Role and Regulation of the RECQL4 Family during Genomic Integrity Maintenance
Abstract
1. RECQL4 Is Critical in Maintaining DNA Integrity and Disease Prevention
1.1. RECQL4 Is Unique Amongst the RecQ Helicases
1.2. Rothmund-Thomson Syndrome
1.3. Baller-Gerold Syndrome
1.4. RAPADILINO
1.5. RECQL4 in Cancer
2. Model Systems to Study RECQL4 Function
2.1. Due to Difficulties Studying RECQL4 Function in Mice, Other Model Systems Have Been Used
2.2. Yeast Has Been Critical in Studying the Functions of RecQ Helicases
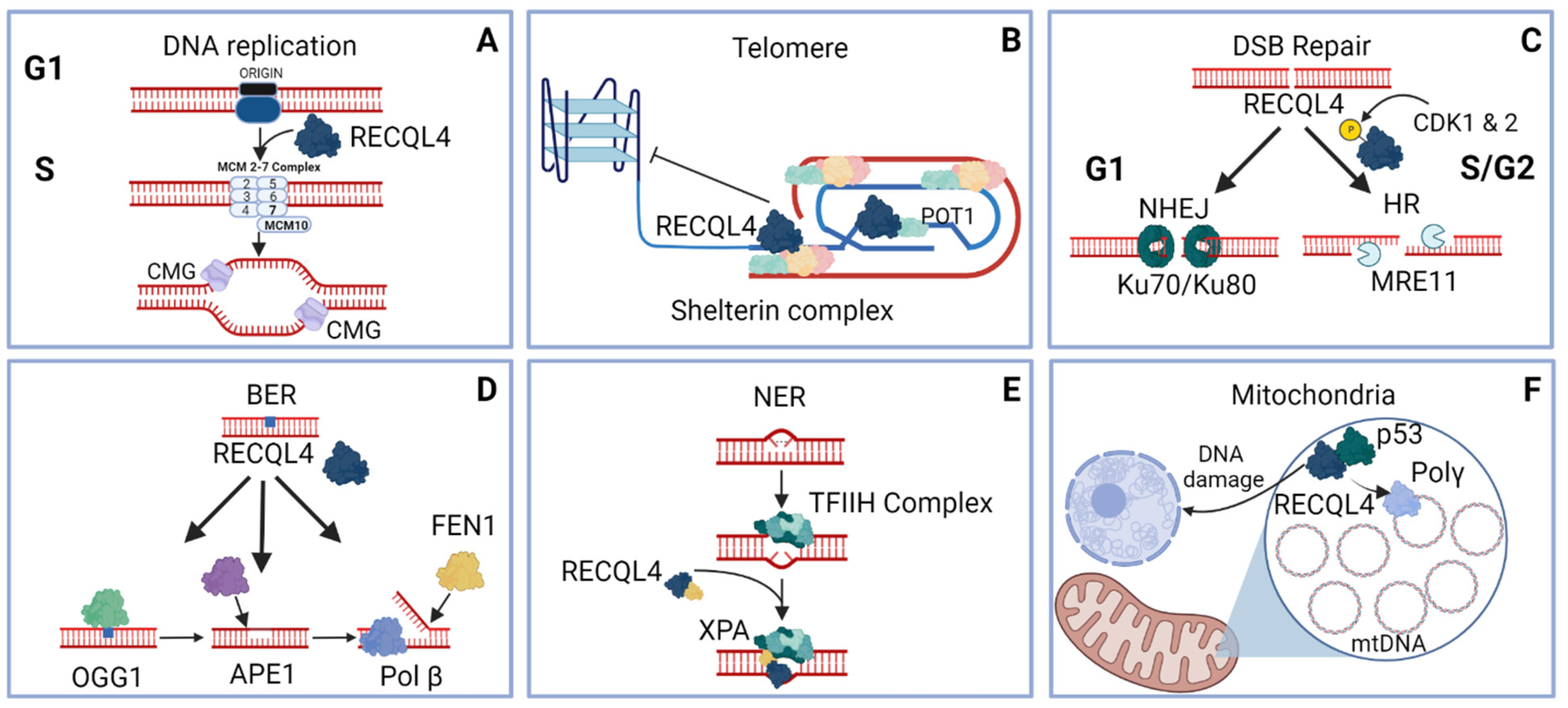
3. Conserved Role for the RECQL4 Family during DNA Replication
3.1. Xenopus RECQL4 Role in Replication Initiation
3.2. Drosophila RECQL4 Role in Replication Initiation
3.3. Human RECQL4 Role in Replication Initiation
4. Role of Hrq1 and RECQL4 during Telomeric DNA Maintenance
4.1. Human RECQL4 Role in Telomeric DNA Maintenance
4.2. Yeast Hrq1 Role during Telomeric DNA Maintenance
5. Role of RECQL4 during DNA Double-Strand Break Repair
5.1. Human and Xenopus RECQL4 Role during Double-Strand Break Repair
5.2. RECQL4 Role during Non-Homologous End Joining
5.3. RECQL4 Role during Homologous Recombination
5.4. Ubiquitylation of RECQL4 during DSB Repair
6. Role of RECQL4 during Base Excision Repair
6.1. RTS Cells Are Sensitive to Oxidizing Agents
6.2. RECQL4 Localizes to Sites of Oxidized DNA Damage
6.3. RECQL4 Interacts with Key BER Proteins
7. Role of RECQL4 during Nucleotide Excision Repair
8. Role of Hrq1 and RECQL4 during DNA Crosslink Repair
9. Role of RECQL4 in Mitochondrial Maintenance
9.1. RECQL4 Localizes to the Mitochondria
9.2. RECQL4 Mitochondrial Localization Is Critical for Sequestering p53 and Enhancing Mitochondrial DNA Replication
9.3. Cells Deficient in RECQL4 Mitochondrial Localization Has Perturbed Bioenergetics
10. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Hickson, I.D. RecQ helicases: Caretakers of the genome. Nat. Rev. Cancer 2003, 3, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Hanada, K.; Hickson, I.D. Molecular genetics of RecQ helicase disorders. Cell Mol. Life Sci. 2007, 64, 2306–2322. [Google Scholar] [CrossRef] [PubMed]
- Bohr, V.A. Rising from the RecQ-age: The role of human RecQ helicases in genome maintenance. Trends Biochem. Sci. 2008, 33, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Chu, W.K.; Hickson, I.D. RecQ helicases: Multifunctional genome caretakers. Nat. Rev. Cancer 2009, 9, 644–654. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, K.A.; Gangloff, S.; Rothstein, R. The RecQ DNA helicases in DNA repair. Annu. Rev. Genet. 2010, 44, 393–417. [Google Scholar] [CrossRef] [PubMed]
- Croteau, D.L.; Popuri, V.; Opresko, P.L.; Bohr, V.A. Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 2014, 83, 519–552. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xie, Y.; Ma, J.; Luo, X.; Nie, P.; Zuo, Z.; Lahrmann, U.; Zhao, Q.; Zheng, Y.; Zhao, Y.; et al. IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 2015, 31, 3359–3361. [Google Scholar] [CrossRef]
- Xu, X.; Liu, Y. Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein, RECQ4. EMBO J. 2009, 28, 568–577. [Google Scholar] [CrossRef]
- Dietschy, T.; Shevelev, I.; Pena-Diaz, J.; Hühn, D.; Kuenzle, S.; Mak, R.; Miah, M.F.; Hess, D.; Fey, M.; Hottiger, M.O.; et al. p300-mediated acetylation of the Rothmund-Thomson-syndrome gene product RECQL4 regulates its subcellular localization. J. Cell Sci. 2009, 122, 1701. [Google Scholar] [CrossRef][Green Version]
- Larizza, L.; Magnani, I.; Roversi, G. Rothmund-Thomson syndrome and RECQL4 defect: Splitting and lumping. Cancer Lett. 2006, 232, 107–120. [Google Scholar] [CrossRef]
- Larizza, L.; Roversi, G.; Volpi, L. Rothmund-Thomson syndrome. Orphanet J. Rare Dis. 2010, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.L.; Plon, S.E. Rothmund-Thomson Syndrome. In GeneReviews(®); Pagon, R.A., Adam, M.P., Ardinger, H.H., Wallace, S.E., Amemiya, A., Bean, L.J., Bird, T.D., Fong, C.-T., Mefford, H.C., Smith, R.J., et al., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Rothmund Uber cataracten in Verbindung mit einer eigentumlichen Hautgeneration. Arch. Klein Exp. Ophthalm 1868, 159–182.
