BRCA1/2 NGS Somatic Testing in Clinical Practice: A Short Report
Abstract
:1. Introduction
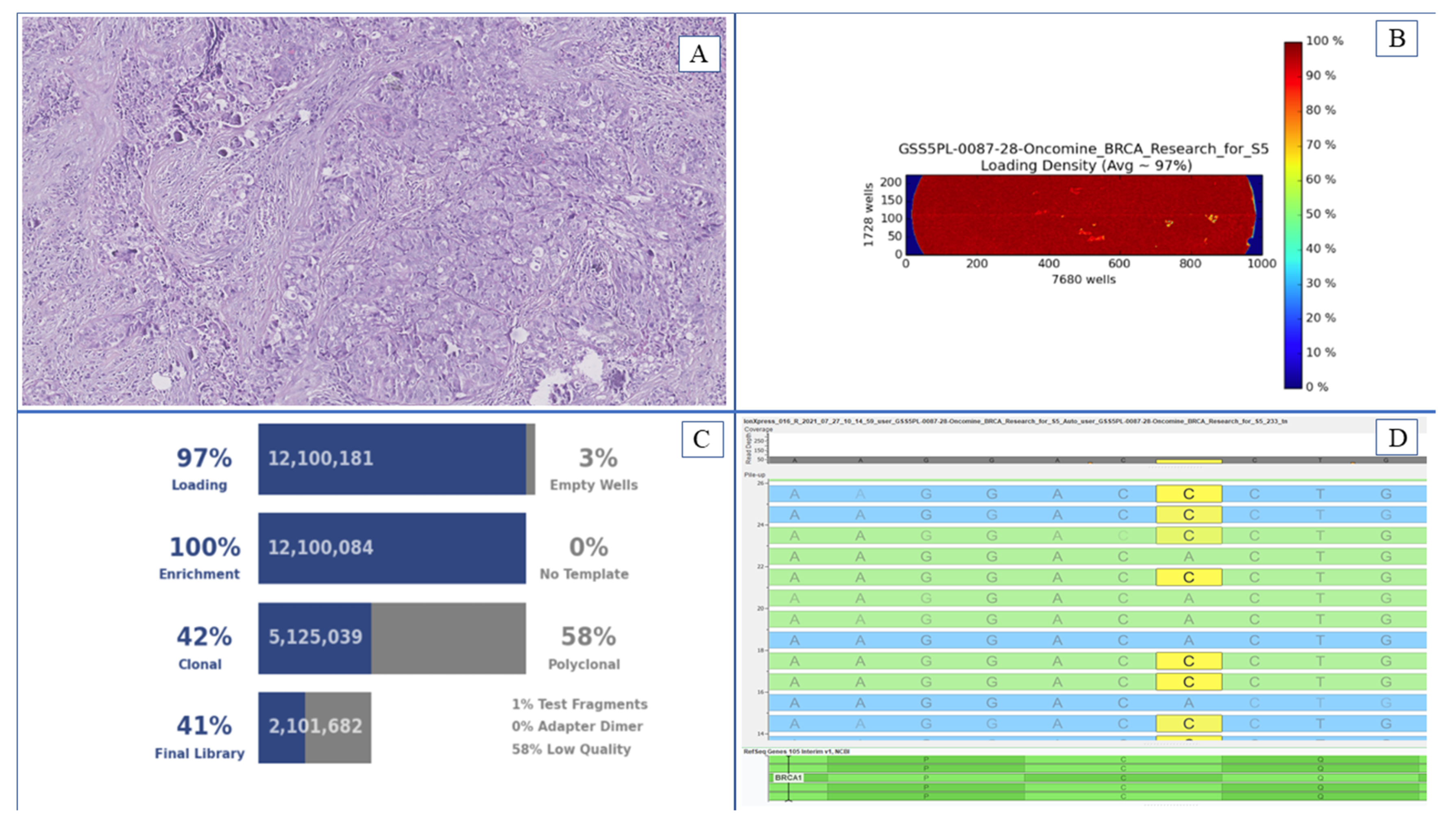
2. Material and Methods
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Kurman, R.J.; Shih, I.M. The Dualistic Model of Ovarian Carcinogenesis: Revisited, Revised, and Expanded. Am. J. Pathol. 2016, 186, 733–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labidi-Galy, S.I.; Papp, E.; Hallberg, D.; Niknafs, N.; Adleff, V.; Noe, M.; Bhattacharya, R.; Novak, M.; Jones, S.; Phallen, J.; et al. High grade serous ovarian carcinomas originate in the fallopian tube. Nat. Commun. 2017, 8, 1093. [Google Scholar] [CrossRef] [PubMed]
- Prat, J.; FIGO Committee on Gynecologic Oncology. Staging classification for cancer of the ovary, fallopian tube, and peritoneum. Int. J. Gynaecol. Obstet. 2014, 124, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Lisio, M.-A.; Fu, L.; Goyeneche, A.; Gao, Z.-H.; Telleria, C. High-Grade Serous Ovarian Cancer: Basic Sciences, Clinical and Therapeutic Standpoints. Int. J. Mol. Sci. 2019, 20, 952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valabrega, G.; Scotto, G.; Tuninetti, V.; Pani, A.; Scaglione, F. Differences in PARP Inhibitors for the Treatment of Ovarian Cancer: Mechanisms of Action, Pharmacology, Safety, and Efficacy. Int. J. Mol. Sci. 2021, 22, 4203. [Google Scholar] [CrossRef] [PubMed]
- Ray-Coquard, I.; Pautier, P.; Pignata, S.; Pérol, D.; González-Martín, A.; Berger, R.; Fujiwara, K.; Vergote, I.; Colombo, N.; Mäenpää, J.; et al. Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2416–2428. [Google Scholar] [CrossRef] [PubMed]
- González-Martín, A.; Pothuri, B.; Vergote, I.; DePont Christensen, R.; Graybill, W.; Mirza, M.R.; McCormick, C.; Lorusso, D.; Hoskins, P.; Freyer, G.; et al. PRIMA/ENGOT-OV26/GOG-3012 Investigators. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2391–2402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, K.; Colombo, N.; Scambia, G.; Kim, B.-G.; Oaknin, A.; Friedlander, M.; Lisyanskaya, A.; Floquet, A.; Leary, A.; Sonke, G.S.; et al. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2018, 379, 2495–2505. [Google Scholar] [CrossRef] [PubMed]
- Swisher, E.M.; Lin, K.K.; Oza, A.; Scott, C.L.; Giordano, H.; Sun, J.; E Konecny, G.; Coleman, R.L.; Tinker, A.V.; O’Malley, D.M.; et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): An international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017, 18, 75–87. [Google Scholar] [CrossRef] [Green Version]
