Diversity, Dispersal and Mode of Reproduction of Amanita exitialis in Southern China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. DNA Extraction, Primer Selection and SNP Markers
2.3. Data and Statistical Analysis
2.3.1. Analyses of Molecular Variance (AMOVA)
2.3.2. STRUCTURE for Population Genetic Structure Analysis
2.3.3. Genetic Relationships among Fruiting Bodies
2.3.4. Mode of Reproduction in Nature
3. Results
3.1. Distributions of Multilocus Genotypes within and among Sites
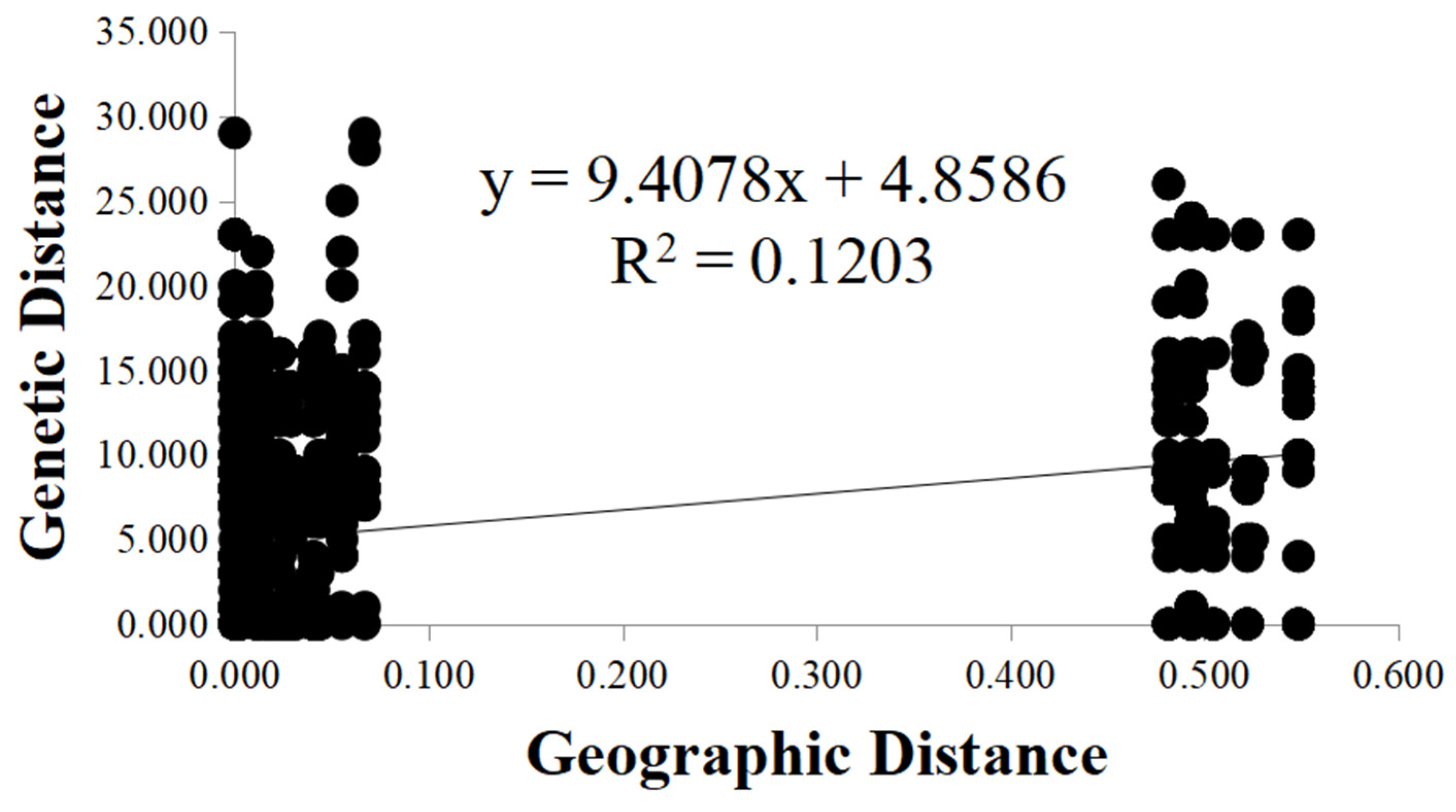
3.2. AMOVA Analysis and Mantel Test Results
3.3. Genetic Clustering
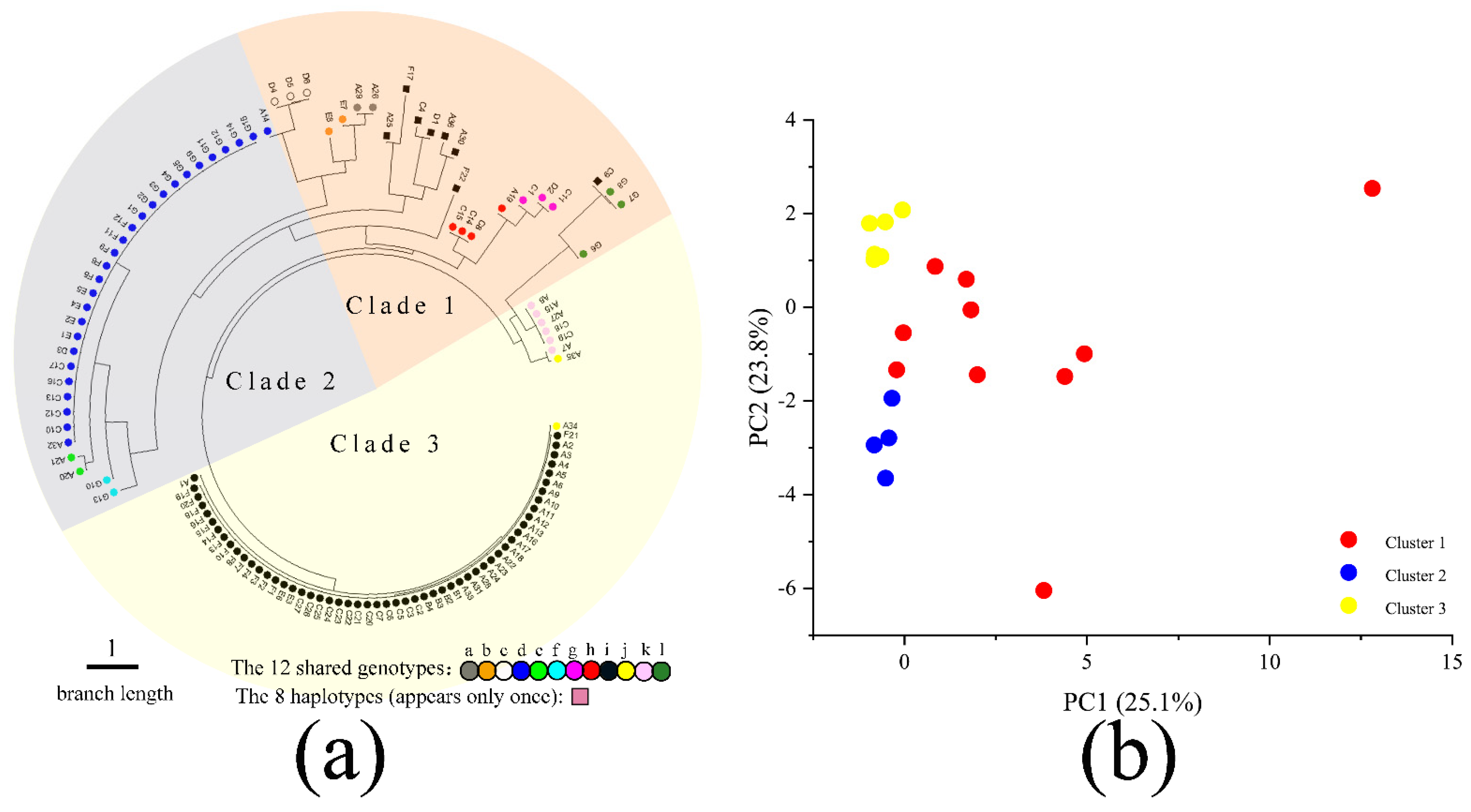
3.4. Neighbor-Joining (NJ) Phylogenetic Tree and PCA Analysis
3.5. Hardy–Weinberg Equilibrium Test
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, W.W.; Pires, S.M.; Liu, Z.T.; Liang, J.J.; Wang, Y.F.; Chen, W.; Liu, C.W.; Liu, J.K.; Han, H.H.; Fu, P.; et al. Vital surveillances: Mushroom poisoning outbreaks—China, 2010–2020. China CDC Wkly. 2021, 3, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhou, L.W.; Yang, Z.L.; Bau, T.; Li, T.H.; Dai, Y.C. Resource diversity of Chinese macrofungi: Edible, medicinal and poisonous species. Fungal Divers. 2019, 98, 1–76. [Google Scholar] [CrossRef]
- Cui, Y.Y.; Cai, Q.; Tang, L.P.; Liu, J.W.; Yang, Z.L. The family Amanitaceae: Molecular phylogeny, higher-rank taxonomy and the species in China. Fungal Divers. 2018, 91, 5–230. [Google Scholar] [CrossRef]