- Thomson, M.S. Poikiloderma congenitale. Br. J. Dermatol. 1936, 221–234. [Google Scholar] [CrossRef]
- Taylor, W.B. Rothmund-Thomson’s syndrome. Arch. Dermatol. 1957, 75, 236–244. [Google Scholar] [CrossRef]
- Ying, K.L.; Oizumi, J.; Curry, C.J. Rothmund-Thomson syndrome associated with trisomy 8 mosaicism. J. Med. Genet. 1990, 27, 258–260. [Google Scholar] [CrossRef]
- Orstavik, K.H.; McFadden, N.; Hagelsteen, J.; Ormerod, E.; van der Hagen, C.B. Instability of lymphocyte chromosomes in a girl with Rothmund-Thomson syndrome. J. Med. Genet. 1994, 31, 570–572. [Google Scholar] [CrossRef]
- Lindor, N.M.; Devries, E.M.; Michels, V.V.; Schad, C.R.; Jalal, S.M.; Donovan, K.M.; Smithson, W.A.; Kvols, L.K.; Thibodeau, S.N.; Dewald, G.W. Rothmund-Thomson syndrome in siblings: Evidence for acquired in vivo mosaicism. Clin. Genet. 1996, 49, 124–129. [Google Scholar] [CrossRef]
- Kitao, S.; Ohsugi, I.; Ichikawa, K.; Goto, M.; Furuichi, Y.; Shimamoto, A. Cloning of two new human helicase genes of the RecQ family: Biological significance of multiple species in higher eukaryotes. Genomics 1998, 54, 443–452. [Google Scholar] [CrossRef]
- Kitao, S.; Shimamoto, A.; Goto, M.; Miller, R.W.; Smithson, W.A.; Lindor, N.M.; Furuichi, Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 1999, 22, 82–84. [Google Scholar] [CrossRef]
- Ajeawung, N.F.; Nguyen, T.T.M.; Lu, L.; Kucharski, T.J.; Rousseau, J.; Molidperee, S.; Atienza, J.; Gamache, I.; Jin, W.; Plon, S.E.; et al. Mutations in ANAPC1, Encoding a Scaffold Subunit of the Anaphase-Promoting Complex, Cause Rothmund-Thomson Syndrome Type 1. Am. J. Hum. Genet. 2019, 105, 625–630. [Google Scholar] [CrossRef]
- Zirn, B.; Bernbeck, U.; Alt, K.; Oeffner, F.; Gerhardinger, A.; Has, C. Rothmund–Thomson syndrome type 1 caused by biallelic ANAPC1 gene mutations. Ski. Health Dis. 2021, 1, e12. [Google Scholar] [CrossRef]
- Baller, F. Radiusaplasie und Inzucht. Z. Menschl. Vererb. Konst. 1950, 29, 782–790. [Google Scholar]
- Gerold, M. Frakturheilung bei einem seltenen Fall kongenitaler Anomalie der oberen Gliedmassen [Healing of a fracture in an unusual case of congenital anomaly of the upper extremities]. Zentralbl. Chir. 1959, 84, 831–834. [Google Scholar] [PubMed]
- Mégarbané, A.; Melki, I.; Souraty, N.; Gerbaka, J.; Ghouzzi, V.; Bonaventure, J.; Mornand, A.; Loiselet, J. Overlap between Baller-Gerold and Rothmund-Thomson syndrome. Clin. Dysmorphol. 2000, 9, 303–305. [Google Scholar] [CrossRef]
- Van Maldergem, L.; Siitonen, H.A.; Jalkh, N.; Chouery, E.; De Roy, M.; Delague, V.; Muenke, M.; Jabs, E.W.; Cai, J.; Wang, L.L.; et al. Revisiting the craniosynostosis-radial ray hypoplasia association: Baller-Gerold syndrome caused by mutations in the RECQL4 gene. J. Med. Genet. 2006, 43, 148–152. [Google Scholar] [CrossRef]
- Piard, J.; Aral, B.; Vabres, P.; Holder-Espinasse, M.; Mégarbané, A.; Gauthier, S.; Capra, V.; Pierquin, G.; Callier, P.; Baumann, C.; et al. Search for ReCQL4 mutations in 39 patients genotyped for suspected Rothmund-Thomson/Baller-Gerold syndromes. Clin. Genet. 2015, 87, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Mo, D.; Zhao, Y.; Balajee, A.S. Human RecQL4 helicase plays multifaceted roles in the genomic stability of normal and cancer cells. Cancer Lett. 2018, 413, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gripp, K.W.; Stolle, C.A.; Celle, L.; McDonald-McGinn, D.M.; Whitaker, L.A.; Zackai, E.H. TWIST gene mutation in a patient with radial aplasia and craniosynostosis: Further evidence for heterogeneity of Baller-Gerold syndrome. Am. J. Med. Genet. 1999, 82, 170–176. [Google Scholar] [CrossRef]
- Seto, M.L.; Lee, S.J.; Sze, R.W.; Cunningham, M.L. Another TWIST on Baller-Gerold syndrome. Am. J. Med. Genet. 2001, 104, 323–330. [Google Scholar] [CrossRef]
