Identification of miR-199-5p and miR-199-3p Target Genes: Paxillin Facilities Cancer Cell Aggressiveness in Head and Neck Squamous Cell Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of Putative Targets Controlled by miR-199-5p and miR-199-3p in HNSCC Cells
2.2. Gene set Enrichment Analysis (GSEA)
2.3. HNSCC Cell Lines
2.4. Transfection of Mature miRNAs and Small-Interfering RNAs (siRNAs) into HNSCC Cells
2.5. RNA Extraction and Quantitative Reverse-Transcription PCR (qRT-PCR)
2.6. Immunohistochemistry
2.7. Western Blotting
2.8. Dual Luciferase Reporter Assays
2.9. Cell Proliferation, Migration, and Invasion Assays in HNSCC Cells
2.10. Statistical Analysis
3. Results
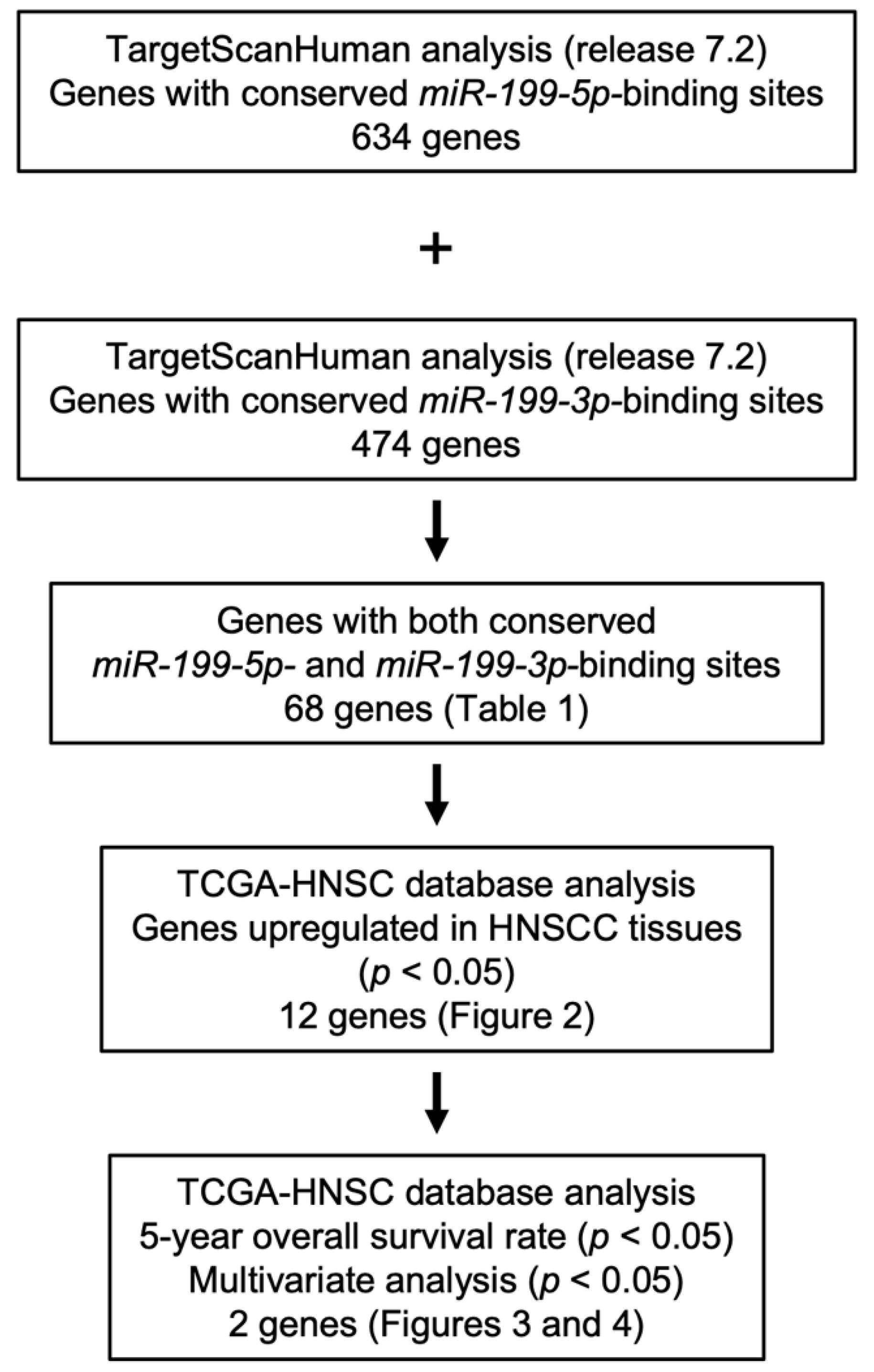
3.1. Identification of miR-199-5p and miR-199-3p Coordinately Regulated Genes in HNSCC Cells
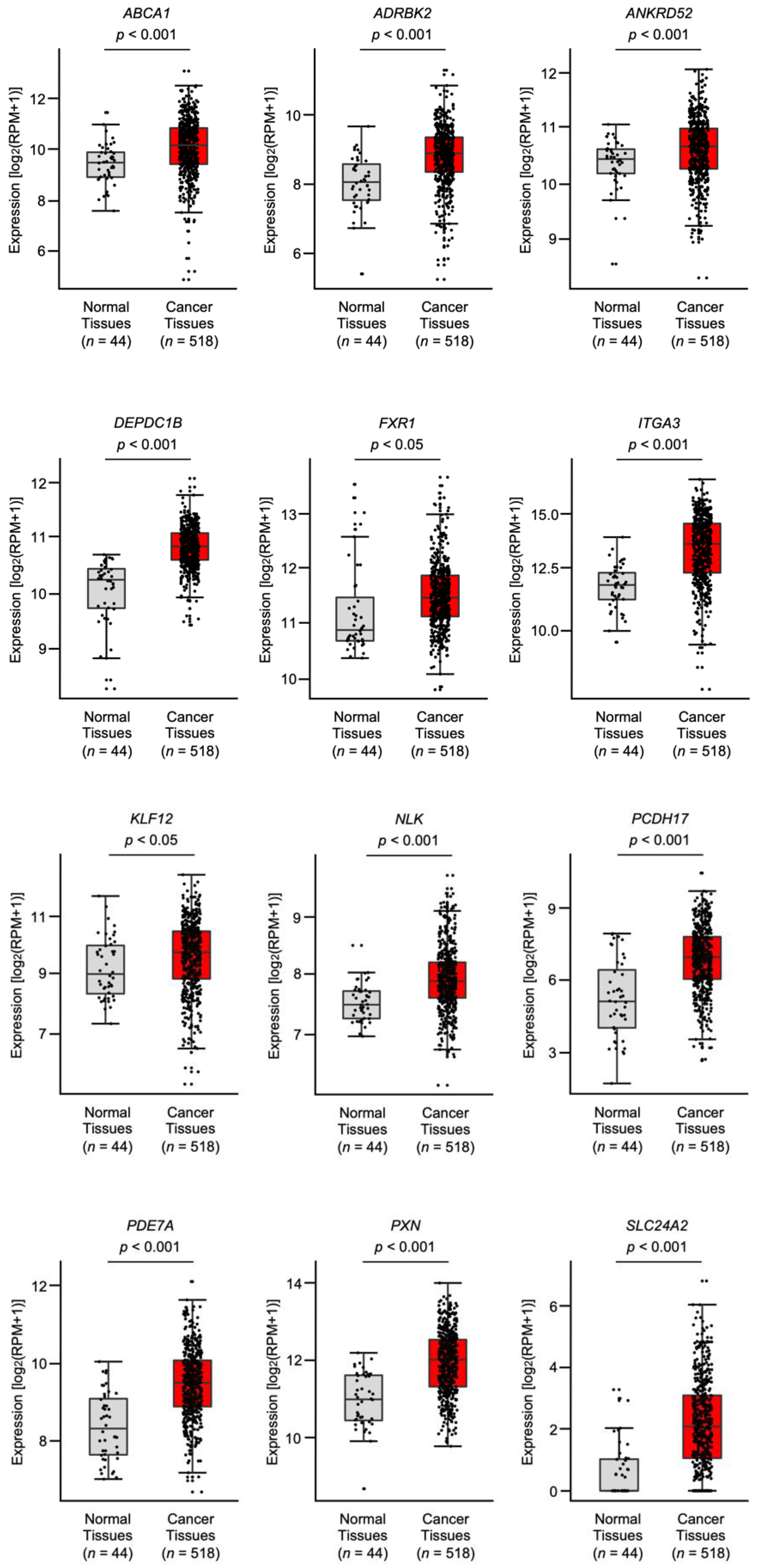
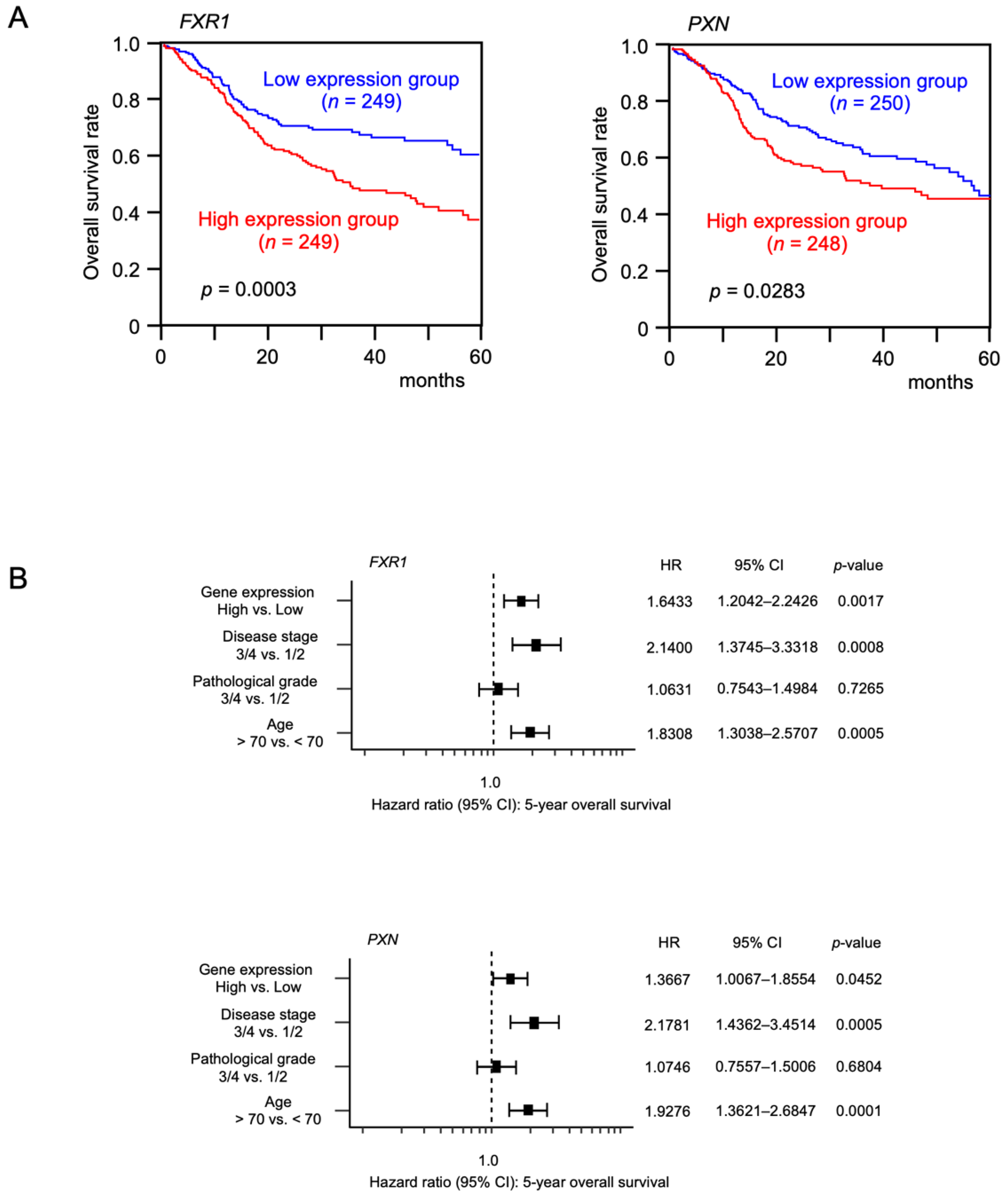
3.2. Expression and Clinical Significance of the Putative Target Genes in Patients with HNSCC according to TCGA Analysis
3.3. FXR1- and PXN-Mediated Pathways in HNSCC Cells
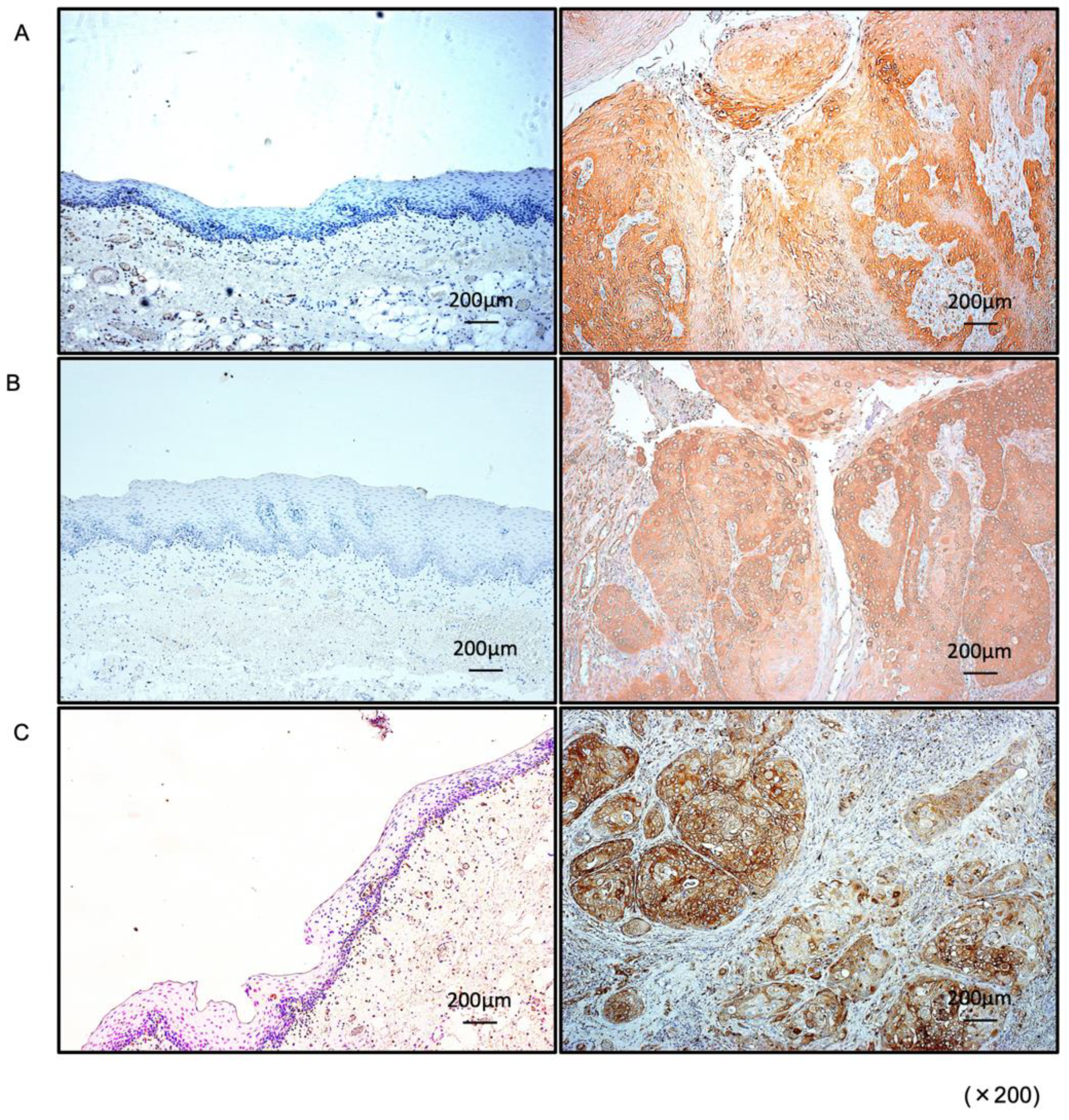
3.4. Expression of PXN in HNSCC Clinical Specimens
3.5. Regulation of PXN Expression by miR-199-5p and miR-199-3p in HNSCC Cells
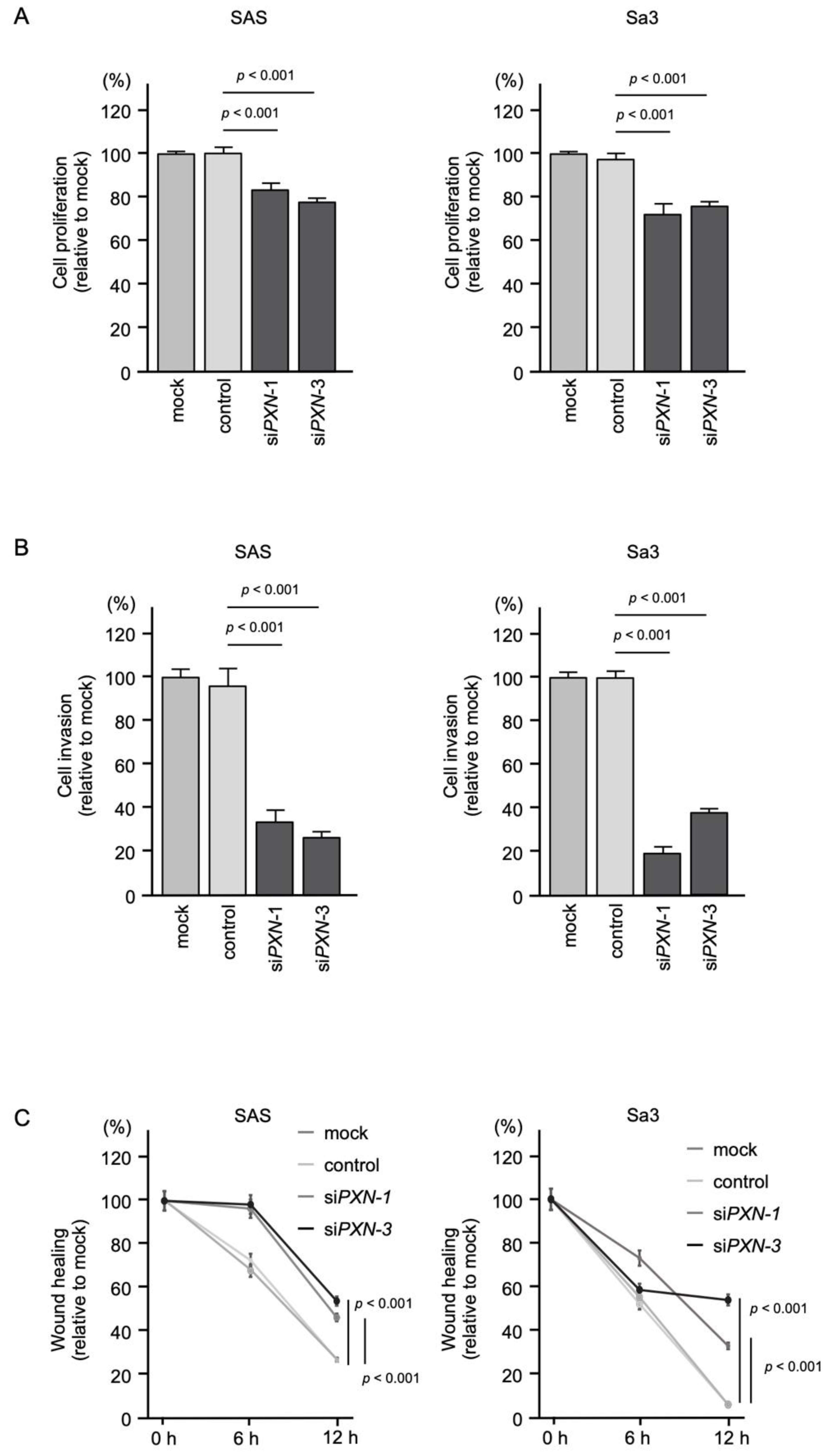
3.6. Effects of PXN Knockdown on the Proliferation, Invasion and Migration of HNSCC Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar] [CrossRef]
- Cramer, J.D.; Burtness, B.; Le, Q.T.; Ferris, R.L. The changing therapeutic landscape of head and neck cancer. Nat. Rev. Clin. Oncol. 2019, 16, 669–683. [Google Scholar] [CrossRef]
- Mehanna, H.; Paleri, V.; West, C.M.; Nutting, C. Head and neck cancer—Part 1: Epidemiology, presentation, and preservation. Clin. Otolaryngol. 2011, 36, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Sha, J.; Bai, Y.; Ngo, H.X.; Okui, T.; Kanno, T. Overview of Evidence-Based Chemotherapy for Oral Cancer: Focus on Drug Resistance Related to the Epithelial-Mesenchymal Transition. Biomolecules 2021, 11, 893. [Google Scholar] [CrossRef]
- Chow, L.Q.M. Head and Neck Cancer. N. Engl. J. Med. 2020, 382, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Leemans, C.R.; Braakhuis, B.J.; Brakenhoff, R.H. The molecular biology of head and neck cancer. Nat. Rev. Cancer 2011, 11, 9–22. [Google Scholar] [CrossRef]
