Genome-Wide Identification and Analysis of the WRKY Gene Family and Cold Stress Response in Acer truncatum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Sequence Retrieval of the WRKY Gene Family in A. truncatum
2.3. The Gene Structure and Chromosomal Location
2.4. Protein Motif Composition Analysis and Functional Annotation
2.5. Sequence Alignment and Phylogenetic Tree Construction
2.6. Analysis of AtruWRKY Gene Expression in Different Tissues/Organs
2.7. Real-Time qRT-PCR Experimental Validation
3. Results
3.1. Identification of AtruWRKY in A. truncatum
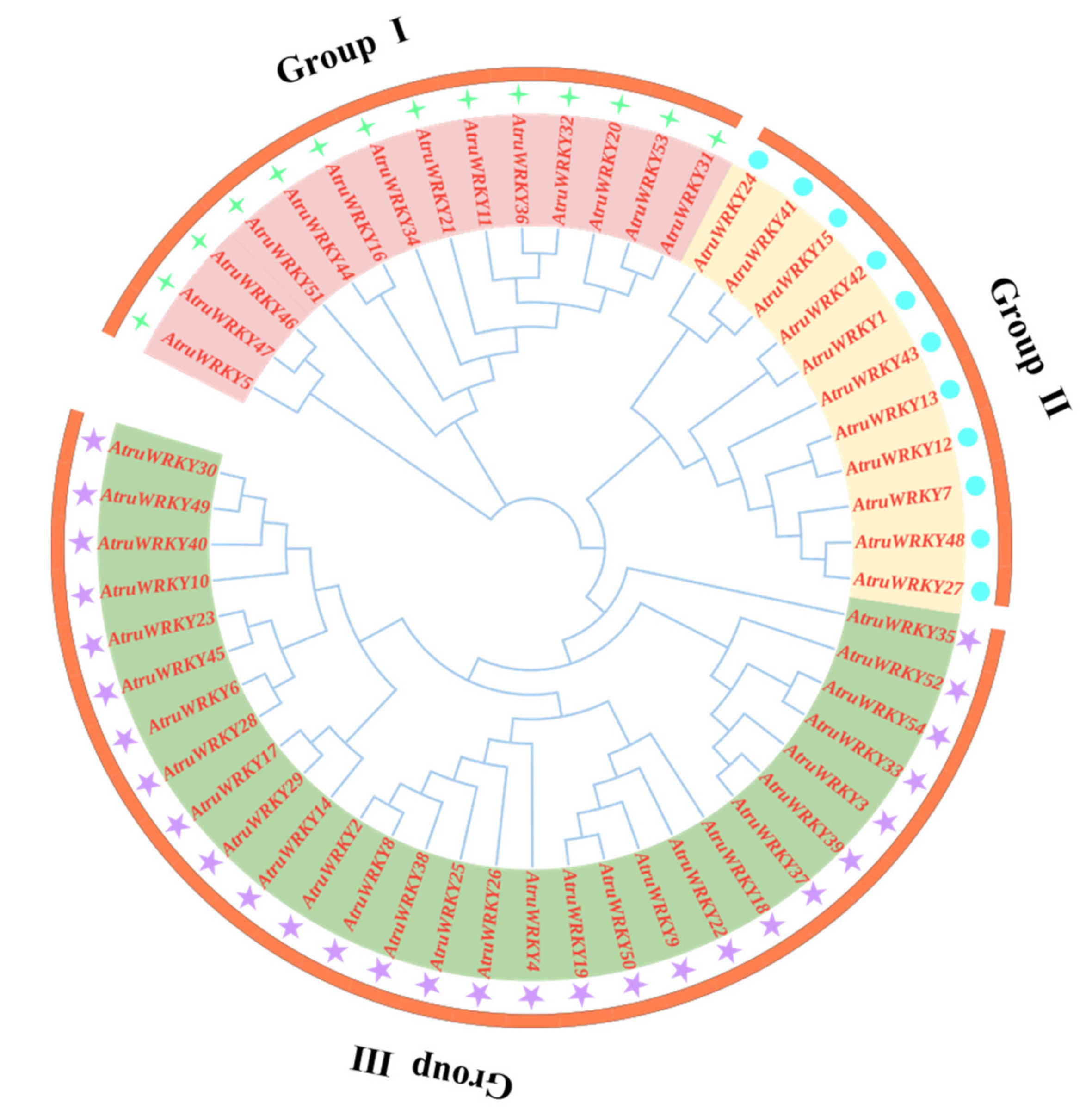
3.2. Phylogenetic Tree Construction and Conserved Motifs
3.3. Chromosomal Location and Gene Structure
3.4. Synteny Analysis of AtruWRKY Genes
3.5. AtruWRKY Expression Profiles in Five Tissues
3.6. Functional Annotation
3.7. Expression Analysis of AtruWRKY Genes under Cold Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hobert, O. Gene regulation by transcription factors and MicroRNAs. Science 2008, 319, 1785–1786. [Google Scholar] [CrossRef] [PubMed]
- Kaplan-Levy, R.N.; Brewer, P.B.; Quon, T.; Smyth, D.R. The trihelix family of transcription factors—Light, stress and development. Trends Plant Sci. 2012, 17, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Abondio, P.; Sazzini, M.; Garagnani, P.; Boattini, A.; Monti, D.; Franceschi, C.; Luiselli, D.; Giuliani, C. The genetic variability of APOE in different human populations and its implications for longevity. Genes 2019, 10, 222. [Google Scholar] [CrossRef] [Green Version]
- Ding, Z.J.; Yan, J.Y.; Li, C.X.; Li, G.X.; Wu, Y.R.; Zheng, S.J. Transcription factor WRKY 46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J. 2015, 84, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.J.; Yan, J.Y.; Li, G.X.; Wu, Z.C.; Zhang, S.Q.; Zheng, S.J. WRKY 41 controls Arabidopsis seed dormancy via direct regulation of ABI 3 transcript levels not downstream of ABA. Plant J. 2014, 79, 810–823. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Jiang, J.J.; Ma, S.H.; Ye, N.H.; Jiang, M.; Cao, J.S.; Zhang, J.H. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef]