- Konstantinopoulos, P.A.; Norquist, B.; Lacchetti, C.; Armstrong, D.; Grisham, R.N.; Goodfellow, P.J.; Kohn, E.C.; Levine, D.A.; Liu, J.F.; Lu, K.H.; et al. Germline and Somatic Tumor Testing in Epithelial Ovarian Cancer: ASCO Guideline. J. Clin. Oncol. 2020, 38, 1222–1245. [Google Scholar] [CrossRef] [PubMed]
- Colombo, N.; Sessa, C.; du Bois, A.; Ledermann, J.; McCluggage, W.G.; McNeish, I.; Morice, P.; Pignata, S.; Ray-Coquard, I.; Vergote, I.; et al. ESMO-ESGO Ovarian Cancer Consensus Conference Working Group. ESMO-ESGO consensus conference recommendations on ovarian cancer: Pathology and molecular biology, early and advanced stages, borderline tumours and recurrent disease. Ann Oncol. 2019, 30, 672–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gori, S.; Barberis, M.; Bella, M.A.; Buttitta, F.; Capoluongo, E.; Carrera, P.; Colombo, N.; Cortesi, L.; Genuardi, M.; Gion, M.; et al. Recommendations for the implementation of BRCA testing in ovarian cancer patients and their relatives. Crit. Rev. Oncol. Hematol. 2019, 140, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Malapelle, U.; Mayo de-Las-Casas, C.; Rocco, D.; Garzon, M.; Pisapia, P.; Jordana-Ariza, N.; Russo, M.; Sgariglia, R.; De Luca, C.; Pepe, F.; et al. Development of a gene panel for next-generation sequencing of clinically relevant mutations in cell-free DNA from cancer patients. Br. J. Cancer 2017, 116, 802–810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Luca, C.; Pepe, F.; Iaccarino, A.; Pisapia, P.; Righi, L.; Listì, A.; Greco, L.; Gragnano, G.; Campione, S.; De Dominicis, G.; et al. RNA-Based Assay for Next-Generation Sequencing of Clinically Relevant Gene Fusions in Non-Small Cell Lung Cancer. Cancers 2021, 13, 139. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, C.; Guerini-Rocco, E.; Buttitta, F.; Iapicca, P.; You, W.; Mauri, M.; Felicioni, L.; Troncone, G.; Malapelle, U.; Scarpa, A.; et al. Reliability and reproducibility among different platforms for tumour BRCA testing in ovarian cancer: A study of the Italian NGS Network. J. Clin. Pathol. 2020, 74, 668–672. [Google Scholar] [CrossRef] [PubMed]
| Patient | Age | Neoplastic Cells (%) | DNA Concentration (ng/µL) | Reads | Mapped Reads | Percent Read on Target (%) | Average Reads per Amplicon | Uniformity of Amplicon Coverage (%) | Mean Read Length (bp) | Molecular Result | Gene |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 74 | 70.00 | 19.90 | 1,103,756.00 | 1,100,608.00 | 98.15 | 3997.00 | 99.63 | 105.00 | WT | |
| 2 | 55 | 70.00 | 60.00 | 447,320.00 | 445,834.00 | 99.87 | 1615.00 | 98.90 | 104.00 | WT | |