- Cohen, E.E.W.; Soulières, D.; Le Tourneau, C.; Dinis, J.; Licitra, L.; Ahn, M.J.; Soria, A.; Machiels, J.P.; Mach, N.; Mehra, R.; et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): A randomised, open-label, phase 3 study. Lancet 2019, 393, 156–167. [Google Scholar] [CrossRef]
- Betel, D.; Wilson, M.; Gabow, A.; Marks, D.S.; Sander, C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008, 36, D149–D153. [Google Scholar] [CrossRef] [Green Version]
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical utility of circulating non-coding RNAs—An update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef]
- Nelson, K.M.; Weiss, G.J. MicroRNAs and cancer: Past, present, and potential future. Mol. Cancer Ther. 2008, 7, 3655–3660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krek, A.; Grun, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef] [Green Version]
- Koshizuka, K.; Hanazawa, T.; Arai, T.; Okato, A.; Kikkawa, N.; Seki, N. Involvement of aberrantly expressed microRNAs in the pathogenesis of head and neck squamous cell carcinoma. Cancer Metastasis Rev. 2017, 36, 525–545. [Google Scholar] [CrossRef]
- Koshizuka, K.; Hanazawa, T.; Kikkawa, N.; Arai, T.; Okato, A.; Kurozumi, A.; Kato, M.; Katada, K.; Okamoto, Y.; Seki, N. Regulation of ITGA3 by the anti-tumor miR-199 family inhibits cancer cell migration and invasion in head and neck cancer. Cancer Sci. 2017, 108, 1681–1692. [Google Scholar] [CrossRef] [Green Version]
- Koma, A.; Asai, S.; Minemura, C.; Oshima, S.; Kinoshita, T.; Kikkawa, N.; Koshizuka, K.; Moriya, S.; Kasamatsu, A.; Hanazawa, T.; et al. Impact of Oncogenic Targets by Tumor-Suppressive miR-139-5p and miR-139-3p Regulation in Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2021, 22, 9947. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Anaya, J. Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. Peer J. Comput. Sci. 2016, 2, e67. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef] [PubMed]
- Oshima, S.; Asai, S.; Seki, N.; Minemura, C.; Kinoshita, T.; Goto, Y.; Kikkawa, N.; Moriya, S.; Kasamatsu, A.; Hanazawa, T.; et al. Identification of Tumor Suppressive Genes Regulated by miR-31-5p and miR-31-3p in Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2021, 22, 6199. [Google Scholar] [CrossRef]