- Xue, C.L.; Li, H.T.; Liu, Z.G.; Wang, L.L.; Zhao, Y.T.; Wei, X.M.; Fang, H.; Liu, M.J.; Zhao, J. Genome-wide analysis of the WRKY gene family and their positive responses to phytoplasma invasion in Chinese jujube. BMC Genom. 2019, 20, 464–478. [Google Scholar] [CrossRef] [Green Version]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Viana, V.E.; Carlos, d.M.L.; Busanello, C.; Pegoraro, C.; Costa, d.O.A. When rice gets the chills: Comparative transcriptome profiling at germination shows WRKY transcription factor responses. Plant Biol. 2021, 23, 100–112. [Google Scholar] [CrossRef]
- Bao, W.Q.; Wang, X.W.; Chen, M.; Chai, T.Y.; Wang, H. A WRKY transcription factor, PcWRKY33, from Polygonum cuspidatum reduces salt tolerance in transgenic Arabidopsis thaliana. Plant Cell Rep. 2018, 37, 1033–1048. [Google Scholar] [CrossRef]
- Qin, Y.X.; Tian, Y.C.; Liu, X.Z. A wheat salinity-induced WRKY transcription factor TaWRKY93 confers multiple abiotic stress tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2015, 464, 428–433. [Google Scholar] [CrossRef]
- Zhang, L.L.; Wan, X.B.; Xu, Y.; Niyitanga, S.; Qi, J.M.; Zhang, L.W. De novo assembly of transcriptome and genome-wide identification reveal GA3 stress-responsive WRKY transcription factors involved in fiber formation in jute (Corchorus capsularis). BMC Plant Biol. 2020, 20, 403–418. [Google Scholar] [CrossRef]
- Xie, L.H.; Yan, T.X.; Li, L.; Chen, M.H.; Ma, Y.N.; Hao, X.L.; Fu, X.Q.; Shen, Q.; Huang, Y.W.; Qin, W.; et al. The WRKY transcription factor AaGSW2 promotes glandular trichome initiation in Artemisia annua. J. Exp. Bot. 2021, 72, 1691–1701. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Jiang, Y.N.; Guo, Y.; Huang, J.B.; Zhou, M.H.; Tang, Y.Y.; Sui, J.M.; Wang, J.S.; Qiao, L.X. A novel salt inducible WRKY transcription factor gene, AhWRKY75, confers salt tolerance in transgenic peanut. Plant Physiol. Bioch. 2021, 160, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.J.; Wang, L.; Su, W.H.; Ren, Y.J.; You, C.H.; Zhang, C.; Que, Y.X.; Su, Y.C. A class III WRKY transcription factor in sugarcane was involved in biotic and abiotic stress responses. Sci. Rep. 2020, 10, 20964. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.M.; Wang, Y.F.; Xu, P.; Zhang, Z.B. Overexpression of a WRKY transcription factor TaWRKY2 enhances drought stress tolerance in transgenic wheat. Front. Plant Sci. 2018, 9, 997. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.L.; Lin, Q.B.; Lan, J.; Zhang, T.Y.; Liu, X.; Miao, R.; Mou, C.L.; Nguyen, T.; Wang, J.C.; Zhang, X.; et al. WRKY transcription factor OsWRKY29 represses seed dormancy in rice by weakening abscisic acid response. Front. Plant Sci. 2020, 11, 691. [Google Scholar] [CrossRef]