| 3 | 51 | 60.00 | 41.00 | 990,443.00 | 987,075.00 | 99.87 | 2578.00 | 99.27 | 108.00 | WT | |
| 4 | 60 | 70.00 | 5.79 | 999,781.00 | 996,779.00 | 99.08 | 3617.00 | 98.53 | 104.00 | WT | |
| 5 | 42 | 70.00 | 60.00 | 1,225,736.00 | 1,221,759.00 | 99.04 | 4433.00 | 99.63 | 105.00 | p.R1495M | BRCA1 |
| 6 | 66 | 70.00 | 2.41 | 940,118.00 | 937,071.00 | 98.71 | 3388.00 | 97.70 | 101.00 | p.Q534X | BRCA1 |
| 7 | 69 | 70.00 | 25.40 | 1,134,001.00 | 1,129,619.00 | 98.79 | 4088.00 | 98.27 | 104.00 | WT | |
| 8 | 91 | 80.00 | 60.00 | 1,107,249.00 | 1,105,435.00 | 99.09 | 4012.00 | 98.90 | 105.00 | WT | |
| 9 | 72 | 70.00 | 60.00 | 978,740.00 | 977,513.00 | 98.93 | 3542.00 | 96.55 | 104.00 | p.K830PfsTer18 | BRCA1 |
| 10 | 66 | 70.00 | 60.00 | 1,015,943.00 | 1,014,574.00 | 99.33 | 3691.00 | 99.27 | 107.00 | WT | |
| 11 | 53 | 70.00 | 38.90 | 529,337.00 | 528,001.00 | 98.76 | 1910.00 | 99.63 | 106.00 | WT | |
| 12 | 71 | 50.00 | 25.30 | 1,111,471.00 | 1,109,964.00 | 99.28 | 4036.00 | 99.27 | 107.00 | WT | |
| 13 | 63 | 50.00 | 6.19 | 1,091,731.00 | 1,090,470.00 | 99.32 | 3967.00 | 98.53 | 106.00 | WT | |
| 14 | 61 | 70.00 | 60.00 | 1,120,367.00 | 1,118,600.00 | 99.15 | 4063.00 | 98.90 | 112.00 | WT | |
| 15 | 61 | 60.00 | 53.00 | 1,140,727.00 | 1,139,018.00 | 99.06 | 4133.00 | 98.99 | 110.00 | WT | |
| 16 | 25 | 50.00 | 60.00 | 1,052,429.00 | 1,051,081.00 | 99.06 | 3814.00 | 98.90 | 109.00 | WT | |
| 17 | 64 | 60.00 | 17.30 | 538,487.00 | 581,558.00 | 99.05 | 2110.00 | 99.63 | 109.00 | WT | |
| 18 | 68 | 80.00 | 60.00 | 574,298.00 | 572,675.00 | 98.75 | 2065.00 | 99.63 | 111.00 | WT | |
| 19 | 52 | 30.00 | 6.38 | 626,317.00 | 624,168.00 | 98.66 | 2256.00 | 99.63 | 115.00 | WT | |
| 20 | 69 | 70.00 | 60.00 | 667,821.00 | 665,818.00 | 98.91 | 2410.00 | 100.00 | 112.00 | WT | |
| 21 | 58 | 70.00 | 11.20 | 585,530.00 | 584,005.00 | 98.92 | 2116.00 | 99.63 | 112.00 | p.Q 1756PfsTer74 | BRCA1 |
| 22 | 59 | 80.00 | 60.00 | 516,539.00 | 514,737.00 | 98.62 | 1861.00 | 100.00 | 115.00 | WT | |
| 23 | 76 | 60.00 | 43.80 | 175,439.00 | 175,084.00 | 97.54 | 692.20 | 100.00 | 102.00 | WT | |
| 24 | 76 | 70.00 | 57.00 | 195,477.00 | 19,501.00 | 98.68 | 704.90 | 100.00 | 102.00 | WT | |
| 25 | 47 | 70.00 | 60.00 | 233,219.00 | 232,620.00 | 98.90 | 841.10 | 93.84 | 106.00 | WT | |
| 26 | 69 | 90.00 | 60.00 | 222,662.00 | 222,066.00 | 98.90 | 804.40 | 100.00 | 107.00 | p.IVS2 + 1G > A | BRCA2 |
| 27 | 48 | 70.00 | 60.00 | 230,212.00 | 229,629.00 | 100.00 | 832.50 | 98.63 | 104.00 | WT | |