- Siitonen, H.A.; Sotkasiira, J.; Biervliet, M.; Benmansour, A.; Capri, Y.; Cormier-Daire, V.; Crandall, B.; Hannula-Jouppi, K.; Hennekam, R.; Herzog, D.; et al. The mutation spectrum in RECQL4 diseases. Eur. J. Hum. Genet. 2009, 17, 151–158. [Google Scholar] [CrossRef]
- Kääriäinen, H.; Ryöppy, S.; Norio, R. RAPADILINO syndrome with radial and patellar aplasia/hypoplasia as main manifestations. Am. J. Med. Genet. 1989, 33, 346–351. [Google Scholar] [CrossRef]
- Siitonen, H.A.; Kopra, O.; Kääriäinen, H.; Haravuori, H.; Winter, R.M.; Säämänen, A.-M.; Peltonen, L.; Kestilä, M. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum. Mol. Genet. 2003, 12, 2837–2844. [Google Scholar] [CrossRef]
- Miozzo, M.; Castorina, P.; Riva, P.; Dalprà, L.; Fuhrman Conti, A.M.; Volpi, L.; Hoe, T.S.; Khoo, A.; Wiegant, J.; Rosenberg, C.; et al. Chromosomal instability in fibroblasts and mesenchymal tumors from 2 sibs with Rothmund-Thomson syndrome. Int. J. Cancer 1998, 77, 504–510. [Google Scholar] [CrossRef]
- Beghini, A.; Castorina, P.; Roversi, G.; Modiano, P.; Larizza, L. RNA processing defects of the helicase gene RECQL4 in a compound heterozygous Rothmund-Thomson patient. Am. J. Med. Genet. A 2003, 120A, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Colombo, E.A.; Locatelli, A.; Cubells Sánchez, L.; Romeo, S.; Elcioglu, N.H.; Maystadt, I.; Esteve Martínez, A.; Sironi, A.; Fontana, L.; Finelli, P.; et al. Rothmund-Thomson Syndrome: Insights from New Patients on the Genetic Variability Underpinning Clinical Presentation and Cancer Outcome. Int. J. Mol. Sci. 2018, 19, 1103. [Google Scholar] [CrossRef]
- Choi, Y.W.; Bae, S.M.; Kim, Y.W.; Lee, H.N.; Kim, Y.W.; Park, T.C.; Ro, D.Y.; Shin, J.C.; Shin, S.J.; Seo, J.S.; et al. Gene expression profiles in squamous cell cervical carcinoma using array-based comparative genomic hybridization analysis. Int. J. Gynecol. Cancer 2007, 17, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Meador, J.A.; Calaf, G.M.; Proietti De-Santis, L.; Zhao, Y.; Bohr, V.A.; Balajee, A.S. Human RecQL4 helicase plays critical roles in prostate carcinogenesis. Cancer Res. 2010, 70, 9207–9217. [Google Scholar] [CrossRef]
- Fang, H.; Nie, L.; Chi, Z.; Liu, J.; Guo, D.; Lu, X.; Hei, T.K.; Balajee, A.S.; Zhao, Y. RecQL4 helicase amplification is involved in human breast tumorigenesis. PLoS ONE 2013, 8, e69600. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Jin, W.; Liu, H.; Wang, L.L. RECQ DNA helicases and osteosarcoma. Adv. Exp. Med. Biol. 2014, 804, 129–145. [Google Scholar] [CrossRef]
- Yong, Z.W.E.; Zaini, Z.M.; Kallarakkal, T.G.; Karen-Ng, L.P.; Rahman, Z.A.A.; Ismail, S.M.; Sharifah, N.A.; Mustafa, W.M.W.; Abraham, M.T.; Tay, K.K.; et al. Genetic alterations of chromosome 8 genes in oral cancer. Sci. Rep. 2014, 4, 6073. [Google Scholar] [CrossRef]
- Arora, A.; Agarwal, D.; Abdel-Fatah, T.M.; Lu, H.; Croteau, D.L.; Moseley, P.; Aleskandarany, M.A.; Green, A.R.; Ball, G.; Rakha, E.A.; et al. RECQL4 helicase has oncogenic potential in sporadic breast cancers. J. Pathol. 2016, 238, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Mo, D.; Fang, H.; Niu, K.; Liu, J.; Wu, M.; Li, S.; Zhu, T.; Aleskandarany, M.A.; Arora, A.; Lobo, D.N.; et al. Human Helicase RECQL4 Drives Cisplatin Resistance in Gastric Cancer by Activating an AKT-YB1-MDR1 Signaling Pathway. Cancer Res. 2016, 76, 3057–3066. [Google Scholar] [CrossRef] [PubMed]
- De, S.; Kumari, J.; Mudgal, R.; Modi, P.; Gupta, S.; Futami, K.; Goto, H.; Lindor, N.M.; Furuichi, Y.; Mohanty, D.; et al. RECQL4 is essential for the transport of p53 to mitochondria in normal human cells in the absence of exogenous stress. J. Cell Sci. 2012, 125, 2509–2522. [Google Scholar] [CrossRef] [PubMed]
- Ughachukwu, P.; Unekwe, P. Efflux pump-mediated resistance in chemotherapy. Ann. Med. Health Sci. Res. 2012, 2, 191–198. [Google Scholar] [CrossRef]