- Cai, Q.; Cui, Y.Y.; Yang, Z.L. Lethal amanita species in China. Mycological 2016, 108, 993–1009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.H. Poisonous Mushrooms: Recognition and Poisoning Treatment; Better Health Channel: Beijing, China, 2016; pp. 1–308. [Google Scholar]
- Chen, Z.H. New advances in researches on poisonous mushrooms since 2000. Mycosystema 2014, 33, 493–516. [Google Scholar] [CrossRef]
- Unluoglu, I.; Tayfur, M. Mushroom poisoning: An analysis of the data between 1996 and 2000. Eur. J. Emerg. Med. 2003, 10, 23–26. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.M.; Hall, M.J.; Falkland, M.M.; Strasser, S.I.; Buckley, N.A. Amanita phalloides poisoning and treatment with silibinin in the Australian Capital Territory and New South Wales. Med. J. Aust. 2013, 198, 43–47. [Google Scholar] [CrossRef] [Green Version]
- Ward, J.; Kapadia, K.; Brush, E.; Salhanick, S.D. Amatoxin poisoning: Case reports and review of current therapies. J. Emerg. Med. 2013, 44, 116–121. [Google Scholar] [CrossRef]
- Liu, M.; Gan, L.X.; Zhang, L.P.; Li, T.H.; Deng, W.Q. Optimization of Ultrasonic Extraction of α-Amanitin from Amanita exitialis by Box-Benhnken Response Surface Design. Acta Edulis Fungi 2021, 28, 93–101. (In Chinese) [Google Scholar]
