miR-34a, miR-1539, miR-30a, miR-142-3p | tissue | human | may play a role in the oncogenesis of MCV-negative tumors. |
| upregulated | miR-181d |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tamas, T.; Baciut, M.; Nutu, A.; Bran, S.; Armencea, G.; Stoia, S.; Manea, A.; Crisan, L.; Opris, H.; Onisor, F.; et al. Is miRNA Regulation the Key to Controlling Non-Melanoma Skin Cancer Evolution? Genes 2021, 12, 1929. https://doi.org/10.3390/genes12121929
Tamas T, Baciut M, Nutu A, Bran S, Armencea G, Stoia S, Manea A, Crisan L, Opris H, Onisor F, et al. Is miRNA Regulation the Key to Controlling Non-Melanoma Skin Cancer Evolution? Genes. 2021; 12(12):1929. https://doi.org/10.3390/genes12121929
Chicago/Turabian StyleTamas, Tiberiu, Mihaela Baciut, Andreea Nutu, Simion Bran, Gabriel Armencea, Sebastian Stoia, Avram Manea, Liana Crisan, Horia Opris, Florin Onisor, and et al. 2021. "Is miRNA Regulation the Key to Controlling Non-Melanoma Skin Cancer Evolution?" Genes 12, no. 12: 1929. https://doi.org/10.3390/genes12121929
APA StyleTamas, T., Baciut, M., Nutu, A., Bran, S., Armencea, G., Stoia, S., Manea, A., Crisan, L., Opris, H., Onisor, F., Baciut, G., Crisan, B., Opris, D., Bumbu, B., Tamas, A., & Dinu, C. (2021). Is miRNA Regulation the Key to Controlling Non-Melanoma Skin Cancer Evolution? Genes, 12(12), 1929. https://doi.org/10.3390/genes12121929