- Ng, A.J.M.; Walia, M.K.; Smeets, M.F.; Mutsaers, A.J.; Sims, N.A.; Purton, L.E.; Walsh, N.C.; Martin, T.J.; Walkley, C.R. The DNA helicase recql4 is required for normal osteoblast expansion and osteosarcoma formation. PLoS Genet. 2015, 11, e1005160. [Google Scholar] [CrossRef]
- Balajee, A.S. Human recql4 as a novel molecular target for cancer therapy. Cytogenet Genome Res. 2021, 161, 305–327. [Google Scholar] [CrossRef]
- Matsuno, K.; Kumano, M.; Kubota, Y.; Hashimoto, Y.; Takisawa, H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase α in the initiation of DNA replication. Mol. Cell. Biol. 2006, 26, 4843–4852. [Google Scholar] [CrossRef]
- Xu, X.; Rochette, P.J.; Feyissa, E.A.; Su, T.V.; Liu, Y. MCM10 mediates RECQ4 association with MCM2-7 helicase complex during DNA replication. EMBO J. 2009, 28, 3005–3014. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Rossi, M.L.; Singh, D.K.; Dunn, C.; Ramamoorthy, M.; Croteau, D.L.; Liu, Y.; Bohr, V.A. RECQL4, the protein mutated in Rothmund-Thomson syndrome, functions in telomere maintenance. J. Biol. Chem. 2012, 287, 196–209. [Google Scholar] [CrossRef]
- Singh, D.K.; Popuri, V.; Kulikowicz, T.; Shevelev, I.; Ghosh, A.K.; Ramamoorthy, M.; Rossi, M.L.; Janscak, P.; Croteau, D.L.; Bohr, V.A. The human RecQ helicases BLM and RECQL4 cooperate to preserve genome stability. Nucleic Acids Res. 2012, 40, 6632–6648. [Google Scholar] [CrossRef]
- Shamanna, R.A.; Singh, D.K.; Lu, H.; Mirey, G.; Keijzers, G.; Salles, B.; Croteau, D.L.; Bohr, V.A. RECQ helicase RECQL4 participates in non-homologous end joining and interacts with the Ku complex. Carcinogenesis 2014, 35, 2415–2424. [Google Scholar] [CrossRef]
- Lu, H.; Shamanna, R.A.; Keijzers, G.; Anand, R.; Rasmussen, L.J.; Cejka, P.; Croteau, D.L.; Bohr, V.A. RECQL4 Promotes DNA End Resection in Repair of DNA Double-Strand Breaks. Cell Rep. 2016, 16, 161–173. [Google Scholar] [CrossRef]
- Petkovic, M.; Dietschy, T.; Freire, R.; Jiao, R.; Stagljar, I. The human Rothmund-Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J. Cell Sci. 2005, 118, 4261–4269. [Google Scholar] [CrossRef][Green Version]
- Schurman, S.H.; Hedayati, M.; Wang, Z.; Singh, D.K.; Speina, E.; Zhang, Y.; Becker, K.; Macris, M.; Sung, P.; Wilson, D.M.; et al. Direct and indirect roles of RECQL4 in modulating base excision repair capacity. Hum. Mol. Genet. 2009, 18, 3470–3483. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Han, X.; Akbari, M.; Croteau, D.L.; Rasmussen, L.J.; Bohr, V.A. Interaction between RECQL4 and OGG1 promotes repair of oxidative base lesion 8-oxoG and is regulated by SIRT1 deacetylase. Nucleic Acids Res. 2020, 48, 6530–6546. [Google Scholar] [CrossRef] [PubMed]
- Woo, L.L.; Futami, K.; Shimamoto, A.; Furuichi, Y.; Frank, K.M. The Rothmund-Thomson gene product RECQL4 localizes to the nucleolus in response to oxidative stress. Exp. Cell Res. 2006, 312, 3443–3457. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Luo, J. RecQ4 facilitates UV light-induced DNA damage repair through interaction with nucleotide excision repair factor xeroderma pigmentosum group A (XPA). J. Biol. Chem. 2008, 283, 29037–29044. [Google Scholar] [CrossRef]
- Ichikawa, K.; Noda, T.; Furuichi, Y. Preparation of the gene targeted knockout mice for human premature aging diseases, Werner syndrome, and Rothmund-Thomson syndrome caused by the mutation of DNA helicases. Nihon Shinkei Seishin Yakurigaku Zasshi 2002, 119, 219–226. [Google Scholar] [CrossRef]
- Wu, J.; Capp, C.; Feng, L.; Hsieh, T. Drosophila homologue of the Rothmund-Thomson syndrome gene: Essential function in DNA replication during development. Dev. Biol. 2008, 323, 130–142. [Google Scholar] [CrossRef]
- Hoki, Y.; Araki, R.; Fujimori, A.; Ohhata, T.; Koseki, H.; Fukumura, R.; Nakamura, M.; Takahashi, H.; Noda, Y.; Kito, S.; et al. Growth retardation and skin abnormalities of the Recql4-deficient mouse. Hum. Mol. Genet. 2003, 12, 2293–2299. [Google Scholar] [CrossRef]