- Hong, M.; Tao, S.; Zhang, L.; Diao, L.T.; Huang, X.; Huang, S.; Xie, S.J.; Xiao, Z.D.; Zhang, H. RNA sequencing: New technologies and applications in cancer research. J. Hematol. Oncol. 2020, 13, 166. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Koshizuka, K.; Hanazawa, T.; Kikkawa, N.; Okato, A.; Idichi, T.; Arai, T.; Sugawara, S.; Katada, K.; Okamoto, Y.; et al. Passenger strand of miR-145-3p acts as a tumor-suppressor by targeting MYO1B in head and neck squamous cell carcinoma. Int. J. Oncol. 2018, 52, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Okada, R.; Koshizuka, K.; Yamada, Y.; Moriya, S.; Kikkawa, N.; Kinoshita, T.; Hanazawa, T.; Seki, N. Regulation of Oncogenic Targets by miR-99a-3p (Passenger Strand of miR-99a-Duplex) in Head and Neck Squamous Cell Carcinoma. Cells 2019, 8, 1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitra, R.; Adams, C.M.; Jiang, W.; Greenawalt, E.; Eischen, C.M. Pan-cancer analysis reveals cooperativity of both strands of microRNA that regulate tumorigenesis and patient survival. Nat. Commun. 2020, 11, 968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Ye, B.; Wang, P.; Yao, F.; Zhang, C.; Yu, G. Overview of microRNA-199a Regulation in Cancer. Cancer Manag. Res. 2019, 11, 10327–10335. [Google Scholar] [CrossRef] [Green Version]
- Wei, D.; Wang, W.; Shen, B.; Zhou, Y.; Yang, X.; Lu, G.; Yang, J.; Shao, Y. MicroRNA-199a-5p suppresses migration and invasion in oral squamous cell carcinoma through inhibiting the EMT-related transcription factor SOX4. Int. J. Mol. Med. 2019, 44, 185–195. [Google Scholar] [CrossRef] [Green Version]
- Wei, D.; Shen, B.; Wang, W.; Zhou, Y.; Yang, X.; Lu, G.; Yang, J.; Shao, Y. MicroRNA-199a-5p functions as a tumor suppressor in oral squamous cell carcinoma via targeting the IKKβ/NF-κB signaling pathway. Int. J. Mol. Med. 2019, 43, 1585–1596. [Google Scholar] [CrossRef] [Green Version]
- Jin, X.; Zhai, B.; Fang, T.; Guo, X.; Xu, L. FXR1 is elevated in colorectal cancer and acts as an oncogene. Tumour Biol. 2016, 37, 2683–2690. [Google Scholar] [CrossRef]
- Qian, J.; Chen, H.; Ji, X.; Eisenberg, R.; Chakravarthy, A.B.; Mayer, I.A.; Massion, P.P. A 3q gene signature associated with triple negative breast cancer organ specific metastasis and response to neoadjuvant chemotherapy. Sci. Rep. 2017, 7, 45828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comtesse, N.; Keller, A.; Diesinger, I.; Bauer, C.; Kayser, K.; Huwer, H.; Lenhof, H.P.; Meese, E. Frequent overexpression of the genes FXR1, CLAPM1 and EIF4G located on amplicon 3q26-27 in squamous cell carcinoma of the lung. Int. J. Cancer 2007, 120, 2538–2544. [Google Scholar] [CrossRef]