- Chen, Z.H.; Zhang, P.; Zhang, Z.G. Investigation and analysis of 102 mushroom poisoning cases in Southern China from 1994 to 2012. Fungal Divers. 2014, 64, 123–131. [Google Scholar] [CrossRef]
- Li, P.; Deng, W.Q.; Li, T.H. Molecular cloning of α-amanitin and characterization of its expression pattern in different parts and development stages of Amanita exitialis fruitbody. Mycol. Prog. 2014, 13, 988. [Google Scholar] [CrossRef]
- Chen, Z.H.; Hu, J.S.; Zhang, Z.G.; Zhang, P.; Li, D.P. Determination and analysis of the main amatoxins and phallotoxins in 28 species of Amanita from China. Mycosystema 2003, 22, 565–573. (In Chinese) [Google Scholar] [PubMed]
- Parnmen, S.; Sikaphan, S.; Leudang, S.; Boonpratuang, T.; Rangsiruji, A.; Naksuwankul, K. Molecular identification of poisonous mushrooms using nuclear ITS region and peptide toxins: A retrospective study on fatal cases in Thailand. J. Toxicol. Sci. 2016, 41, 65–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Li, H.J.; Zhang, H.S.; Zhang, Y.Z.; Sun, C.Y. Investigating and analyzing three cohorts of mushroom poisoning caused by Amanita exitialis in Yunnan, China. Hum. Exp. Toxicol. 2017, 37, 665–678. [Google Scholar] [CrossRef]