- Mann, M.B.; Hodges, C.A.; Barnes, E.; Vogel, H.; Hassold, T.J.; Luo, G. Defective sister-chromatid cohesion, aneuploidy and cancer predisposition in a mouse model of type II Rothmund-Thomson syndrome. Hum. Mol. Genet. 2005, 14, 813–825. [Google Scholar] [CrossRef]
- Gangloff, S.; McDonald, J.P.; Bendixen, C.; Arthur, L.; Rothstein, R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: A potential eukaryotic reverse gyrase. Mol. Cell. Biol. 1994, 14, 8391–8398. [Google Scholar] [CrossRef]
- Watt, P.M.; Louis, E.J.; Borts, R.H.; Hickson, I.D. Sgs1: A eukaryotic homolog of E. coil RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell 1995, 81, 253–260. [Google Scholar] [CrossRef]
- Barea, F.; Tessaro, S.; Bonatto, D. In silico analyses of a new group of fungal and plant RecQ4-homologous proteins. Comput Biol Chem 2008, 32, 349–358. [Google Scholar] [CrossRef]
- Rogers, C.M.; Wang, J.C.-Y.; Noguchi, H.; Imasaki, T.; Takagi, Y.; Bochman, M.L. Yeast Hrq1 shares structural and functional homology with the disease-linked human RecQ4 helicase. Nucleic Acids Res. 2017, 45, 5217–5230. [Google Scholar] [CrossRef]
- Prokhorova, T.A.; Mowrer, K.; Gilbert, C.H.; Walter, J.C. DNA replication of mitotic chromatin in Xenopus egg extracts. Proc. Natl. Acad. Sci. USA 2003, 100, 13241–13246. [Google Scholar] [CrossRef]
- Gillespie, P.J.; Gambus, A.; Blow, J.J. Preparation and use of Xenopus egg extracts to study DNA replication and chromatin associated proteins. Methods 2012, 57, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Hoogenboom, W.S.; Klein Douwel, D.; Knipscheer, P. Xenopus egg extract: A powerful tool to study genome maintenance mechanisms. Dev. Biol. 2017, 428, 300–309. [Google Scholar] [CrossRef]
- Sangrithi, M.N.; Bernal, J.A.; Madine, M.; Philpott, A.; Lee, J.; Dunphy, W.G.; Venkitaraman, A.R. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell 2005, 121, 887–898. [Google Scholar] [CrossRef] [PubMed]
- Germani, F.; Bergantinos, C.; Johnston, L.A. Mosaic analysis in drosophila. Genetics 2018, 208, 473–490. [Google Scholar] [CrossRef]
- Capp, C.; Wu, J.; Hsieh, T. Drosophila RecQ4 has a 3′-5’ DNA helicase activity that is essential for viability. J. Biol. Chem. 2009, 284, 30845–30852. [Google Scholar] [CrossRef] [PubMed]
- Im, J.-S.; Ki, S.-H.; Farina, A.; Jung, D.-S.; Hurwitz, J.; Lee, J.-K. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc. Natl. Acad. Sci. USA 2009, 106, 15628–15632. [Google Scholar] [CrossRef]
- Lu, H.; Shamanna, R.A.; de Freitas, J.K.; Okur, M.; Khadka, P.; Kulikowicz, T.; Holland, P.P.; Tian, J.; Croteau, D.L.; Davis, A.J.; et al. Cell cycle-dependent phosphorylation regulates RECQL4 pathway choice and ubiquitination in DNA double-strand break repair. Nat. Commun. 2017, 8, 2039. [Google Scholar] [CrossRef]
- Walne, A.J.; Vulliamy, T.; Beswick, R.; Kirwan, M.; Dokal, I. Mutations in C16orf57 and normal-length telomeres unify a subset of patients with dyskeratosis congenita, poikiloderma with neutropenia and Rothmund-Thomson syndrome. Hum. Mol. Genet. 2010, 19, 4453–4461. [Google Scholar] [CrossRef]
- Vennos, E.; Collins, M.; James, W. Rothmund-Thomson syndrome: World literature Review of the world literature. J. Am. Acad. Dermatol. 1992, 27, 750–762. [Google Scholar] [CrossRef]
- Opresko, P.L.; Otterlei, M.; Graakjaer, J.; Bruheim, P.; Dawut, L.; Kølvraa, S.; May, A.; Seidman, M.M.; Bohr, V.A. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol. Cell 2004, 14, 763–774. [Google Scholar] [CrossRef]
- Ghosh, A.; Rossi, M.L.; Aulds, J.; Croteau, D.; Bohr, V.A. Telomeric D-loops containing 8-oxo-2′-deoxyguanosine are preferred substrates for Werner and Bloom syndrome helicases and are bound by POT1. J. Biol. Chem. 2009, 284, 31074–31084. [Google Scholar] [CrossRef]
- Chang, S.; Multani, A.S.; Cabrera, N.G.; Naylor, M.L.; Laud, P.; Lombard, D.; Pathak, S.; Guarente, L.; DePinho, R.A. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat. Genet. 2004, 36, 877–882. [Google Scholar] [CrossRef]