- Qie, S.; Majumder, M.; Mackiewicz, K.; Howley, B.V.; Peterson, Y.K.; Howe, P.H.; Palanisamy, V.; Diehl, J.A. Fbxo4-mediated degradation of Fxr1 suppresses tumorigenesis in head and neck squamous cell carcinoma. Nat. Commun. 2017, 8, 1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majumder, M.; House, R.; Palanisamy, N.; Qie, S.; Day, T.A.; Neskey, D.; Diehl, J.A.; Palanisamy, V. RNA-Binding Protein FXR1 Regulates p21 and TERC RNA to Bypass p53-Mediated Cellular Senescence in OSCC. PLoS Genet. 2016, 12, e1006306. [Google Scholar] [CrossRef] [Green Version]
- Majumder, M.; Palanisamy, V. RNA binding protein FXR1-miR301a-3p axis contributes to p21WAF1 degradation in oral cancer. PLoS Genet. 2020, 16, e1008580. [Google Scholar] [CrossRef] [Green Version]
- Brown, M.C.; Turner, C.E. Paxillin: Adapting to change. Physiol. Rev. 2004, 84, 1315–1339. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.K.; Hanson, D.A.; Schlaepfer, D.D. Focal adhesion kinase: In command and control of cell motility. Nat. Rev. Mol. Cell Biol. 2005, 6, 56–68. [Google Scholar] [CrossRef]
- Wozniak, M.A.; Modzelewska, K.; Kwong, L.; Keely, P.J. Focal adhesion regulation of cell behavior. Biochim. Biophys. Acta 2004, 1692, 103–119. [Google Scholar] [CrossRef]
- Zhou, J.; Yi, Q.; Tang, L. The roles of nuclear focal adhesion kinase (FAK) on Cancer: A focused review. J. Exp. Clin. Cancer Res. 2019, 38, 250. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zhao, H.; Xiao, Y.; Shen, P.; Tan, L.; Zhang, S.; Liu, Q.; Gao, Z.; Zhao, J.; Zhao, Y.; et al. Pan-cancer analysis reveals an immunological role and prognostic potential of PXN in human cancer. Aging 2021, 13, 16248–16266. [Google Scholar] [CrossRef] [PubMed]
- Deakin, N.O.; Pignatelli, J.; Turner, C.E. Diverse roles for the paxillin family of proteins in cancer. Genes Cancer 2012, 3, 362–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.; Zhang, H.; Hu, X.; Li, W. Knockdown of long non-coding RNA XIST inhibits cell viability and invasion by regulating miR-137/PXN axis in non-small cell lung cancer. Int. J. Biol. Macromol. 2018, 111, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Jia, Y.; Chen, H.; Chen, W.; Zhou, X. Long non-coding RNA PXN-AS1 suppresses pancreatic cancer progression by acting as a competing endogenous RNA of miR-3064 to upregulate PIP4K2B expression. J. Exp. Clin. Cancer Res. 2019, 38, 390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, J.H.; Liu, X.N.; Wang, T.T.; Pan, W.; Tao, Q.F.; Zhou, W.P.; Wang, F.; Sun, S.H. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat. Cell Biol. 2017, 19, 820–832. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, P.; Sun, H.; Liu, Y. LINC01094/miR-577 axis regulates the progression of ovarian cancer. J. Ovarian Res. 2020, 13, 122. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.S.; Long, H.; Zhao, L.; Li, H.X.; Zhang, X.N. LncRNA HOTTIP promotes proliferation and inhibits apoptosis of gastric carcinoma cells via adsorbing miR-615-3p. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 6692–6698. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhang, C.; Lian, H.; Xie, L.; Xue, J.; Yin, N.; Guan, F. LncRNA linc00460 sponges miR-1224-5p to promote esophageal cancer metastatic potential and epithelial-mesenchymal transition. Pathol. Res. Pract. 2020, 216, 153026. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.Y.; Zhang, K.; Liu, G.Q. LncRNA LINP1 promotes malignant progression of pancreatic cancer by adsorbing microRNA-491-3p. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 9315–9324. [Google Scholar] [CrossRef]
- Li, P.; Wang, L.; Li, P.; Hu, F.; Cao, Y.; Tang, D.; Ye, G.; Li, H.; Wang, D. Silencing lncRNA XIST exhibits antiproliferative and proapoptotic effects on gastric cancer cells by up-regulating microRNA-132 and down-regulating PXN. Aging 2020, 13, 14469–14481. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Wang, Y.; Zhang, Y.; Zhang, J.; Zhang, L.; Li, J. LncRNA DLX6-AS1 Contributes to Epithelial-Mesenchymal Transition and Cisplatin Resistance in Triple-negative Breast Cancer via Modulating Mir-199b-5p/Paxillin Axis. Cell Transplant. 2020, 29, 963689720929983. [Google Scholar] [CrossRef]
- Liu, Z.; Mao, L.; Wang, L.; Zhang, H.; Hu, X. miR-218 functions as a tumor suppressor gene in cervical cancer. Mol. Med. Rep. 2020, 21, 209–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; He, J.; Shao, M.; Cui, B.; Peng, F.; Li, J.; Ran, Y.; Jin, D.; Kong, J.; Chang, J.; et al. Downregulation of miR-218-5p promotes invasion of oral squamous cell carcinoma cells via activation of CD44-ROCK signaling. Biomed. Pharmacother. 2018, 106, 646–654. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.W.; Chuang, C.Y.; Lin, W.L.; Sung, W.W.; Cheng, Y.W.; Lee, H. Paxillin promotes tumor progression and predicts survival and relapse in oral cavity squamous cell carcinoma by microRNA-218 targeting. Carcinogenesis 2014, 35, 1823–1829. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Hanazawa, T.; Nohata, N.; Kikkawa, N.; Enokida, H.; Yoshino, H.; Yamasaki, T.; Hidaka, H.; Nakagawa, M.; Okamoto, Y.; et al. Tumor suppressive microRNA-218 inhibits cancer cell migration and invasion through targeting laminin-332 in head and neck squamous cell carcinoma. Oncotarget 2012, 3, 1386–1400. [Google Scholar] [CrossRef] [Green Version]