- Li, H.J.; Zhang, H.S.; Zhang, Y.Z.; Zhang, K.P.; Zhou, J.; Yin, Y.; Jiang, S.F.; Ma, P.B.; He, Q.; Zhang, Y.T.; et al. Preplanned Studies: Mushroom Poisoning Outbreaks—China, 2019. China CDC Wkly. 2020, 2, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.J.; Yao, Q.M.; Li, H.J.; Pu, Y.; Li, C.H.; Yu, C.M. Detection and analysis of cyclopeptides in Amanita exitialis from Chuxiong Prefecture, Yunnan Province. Mycosystema 2020, 39, 1766–1773. (In Chinese) [Google Scholar]
- Hu, J.S. The Determination, Isolation, Identification and Function of Main Peptide Toxins in Amanita exitialis. Ph.D. Thesis, Hunan Normal University, Changsha, China, 2013. [Google Scholar]
- Zhang, P.; Chen, Z.H.; Hu, J.S.; Wei, B.Y.; Zhang, Z.G.; Hu, W.Q. Production and characterization of Amanitin toxins from a pure culture of Amanita exitialis. FEMS Microbiol. Lett. 2005, 252, 223–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.L.; Li, T.H. Notes on three white Amanitae of section Phalloideae (Amanitaceae) from China. Mycotaxon 2001, 78, 439–448. [Google Scholar]
- Bhatt, R.; Tulloss, R.E.; Semwal, K.C.; Bhatt, V.K.; Moncalvo, J.M.; Stephenson, S. Amanitaceae reported from India. A critically annotated checklist. Mycotaxon 2003, 88, 249–270. [Google Scholar]
- Li, P.; Deng, W.Q.; Li, T.H.; Song, B.; Shen, Y.H. Illumina-based de novo transcriptome sequencing and analysis of Amanita exitialis basidiocarps. Gene 2013, 532, 63–71. [Google Scholar] [CrossRef]
- Luo, H. Advances of omics research on poisonous mushrooms. Mycosystema 2019, 38, 2087–2098. [Google Scholar]