- Takai, H.; Smogorzewska, A.; de Lange, T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003, 13, 1549–1556. [Google Scholar] [CrossRef]
- Ferrarelli, L.K.; Popuri, V.; Ghosh, A.K.; Tadokoro, T.; Canugovi, C.; Hsu, J.K.; Croteau, D.L.; Bohr, V.A. The RECQL4 protein, deficient in Rothmund-Thomson syndrome is active on telomeric D-loops containing DNA metabolism blocking lesions. DNA Repair 2013, 12, 518–528. [Google Scholar] [CrossRef]
- Bochman, M.L.; Paeschke, K.; Chan, A.; Zakian, V.A. Hrq1, a homolog of the human RecQ4 helicase, acts catalytically and structurally to promote genome integrity. Cell Rep. 2014, 6, 346–356. [Google Scholar] [CrossRef]
- Nickens, D.G.; Rogers, C.M.; Bochman, M.L. The Saccharomyces cerevisiae Hrq1 and Pif1 DNA helicases synergistically modulate telomerase activity in vitro. J. Biol. Chem. 2018, 293, 14481–14496. [Google Scholar] [CrossRef]
- Featherstone, C.; Jackson, S.P. DNA double-strand break repair. Curr. Biol. 1999, 9, R759–R761. [Google Scholar] [CrossRef]
- Jeggo, P.A.; Löbrich, M. DNA double-strand breaks: Their cellular and clinical impact? Oncogene 2007, 26, 7717–7719. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.J.; Paterson, M.C. Enhanced radiosensitivity and defective DNA repair in cultured fibroblasts derived from Rothmund Thomson syndrome patients. Mutat. Res. 1982, 94, 213–228. [Google Scholar] [CrossRef]
- Kumata, Y.; Tada, S.; Yamanada, Y.; Tsuyama, T.; Kobayashi, T.; Dong, Y.-P.; Ikegami, K.; Murofushi, H.; Seki, M.; Enomoto, T. Possible involvement of RecQL4 in the repair of double-strand DNA breaks in Xenopus egg extracts. Biochim. Biophys. Acta 2007, 1773, 556–564. [Google Scholar] [CrossRef]
- Singh, D.K.; Karmakar, P.; Aamann, M.; Schurman, S.H.; May, A.; Croteau, D.L.; Burks, L.; Plon, S.E.; Bohr, V.A. The involvement of human RECQL4 in DNA double-strand break repair. Aging Cell 2010, 9, 358–371. [Google Scholar] [CrossRef]
- Tan, Q.; Niu, K.; Zhu, Y.; Chen, Z.; Li, Y.; Li, M.; Wei, D.; Balajee, A.S.; Fang, H.; Zhao, Y. RNF8 ubiquitinates RecQL4 and promotes its dissociation from DNA double strand breaks. Oncogenesis 2021, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Bozzella, M.; Seluanov, A.; Gorbunova, V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle 2008, 7, 2902–2906. [Google Scholar] [CrossRef]
- Werner, S.R.; Prahalad, A.K.; Yang, J.; Hock, J.M. RECQL4-deficient cells are hypersensitive to oxidative stress/damage: Insights for osteosarcoma prevalence and heterogeneity in Rothmund-Thomson syndrome. Biochem. Biophys. Res. Commun. 2006, 345, 403–409. [Google Scholar] [CrossRef]
- Schärer, O.D. Nucleotide excision repair in eukaryotes. Cold Spring Harb. Perspect. Biol. 2013, 5, a012609. [Google Scholar] [CrossRef] [PubMed]
- Van Houten, B.; Kong, M. Eukaryotic Nucleotide Excision Repair. In Encyclopedia of Cell Biology; Elsevier: Amsterdam, The Netherlands, 2016; pp. 435–441. [Google Scholar]
- Berg, E.; Chuang, T.-Y.; Cripps, D. Madison A case report, phototesting, and literature review. J. Am. Acad. Dermatol. 1987, 17, 332–338. [Google Scholar] [CrossRef]
- Shinya, A.; Nishigori, C.; Moriwaki, S.; Takebe, H.; Kubota, M.; Ogino, A.; Imamura, S. A Case of Rothmund-Thomson Syndrome With Reduced DNA Repair Capacity. Arch. Dermatolol. 1993, 129, 332–336. [Google Scholar] [CrossRef]
- Vasseur, F.; Dalaporte, E.; Zabot, M.T.; Sturque, M.N.; Barrut, D.; Savary, J.B.; Thomas, L.; Thomas, P. Excision Repair Defect in Rothmund Thomson Syndrome.pdf. Acta Derm. Venereol. 1999, 79, 150–152. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Liu, H.; Zhang, Y.; Otta, S.K.; Plon, S.E.; Wang, L.L. Sensitivity of RECQL4-deficient fibroblasts from Rothmund-Thomson syndrome patients to genotoxic agents. Hum. Genet. 2008, 123, 643–653. [Google Scholar] [CrossRef]
- Moldovan, G.-L.; D’Andrea, A.D. How the fanconi anemia pathway guards the genome. Annu. Rev. Genet. 2009, 43, 223–249. [Google Scholar] [CrossRef]