| 28 | 52 | 70.00 | 60.00 | 215,102.00 | 214,804.00 | 99.00 | 778.90 | 100.00 | 105.00 | WT | |
| 29 | 44 | 50.00 | 60.00 | 218,442.00 | 217,974.00 | 99.10 | 791.20 | 100.00 | 107.00 | WT | |
| 30 | 57 | 60.00 | 60.00 | 748,780.00 | 746,568.00 | 98.08 | 2682.00 | 100.00 | 106.00 | WT | |
| 31 | 45 | 50.00 | 32.20 | 633,164.00 | 631,252.00 | 98.12 | 2269.00 | 98.90 | 103.00 | p.N319KfsTer8) | BRCA2 |
| 32 | 73 | 80.00 | 60.00 | 754,043.00 | 702,880.00 | 98.75 | 2542.00 | 100.00 | 105.00 | WT | |
| 33 | 63 | 60.00 | 60.00 | 808,451.00 | 806,608.00 | 98.57 | 2913.00 | 100.00 | 107.00 | WT | |
| 34 | 67 | 70.00 | 33.70 | 482,605.00 | 481,960.00 | 97.97 | 1727.00 | 96.30 | 103.00 | WT | |
| 35 | 39 | 80.00 | 51.00 | 932,611.00 | 931,119.00 | 97.67 | 3331.00 | 99.27 | 102.00 | WT | |
| 36 | 51 | 50.00 | 53.00 | 1,119,066.00 | 1,116,901.00 | 98.26 | 4020.00 | 100.00 | 105.00 | WT | |
| 37 | 66 | 70.00 | 7.15 | 1,020,993.00 | 1,019,060.00 | 98.89 | 3691.00 | 100.00 | 102.00 | p.L1072Ter) | BRCA2 |
| 38 | 56 | 60.00 | 60.00 | 992,136.00 | 990,635.00 | 98.49 | 3574.00 | 100.00 | 102.00 | WT | |
| 39 | 57 | 70.00 | 60.00 | 1,952,836.00 | 1,949,593.00 | 98.64 | 7045.00 | 99.63 | 106.00 | p.T1378Ter | BRCA2 |
| 40 | 77 | 70.00 | 60.00 | 1,584,808.00 | 1,582,139.00 | 98.47 | 5707.00 | 99.27 | 103.00 | WT | |
| 41 | 70 | 20.00 | 13.80 | 689,251.00 | 687,985.00 | 95.00 | 3914.00 | 94.61 | 104.00 | WT | |
| 42 | 60 | 50.00 | 60.00 | 668,595.00 | 667,463.00 | 94.00 | 3757.00 | 94.61 | 105.00 | WT | |
| 43 | 40 | 50.00 | 60.00 | 675,510.00 | 674,213.00 | 95.64 | 3861.00 | 95.21 | 104.00 | WT | |
| 44 | 70 | 50.00 | 22.20 | 505.00 | 505.00 | 97.03 | 2.90 | 91.57 | 105.00 | RIP | |
| 45 | 64 | 50.00 | 10.70 | 683,922.00 | 682,904.00 | 93.36 | 3815.00 | 95.21 | 102.00 | WT | |
| 46 | 52 | 10.00 | 51.00 | 619,053.00 | 617,877.00 | 94.31 | 3489.00 | 94.01 | 103.00 | WT | |
| 47 | 81 | 60.00 | 30.30 | 472,447.00 | 470,375.00 | 94.42 | 2654.00 | 93.41 | 109.00 | WT | |
| 48 | 67 | 70.00 | 60.00 | 204,918.00 | 204,318.00 | 93.72 | 1147.00 | 94.01 | 111.00 | WT | |
| 49 | 33 | 60.00 | 60.00 | 574,974.00 | 572,585.00 | 93.02 | 3189.00 | 95.21 | 109.00 | p.Q1811Ter | BRCA1 |
| 50 | 72 | 70.00 | 60.00 | 13,533,583.00 | 13,391,178.00 | 94.89 | 76,092.00 | 95.81 | 105.00 | WT | |
| 51 | 74 | 40.00 | 5.40 | 9,481,551.00 | 9,408,339.00 | 94.13 | 53,031.00 | 95.21 | 104.00 | WT | |
| 52 | 49 | 50.00 | 19.70 | 12,876,898.00 | 12,766,795.00 | 94.17 | 71,989.00 | 96.41 | 103.00 | WT | |
| 53 | 48 | 60.00 | 60.00 | 13,327,661.00 | 13,176,126.00 | 94.45 | 74,523.00 | 96.41 | 106.00 | WT | |