- Zhang, C.H.; Zou, J.P.; Deng, W.Q.; Li, T.H.; Jiang, Z.D. Molecular cloning and the expression pattern of AePOPB involved in the α-amanitin biosynthesis in Amanita exitialis fruiting bodies. Gene 2018, 662, 123–130. [Google Scholar] [CrossRef]
- He, Z.M. Isothermal Amplification Detection for Lethal Amanitas and the Diversity of Toxin Genes in Amanita Mushrooms. Ph.D. Thesis, Hunan Normal University, Changsha, China, 2019. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Johnson, P.H.; Lee, A.S.; Sinsheimer, R.L. Production of specific fragments of φX174 replicative form DNA by a restriction enzyme from Haemophilus parainfluenzae, endonuclease HP. J. Virol. 1973, 11, 596–599. [Google Scholar] [CrossRef] [Green Version]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [Green Version]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27 Pt 1, 209–220. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Rosenberg, N.A. DISTRUCT: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Pickrell, J.K.; Pritchard, J.K. Inference of population splits and mixtures from genome-wide allele frequency data. Nat. Preced. 2012, 1, 1–17. [Google Scholar] [CrossRef]
- Pearson, K. LIII. On lines and planes of closest fit to systems of points in space. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1901, 2, 559–572. [Google Scholar] [CrossRef] [Green Version]