- Kottemann, M.C.; Smogorzewska, A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature 2013, 493, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Ceccaldi, R.; Sarangi, P.; D’Andrea, A.D. The Fanconi anaemia pathway: New players and new functions. Nat. Rev. Mol. Cell Biol. 2016, 17, 337–349. [Google Scholar] [CrossRef]
- Deans, A.J.; West, S.C. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer 2011, 11, 467–480. [Google Scholar] [CrossRef]
- Schärer, O.D. DNA interstrand crosslinks: Natural and drug-induced DNA adducts that induce unique cellular responses. Chembiochem 2005, 6, 27–32. [Google Scholar] [CrossRef]
- Langevin, F.; Crossan, G.P.; Rosado, I.V.; Arends, M.J.; Patel, K.J. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 2011, 475, 53–58. [Google Scholar] [CrossRef]
- Uebelacker, M.; Lachenmeier, D.W. Quantitative determination of acetaldehyde in foods using automated digestion with simulated gastric fluid followed by headspace gas chromatography. J. Autom. Methods Manag. Chem. 2011, 2011, 907317. [Google Scholar] [CrossRef]
- Sonohara, Y.; Yamamoto, J.; Tohashi, K.; Takatsuka, R.; Matsuda, T.; Iwai, S.; Kuraoka, I. Acetaldehyde forms covalent GG intrastrand crosslinks in DNA. Sci. Rep. 2019, 9, 660. [Google Scholar] [CrossRef]
- Choi, D.-H.; Min, M.-H.; Kim, M.-J.; Lee, R.; Kwon, S.-H.; Bae, S.-H. Hrq1 facilitates nucleotide excision repair of DNA damage induced by 4-nitroquinoline-1-oxide and cisplatin in Saccharomyces cerevisiae. J. Microbiol. 2014, 52, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Kumari, J.; Hussain, M.; De, S.; Chandra, S.; Modi, P.; Tikoo, S.; Singh, A.; Sagar, C.; Sepuri, N.B.V.; Sengupta, S. Mitochondrial functions of RECQL4 are required for the prevention of aerobic glycolysis-dependent cell invasion. J. Cell Sci. 2016, 129, 1312–1318. [Google Scholar] [CrossRef] [PubMed]
- Trifunovic, A.; Larsson, N.G. Mitochondrial dysfunction as a cause of ageing. J. Intern. Med. 2008, 263, 167–178. [Google Scholar] [CrossRef]
- Cui, H.; Kong, Y.; Zhang, H. Oxidative stress, mitochondrial dysfunction, and aging. J. Signal. Transduct. 2012, 2012, 646354. [Google Scholar] [CrossRef] [PubMed]
- Lane, R.K.; Hilsabeck, T.; Rea, S.L. The role of mitochondrial dysfunction in age-related diseases. Biochim. Biophys. Acta 2015, 1847, 1387–1400. [Google Scholar] [CrossRef] [PubMed]
- Payne, B.A.I.; Chinnery, P.F. Mitochondrial dysfunction in aging: Much progress but many unresolved questions. Biochim. Biophys. Acta 2015, 1847, 1347–1353. [Google Scholar] [CrossRef]
- Haas, R.H. Mitochondrial dysfunction in aging and diseases of aging. Biology 2019, 8, 48. [Google Scholar] [CrossRef]
- Croteau, D.L.; Rossi, M.L.; Canugovi, C.; Tian, J.; Sykora, P.; Ramamoorthy, M.; Wang, Z.M.; Singh, D.K.; Akbari, M.; Kasiviswanathan, R.; et al. RECQL4 localizes to mitochondria and preserves mitochondrial DNA integrity. Aging Cell 2012, 11, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Chi, Z.; Nie, L.; Peng, Z.; Yang, Q.; Yang, K.; Tao, J.; Mi, Y.; Fang, X.; Balajee, A.S.; Zhao, Y. RecQL4 cytoplasmic localization: Implications in mitochondrial DNA oxidative damage repair. Int. J. Biochem. Cell Biol. 2012, 44, 1942–1951. [Google Scholar] [CrossRef]
- Gupta, S.; De, S.; Srivastava, V.; Hussain, M.; Kumari, J.; Muniyappa, K.; Sengupta, S. RECQL4 and p53 potentiate the activity of polymerase γ and maintain the integrity of the human mitochondrial genome. Carcinogenesis 2014, 35, 34–45. [Google Scholar] [CrossRef]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Turrens, J.F. Mitochondrial formation of reactive oxygen species. J. Physiol. 2003, 552, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Reyes, I.; Cuezva, J.M. The H(+)-ATP synthase: A gate to ROS-mediated cell death or cell survival. Biochim. Biophys. Acta 2014, 1837, 1099–1112. [Google Scholar] [CrossRef]