| 54 | 73 | 60.00 | 60.00 | 1521.00 | 1503.00 | 93.35 | 8.40 | 93.53 | 103.00 | WT | |
| 55 | 53 | 60.00 | 18.80 | 592,625.00 | 590,434.00 | 94.04 | 3325.00 | 96.41 | 122.00 | p.Q2157IfsTer18 | BRCA2 |
| 56 | 76 | 20.00 | 0.60 | 459,548.00 | 458,573.00 | 96.02 | 2637.00 | 96.41 | 106.00 | WT | |
| 57 | 68 | 80.00 | 60.00 | 442,179.00 | 440,884.00 | 94.76 | 2502.00 | 95.81 | 108.00 | WT | |
| 58 | 62 | 50.00 | 8.50 | 382,324.00 | 381,454.00 | 96.15 | 2196.00 | 95.81 | 106.00 | WT | |
| 59 | 54 | 40.00 | 11.60 | 433,234.00 | 431,971.00 | 95.86 | 2480.00 | 95.25 | 111.00 | p.C61G | BRCA1 |
| 60 | 66 | 70.00 | 47.00 | 407,607.00 | 406,640.00 | 95.03 | 2314.00 | 95.03 | 106.00 | WT | |
| 61 | 63 | 20.00 | 3.40 | 276,753.00 | 275,901.00 | 96.05 | 1587.00 | 95.21 | 111.00 | WT | |
| 62 | 62 | 70.00 | 37.80 | 280,573.00 | 279,771.00 | 96.01 | 1609.00 | 95.21 | 104.00 | WT | |
| 63 | 64 | 70.00 | 16.00 | 227,648.00 | 228,989.00 | 95.27 | 1295.00 | 94.01 | 107.00 | WT | |
| 64 | 74 | 40.00 | 4.14 | 275,446.00 | 274,321.00 | 95.03 | 1561.00 | 94.01 | 111.00 | WT | |
| 65 | 61 | 10.00 | 6.90 | 248,198.00 | 247,411.00 | 95.53 | 1415.00 | 95.21 | 112.00 | WT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations All authors have read and agreed to the published version of the manuscript.. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pepe, F.; Pisapia, P.; Russo, G.; Nacchio, M.; Pallante, P.; Vigliar, E.; De Angelis, C.; Insabato, L.; Bellevicine, C.; De Placido, S.; et al. BRCA1/2 NGS Somatic Testing in Clinical Practice: A Short Report. Genes 2021, 12, 1917. https://doi.org/10.3390/genes12121917
Pepe F, Pisapia P, Russo G, Nacchio M, Pallante P, Vigliar E, De Angelis C, Insabato L, Bellevicine C, De Placido S, et al. BRCA1/2 NGS Somatic Testing in Clinical Practice: A Short Report. Genes. 2021; 12(12):1917. https://doi.org/10.3390/genes12121917
Chicago/Turabian StylePepe, Francesco, Pasquale Pisapia, Gianluca Russo, Mariantonia Nacchio, Pierlorenzo Pallante, Elena Vigliar, Carmine De Angelis, Luigi Insabato, Claudio Bellevicine, Sabino De Placido, and et al. 2021. "BRCA1/2 NGS Somatic Testing in Clinical Practice: A Short Report" Genes 12, no. 12: 1917. https://doi.org/10.3390/genes12121917
APA StylePepe, F., Pisapia, P., Russo, G., Nacchio, M., Pallante, P., Vigliar, E., De Angelis, C., Insabato, L., Bellevicine, C., De Placido, S., Troncone, G., & Malapelle, U. (2021). BRCA1/2 NGS Somatic Testing in Clinical Practice: A Short Report. Genes, 12(12), 1917. https://doi.org/10.3390/genes12121917