| Entrez Gene ID | Gene Symbol | Gene Name | miR-199-5p Total Conserved Sites | miR-199-3p Total Conserved Sites |
|---|---|---|---|---|
| 19 | ABCA1 | ATP-binding cassette, sub-family A (ABC1), member 1 | 1 | 1 |
| 340485 | ACER2 | alkaline ceramidase 2 | 1 | 1 |
| 92 | ACVR2A | activin A receptor, type IIA | 1 | 2 |
| 93 | ACVR2B | activin A receptor, type IIB | 3 | 1 |
| 57188 | ADAMTSL3 | ADAMTS-like 3 | 1 | 3 |
| 120 | ADD3 | adducin 3 (γ) | 1 | 1 |
| 157 | ADRBK2 | adrenergic, beta, receptor kinase 2 | 1 | 1 |
| 81573 | ANKRD13C | ankyrin repeat domain 13C | 1 | 1 |
| 283373 | ANKRD52 | ankyrin repeat domain 52 | 1 | 1 |
| 57569 | ARHGAP20 | Rho GTPase activating protein 20 | 1 | 1 |
| 23365 | ARHGEF12 | Rho guanine nucleotide exchange factor (GEF) 12 | 1 | 1 |
| 222255 | ATXN7L1 | ataxin 7-like 1 | 1 | 1 |
| 27443 | CECR2 | cat eye syndrome chromosome region, candidate 2 | 2 | 1 |
| 10659 | CELF2 | CUGBP, Elav-like family member 2 | 1 | 2 |
| 387119 | CEP85L | centrosomal protein 85kDa-like | 1 | 1 |
| 153222 | CREBRF | CREB3 regulatory factor | 1 | 1 |
| 51232 | CRIM1 | cysteine rich transmembrane BMP regulator 1 (chordin-like) | 1 | 1 |
| 1496 | CTNNA2 | catenin (cadherin-associated protein), α2 | 1 | 1 |
| 84301 | DDI2 | DNA-damage inducible 1 homolog 2 (S. cerevisiae) | 1 | 1 |
| 55789 | DEPDC1B | DEP domain containing 1B | 1 | 1 |
| 2066 | ERBB4 | v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4 | 1 | 2 |
| 55137 | FIGN | fidgetin | 1 | 1 |
| 23767 | FLRT3 | fibronectin leucine rich transmembrane protein 3 | 1 | 1 |
| 10690 | FUT9 | fucosyltransferase 9 (α (1,3) fucosyltransferase) | 1 | 1 |
| 8087 | FXR1 | fragile X mental retardation, autosomal homolog 1 | 2 | 1 |
| 2651 | GCNT2 | glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) | 1 | 1 |
| 54891 | INO80D | INO80 complex subunit D | 1 | 1 |
| 3675 | ITGA3 | integrin, α3 (antigen CD49C, α 3 subunit of VLA-3 receptor) | 1 | 1 |
| 8516 | ITGA8 | integrin, α8 | 1 | 1 |
| 11278 | KLF12 | Kruppel-like factor 12 | 1 | 1 |
| 26249 | KLHL3 | kelch-like family member 3 | 1 | 3 |
| 84458 | LCOR | ligand dependent nuclear receptor corepressor | 2 | 2 |