- Holsinger, K.E.; Weir, B.S. Genetics in geographically structured populations: Defining, estimating and interpreting FST. Nat. Rev. Genet. 2009, 10, 639–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, S. Genetic structure of populations. Br. Med. J. 1950, 2, 36. [Google Scholar] [PubMed] [Green Version]
- Summerbell, R.C.; Castle, A.J.; Horgen, P.A.; Anderson, J.B. Inheritance of restriction fragment length polymorphisms in Agaricus brunnescens. Genetics 1989, 123, 293–300. [Google Scholar] [CrossRef]
- Sonnenberg, A.; Baars, J.; Hendrickx, P.; Lavrijssen, B.; Gao, W.; Weijn, A.; Mes, J. Breeding and strain protection in the button mushroom Agaricus bisporus. In Proceedings of the 7th International Conference of the World Society for Mushroom Biology and Mushroom Products, Arcachon, France, 4–7 October 2011; ICMBMP: Arcachon, France, 2011; pp. 7–15. [Google Scholar]
- Xu, J.P. Is natural population of Candida tropicalis sexual, parasexual, and/or asexual? Front. Cell. Infect. Microbiol. 2021, 11, 1066. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.P.; Desmerger, C.; Callac, P. Fine-scale genetic analyses reveal unexpected spatial-temporal heterogeneity in two natural populations of the commercial mushroom Agaricus bisporus. Microbiology 2002, 148, 1253–1262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.P. Analysis of inbreeding depression in Agaricus bisporus. Genetics 1995, 141, 137–145. [Google Scholar] [CrossRef]