- Prasad, R.; Çağlayan, M.; Dai, D.-P.; Nadalutti, C.A.; Zhao, M.-L.; Gassman, N.R.; Janoshazi, A.K.; Stefanick, D.F.; Horton, J.K.; Krasich, R.; et al. DNA polymerase β: A missing link of the base excision repair machinery in mammalian mitochondria. DNA Repair 2017, 60, 77–88. [Google Scholar] [CrossRef]
- Sykora, P.; Kanno, S.; Akbari, M.; Kulikowicz, T.; Baptiste, B.A.; Leandro, G.S.; Lu, H.; Tian, J.; May, A.; Becker, K.A.; et al. DNA polymerase β participates in mitochondrial DNA repair. Mol. Cell. Biol. 2017, 37, e00237-17. [Google Scholar] [CrossRef]


| AP | Apurinic |
| ATM | Ataxia-telangiectasia mutated |
| BER | Base excision repair |
| BGS | Baller-Gerold syndrome |
| BLM | Bloom syndrome protein |
| CPD | Cyclobutane pyrimidine dimers |
| CMG | CDC45-MCM2-7-GINS |
| DSB | Double-strand break |
| DSBR | Double-strand break repair |
| IP | Immunoprecipitation |
| IR | Ionizing radiation |
| MLS | Mitochondrial localization sequence |
| mtDNA | Mitochondria DNA |
| NER | Nucleotide excision repair |
| NHEJ | Non-Homologous End Joining |
| NLS | Nuclear localization sequence |
| RTS | Rothmund-Thomson syndrome |
| xRTS | Xenopus RECQL4 |
| TIFs | Telomere dysfunction-induced foci |
| UV | Ultraviolet |
| WRN | Werner syndrome ATP-dependent helicase |
| Process | Protein | Detection Methods * | Interaction Region | Function | References |
|---|---|---|---|---|---|
| Localization | p300 | Co-IP, Co-Loc, Pull Down | 1–408 aa | RECQL4 cellular localization | [9] |
| Replication | Cut5 | Co-IP | N-terminus | DNA Replication, Xenopus Cut5 with xRTS | [48] |
| MCM7 | Co-IP, MS | ND | DNA Replication | [49] | |
| MCM10 | Co-IP, MS, Pull Down | 1–200 aa | DNA Replication, inhibition of RECQL4 helicase activity | [49] | |
| SLD5 | Co-IP, MS | ND | DNA Replication | [49] | |
| Telomere | TRF1 | Co-Loc | ND | Telomere maintenance, Stimulates RECQL4 helicase activity | [50] |
| TRF2 | Co-IP | ND | Telomere maintenance, Stimulates RECQL4 helicase activity | [50] | |
| WRN | Co-IP | ND | RECQL4 stimulates WRN telomeric D-loop resolution | [50] | |
| DSB Repair | BLM | Co-IP, Y2H | 1–471 aa | Increase RECQL4 retention time at DSB sites, BLM stimulation | [51] |
| KU70 | Co-IP | N-terminus | NHEJ, RECQL4 enhances KU complex DNA binding. KU inhibits RECQL4 helicase activity | [52] | |
| KU80 | Co-IP | ND | NHEJ, RECQL4 enhances KU complex DNA binding. KU inhibits RECQL4 helicase activity | [52] | |
| MRE11 | Co-Loc, IP, Pull Down | N-terminus | HR, DNA end resection | [53] | |
| RAD51 | Co-Loc, Co-IP | ND | DSB Repair | [54] | |
| BER | APE1 | Co-Loc | ND | APE1 endonuclease stimulation | [55] |
| FEN1 | Co-Loc | ND | FEN1 incision stimulation | [55] | |
| OGG1 | Co-IP | N-terminus | Stimulates OGG1 AP lyase activity | [56] | |
| PARP1 | Co-IP, PDS | 833–1208 aa | Base excision repair | [57] | |
| POL β | ND | ND | Stimulates POLβ DNA synthesis activity | [55] | |
| NER | XPA | Co-IP, Co-Loc, Fractionation, Pull Down | ND | Nucleotide excision repair | [58] |
| Mitochondria | p53 | Co-IP, Co-Loc, Fractination | 270–400 aa | Sequestering p53 from nucleus, mtDNA synthesis | [44] |
| TOM20 | Pull Down | 13–18 aa | Mitochondrial import | [44] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luong, T.T.; Bernstein, K.A. Role and Regulation of the RECQL4 Family during Genomic Integrity Maintenance. Genes 2021, 12, 1919. https://doi.org/10.3390/genes12121919
Luong TT, Bernstein KA. Role and Regulation of the RECQL4 Family during Genomic Integrity Maintenance. Genes. 2021; 12(12):1919. https://doi.org/10.3390/genes12121919
Chicago/Turabian StyleLuong, Thong T., and Kara A. Bernstein. 2021. "Role and Regulation of the RECQL4 Family during Genomic Integrity Maintenance" Genes 12, no. 12: 1919. https://doi.org/10.3390/genes12121919
APA StyleLuong, T. T., & Bernstein, K. A. (2021). Role and Regulation of the RECQL4 Family during Genomic Integrity Maintenance. Genes, 12(12), 1919. https://doi.org/10.3390/genes12121919