| 10960 | LMAN2 | lectin, mannose-binding 2 | 1 | 1 |
| 84061 | MAGT1 | magnesium transporter 1 | 1 | 1 |
| 4217 | MAP3K5 | mitogen-activated protein kinase 5 | 1 | 1 |
| 5599 | MAPK8 | mitogen-activated protein kinase 8 | 1 | 1 |
| 90411 | MCFD2 | multiple coagulation factor deficiency 2 | 1 | 1 |
| 54842 | MFSD6 | major facilitator superfamily domain containing 6 | 1 | 1 |
| 51701 | NLK | nemo-like kinase | 3 | 1 |
| 57532 | NUFIP2 | nuclear fragile X mental retardation protein interacting protein 2 | 1 | 1 |
| 10298 | PAK4 | p21 protein (Cdc42/Rac)-activated kinase 4 | 1 | 2 |
| 27253 | PCDH17 | protocadherin 17 | 1 | 1 |
| 5150 | PDE7A | phosphodiesterase 7A | 1 | 1 |
| 57475 | PLEKHH1 | pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 | 1 | 1 |
| 5495 | PPM1B | protein phosphatase, Mg2+/Mn2+ dependent, 1B | 1 | 1 |
| 55607 | PPP1R9A | protein phosphatase 1, regulatory subunit 9A | 1 | 1 |
| 63976 | PRDM16 | PR domain containing 16 | 1 | 1 |
| 5813 | PURA | purine-rich element binding protein A | 1 | 1 |
| 5829 | PXN | paxillin | 1 | 1 |
| 5925 | RB1 | retinoblastoma 1 | 1 | 1 |
| 54502 | RBM47 | RNA binding motif protein 47 | 1 | 1 |
| 5991 | RFX3 | regulatory factor X, 3 (influences HLA class II expression) | 1 | 1 |
| 6096 | RORB | RAR-related orphan receptor B | 1 | 2 |
| 9644 | SH3PXD2A | SH3 and PX domains 2A | 1 | 1 |
| 25769 | SLC24A2 | solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 | 1 | 1 |
| 8303 | SNN | stannin | 1 | 1 |
| 6667 | SP1 | Sp1 transcription factor | 1 | 1 |
| 257397 | TAB3 | TGF-β activated kinase 1/MAP3K7 binding protein 3 | 1 | 1 |
| 57551 | TAOK1 | TAO kinase 1 | 1 | 2 |
| 10099 | TSPAN3 | tetraspanin 3 | 1 | 1 |
| 57695 | USP37 | ubiquitin specific peptidase 37 | 1 | 1 |
| 23063 | WAPAL | wings apart-like homolog (Drosophila) | 1 | 2 |
| 10472 | ZBTB18 | zinc finger and BTB domain containing 18 | 1 | 2 |
| 26137 | ZBTB20 | zinc finger and BTB domain containing 20 | 2 | 2 |
| 6935 | ZEB1 | zinc finger E-box binding homeobox 1 | 1 | 1 |
| 80139 | ZNF703 | zinc finger protein 703 | 1 | 1 |
| 374655 | ZNF710 | zinc finger protein 710 | 1 | 1 |
| 283337 | ZNF740 | zinc finger protein 740 | 1 | 1 |
| A. Significantly Enriched Gene Sets in the High FXR1 Expression Group | ||
|---|---|---|
| Name | Normalized Enrichment Score | FDR q-Value |
| KEGG_Cardiac muscle contraction | 2.009 | 0.001 |
| KEGG_Dilated cardiomyopathy | 1.968 | 0.001 |