- Liang, Y.; Guo, L.D.; Ma, K.P. Population genetic structure of an ectomycorrhizal fungus Amanita manginiana in a subtropical forest over two years. Mycorrhiza 2005, 15, 137–142. [Google Scholar] [CrossRef] [PubMed]

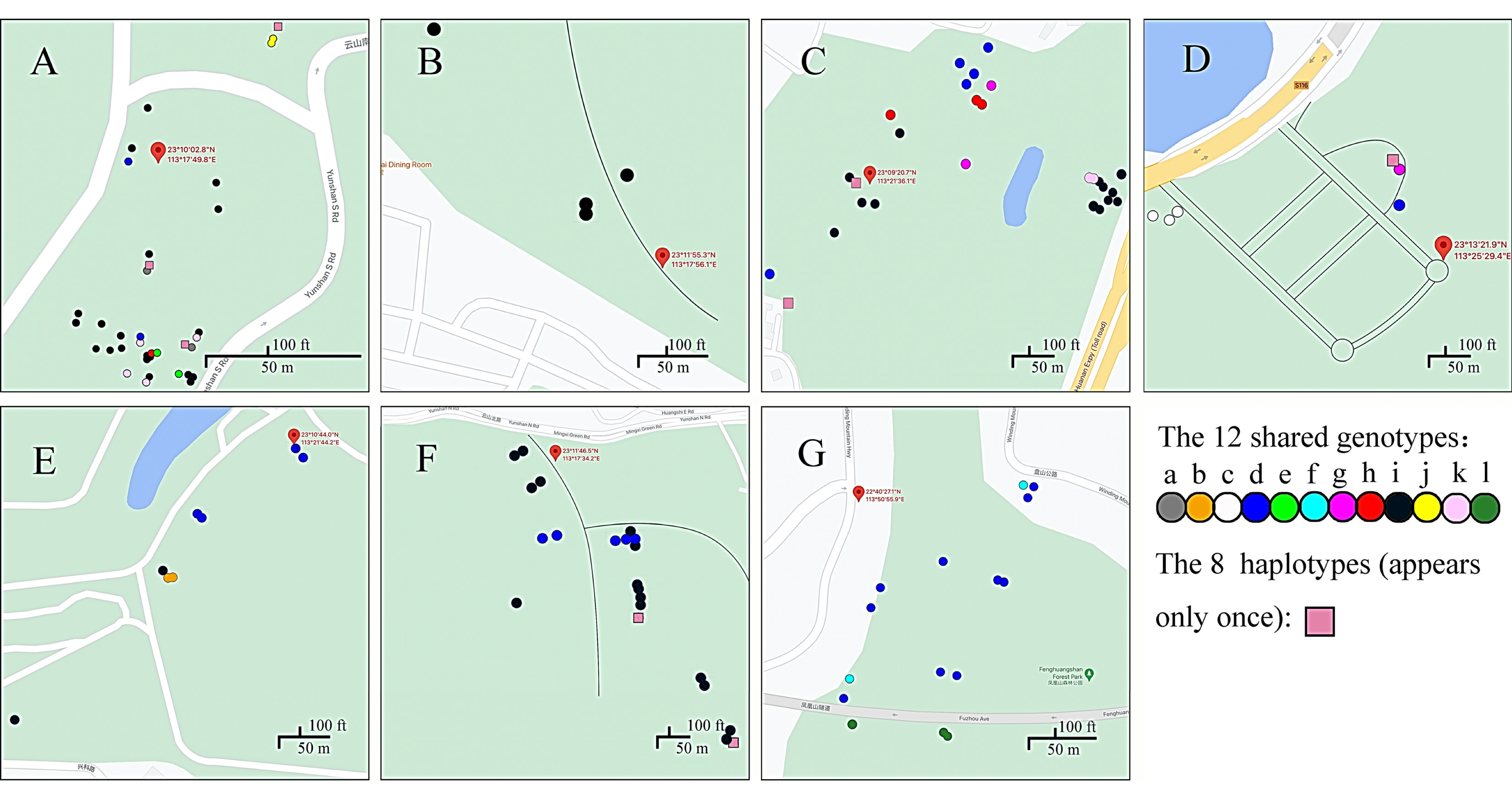
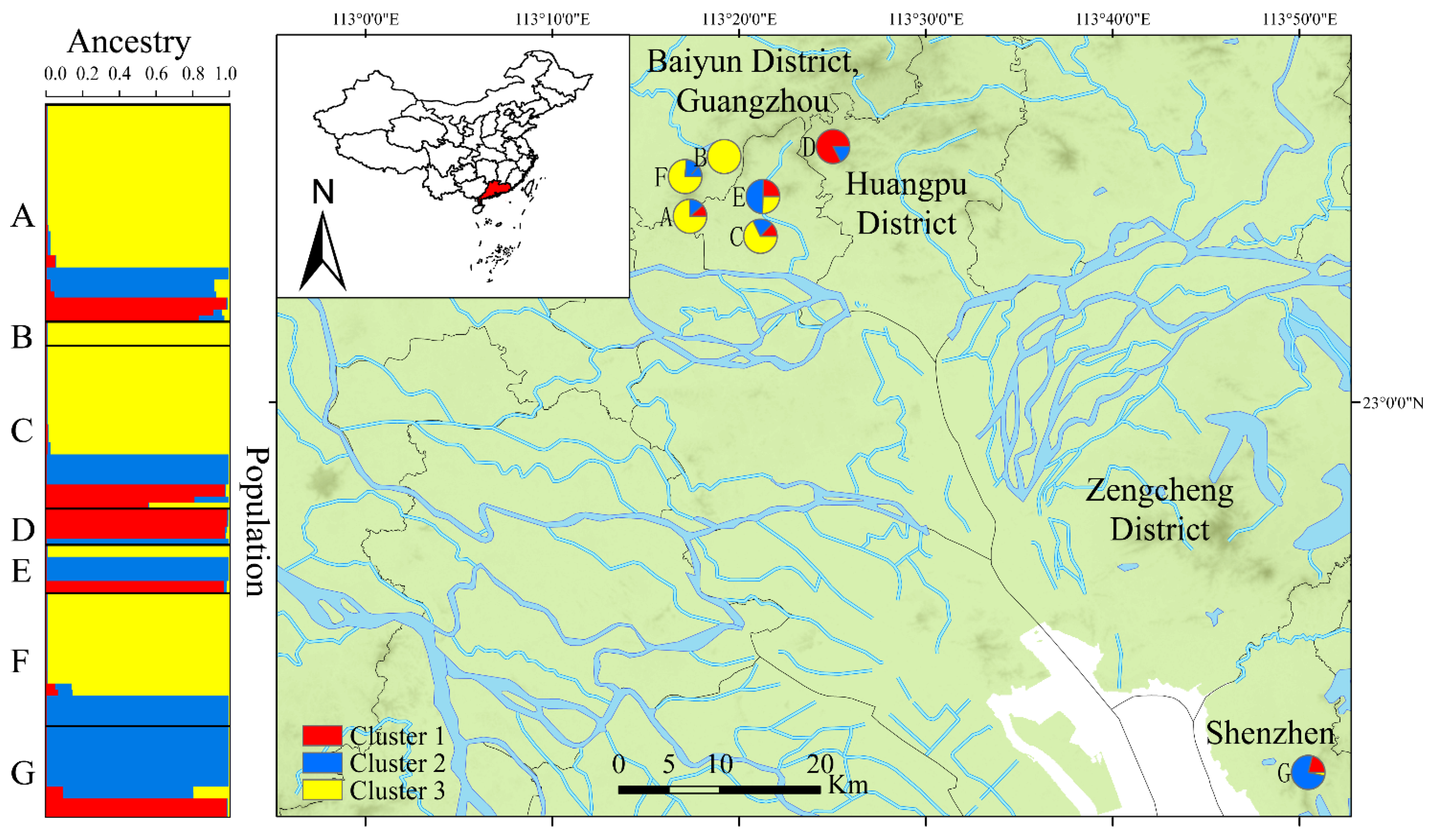
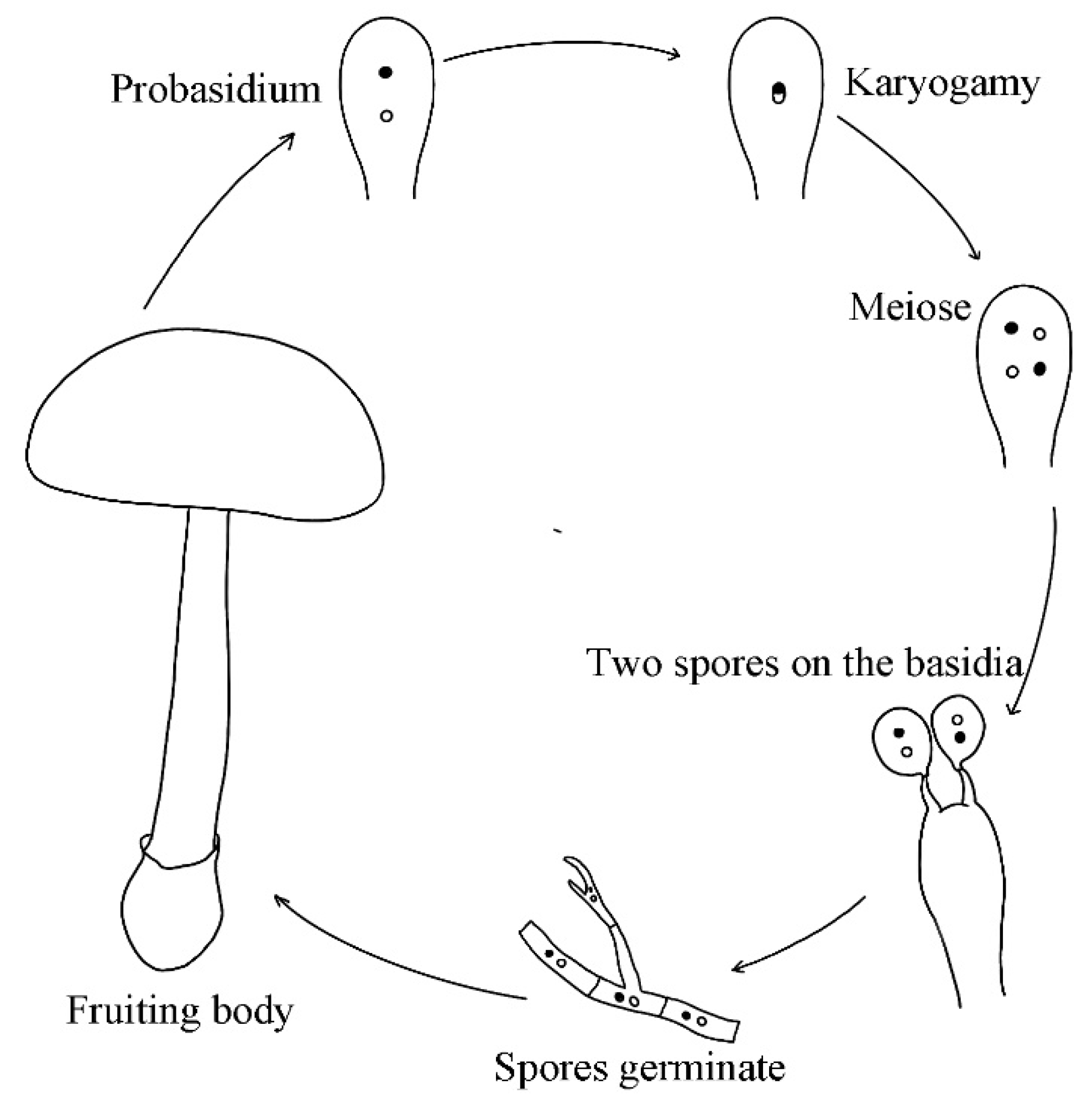
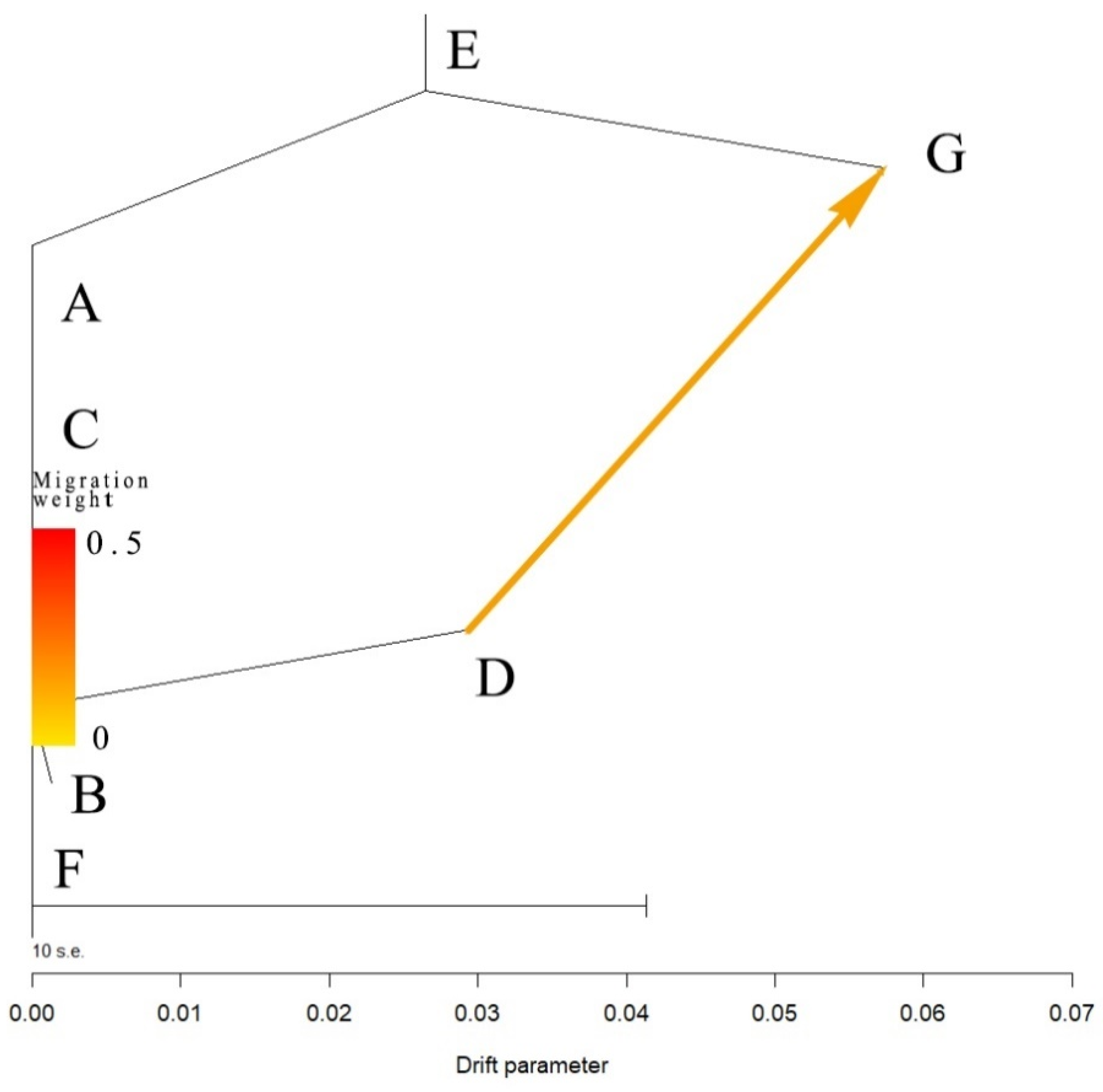
| Code | City | Location | Sample Size | Latitude | Longitude | Altitude (above Sea Level) |
|---|---|---|---|---|---|---|
| A | Guangzhou | Duguting, Baiyunshan Mountain | 36 | 23.1674292 | 113.2971586 | 200–300 m |
| B | Guangzhou | Guangzhou University of Foreign Studies | 4 | 23.1986838 | 113.2989164 | 0–50 m |
| C | Guangzhou | South China Agricultural University | 27 | 23.1557492 | 113.3600235 | 0–50 m |
| D | Guangzhou | Guangzhou Tianluhu Forest Park | 6 | 23.2227467 | 113.4248414 | 200–300 m |
| E | Guangzhou | South China Botanical Garden | 8 | 23.1788847 | 113.3622869 | 0–50 m |
| F | Guangzhou | BeeWorld, Baiyunshan District | 22 | 23.1963429 | 113.2928271 | 100–200 m |
| G | Shenzhen | Fenghuangshan Forest Park | 15 | 22.6742000 | 113.8488670 | 100–200 m |
| Source of Variation | d.f. | Sum of Squares | Variance Components | Percentage of Variation (%) | Fst | Nm |
|---|---|---|---|---|---|---|