| KEGG_Hypertrophic cardiomyopathy HCM | 1.929 | 0.003 |
| KEGG_Maturity onset diabetes of the young | 1.918 | 0.002 |
| B. Significantly enriched gene sets in the high PXN expression group | ||
| Name | normalized enrichment score | FDR q-value |
| KEGG_Focal adhesion | 2.458 | q < 0.001 |
| KEGG_ECM receptor interaction | 2.316 | q < 0.001 |
| KEGG_Cytosolic DNA sensing pathway | 2.117 | 0.001 |
| KEGG_Proteosome | 2.029 | 0.003 |
| KEGG_Hypetrophic cardiomyopathy HCM | 1.995 | 0.005 |
| KEGG_NOD-like receptor signaling pathway | 1.896 | 0.009 |
| KEGG_Viral myocarditis | 1.894 | 0.008 |
| KEGG_Dilated Cardiomyopathy | 1.804 | 0.014 |
| KEGG_Bladder cancer | 1.784 | 0.016 |
| KEGG_Cytokine cytokine receptor interaction | 1.776 | 0.016 |
| KEGG_JAK/STAT signaling pathway | 1.712 | 0.026 |
| KEGG_RIG-I-like receptor signaling pathway | 1.673 | 0.033 |
| KEGG_Small cell lung cancer | 1.644 | 0.038 |
| KEGG_Arryhythmogenic right ventricular cardiomyopathy ARVC | 1.639 | 0.037 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanaka, N.; Minemura, C.; Asai, S.; Kikkawa, N.; Kinoshita, T.; Oshima, S.; Koma, A.; Kasamatsu, A.; Hanazawa, T.; Uzawa, K.; et al. Identification of miR-199-5p and miR-199-3p Target Genes: Paxillin Facilities Cancer Cell Aggressiveness in Head and Neck Squamous Cell Carcinoma. Genes 2021, 12, 1910. https://doi.org/10.3390/genes12121910
Tanaka N, Minemura C, Asai S, Kikkawa N, Kinoshita T, Oshima S, Koma A, Kasamatsu A, Hanazawa T, Uzawa K, et al. Identification of miR-199-5p and miR-199-3p Target Genes: Paxillin Facilities Cancer Cell Aggressiveness in Head and Neck Squamous Cell Carcinoma. Genes. 2021; 12(12):1910. https://doi.org/10.3390/genes12121910
Chicago/Turabian StyleTanaka, Nozomi, Chikashi Minemura, Shunichi Asai, Naoko Kikkawa, Takashi Kinoshita, Sachi Oshima, Ayaka Koma, Atsushi Kasamatsu, Toyoyuki Hanazawa, Katsuhiro Uzawa, and et al. 2021. "Identification of miR-199-5p and miR-199-3p Target Genes: Paxillin Facilities Cancer Cell Aggressiveness in Head and Neck Squamous Cell Carcinoma" Genes 12, no. 12: 1910. https://doi.org/10.3390/genes12121910
APA StyleTanaka, N., Minemura, C., Asai, S., Kikkawa, N., Kinoshita, T., Oshima, S., Koma, A., Kasamatsu, A., Hanazawa, T., Uzawa, K., & Seki, N. (2021). Identification of miR-199-5p and miR-199-3p Target Genes: Paxillin Facilities Cancer Cell Aggressiveness in Head and Neck Squamous Cell Carcinoma. Genes, 12(12), 1910. https://doi.org/10.3390/genes12121910