| Among-population | 6 | 71.906 | 11.984 | 19% | 0.191 *** | 1.056 |
| Within-population | 111 | 282.780 | 2.548 | 81% | ||
| Total | 117 | 354.686 | 100% |
| Locus | Observed Heterozygotes | Expected Heterozygotes | Chi-Square Value |
|---|---|---|---|
| M12 | 73 | 49.640 | 21.849 *** |
| C21 | 106 | 59.000 | 74.881 *** |
| H11-300 | 64 | 53.809 | 5.104 * |
| A31-88 | 0 | 5.847 | 118.000 *** |
| A31-138 | 0 | 1.983 | 118.000 *** |
| A31-425 | 3 | 46.182 | 103.167 *** |
| A31-440 | 71 | 55.436 | 9.301 ** |
| G24-106 | 6 | 11.390 | 26.424 *** |
| G24-163 | 41 | 33.877 | 5.217 * |
| G24-489 | 77 | 51.877 | 27.674 *** |
| G24-600 | 106 | 58.390 | 78.453 *** |
| G24-615 | 5 | 10.487 | 32.305 *** |
| K47-355 | 103 | 58.962 | 65.826 *** |
| K47-556 | 103 | 58.962 | 65.826 *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, J.; Xu, J.; Zhang, P. Diversity, Dispersal and Mode of Reproduction of Amanita exitialis in Southern China. Genes 2021, 12, 1907. https://doi.org/10.3390/genes12121907
Zhong J, Xu J, Zhang P. Diversity, Dispersal and Mode of Reproduction of Amanita exitialis in Southern China. Genes. 2021; 12(12):1907. https://doi.org/10.3390/genes12121907
Chicago/Turabian StyleZhong, Juan, Jianping Xu, and Ping Zhang. 2021. "Diversity, Dispersal and Mode of Reproduction of Amanita exitialis in Southern China" Genes 12, no. 12: 1907. https://doi.org/10.3390/genes12121907