- Bao, F.; Ding, A.Q.; Cheng, T.R.; Wang, J.; Zhang, Q.X. Genome-wide analysis of members of the WRKY gene family and their cold stress response in Prunus mume. Genes 2019, 10, 911. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.J.; Li, X.H.; Liu, Z.W.; Li, H.; Wang, Y.X.; Zhuang, J. Transcriptome-wide identification of Camellia sinensis WRKY transcription factors in response to temperature stress. Molecul. Genet. Genom. 2016, 291, 255–269. [Google Scholar] [CrossRef]
- Wang, R.K.; Liu, P.; Fan, J.S.; Li, L.L. Comparative transcriptome analysis two genotypes of Acer truncatum Bunge seeds reveals candidate genes that influences seed VLCFAs accumulation. Sci. Rep. 2018, 8, 151–161. [Google Scholar] [CrossRef]
- Wang, R.K.; Fan, J.S.; Chang, P.; Zhu, L.; Zhao, M.R.; Li, L.L. Genome survey sequencing of Acer truncatum Bunge to identify genomic information, simple sequence repeat (SSR) markers and complete chloroplast genome. Forests 2019, 10, 87. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.Y.; Wang, Y.N.; Zhu, L.; Bi, C.W.; Li, S.X.; Li, S.S.; Wen, J.; Yan, K.Y.; Li, Q.Z.; Ono, S. Characterization of the complete chloroplast genome of Acer truncatum Bunge (Sapindales: Aceraceae): A new woody oil tree species producing nervonic acid. BioMed Res. Int. 2019, 2019, 7417239. [Google Scholar] [CrossRef]
- Gu, R.H.; Rybalov, L.; Negrin, A.; Morcol, T.; Long, W.W.; Myers, A.K.; Isaac, G.; Yuk, J.; Kennelly, E.J.; Long, C.L. Metabolic profiling of different parts of Acer truncatum from the mongolian plateau using UPLC-QTOF-MS with comparative bioactivity assays. J. Agr. Food Chem. 2019, 67, 1585–1597. [Google Scholar] [CrossRef]
- Ma, X.F.; Wu, L.H.; Ito, Y.; Tian, W.X. Application of preparative high-speed counter-current chromatography for separation of methyl gallate from Acer truncatum Bunge. J. Chromatogr. A 2005, 1076, 212–215. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.J.; Luo, X.G.; Yan, L.H.; Si, C.L.; Wang, N.; He, H.P.; Zhang, T.C. Transcriptome analysis of unsaturated fatty acids biosynthesis shows essential genes in sprouting of Acer truncatum Bunge seeds. Food Biosci. 2020, 41, 100739–100774. [Google Scholar] [CrossRef]
- Li, L.; Manning, W.J.; Tong, L.; Wang, X.K. Chronic drought stress reduced but not protected Shantung maple (Acer truncatum Bunge) from adverse effects of ozone (O3) on growth and physiology in the suburb of Beijing, China. Environ. Pollut. 2015, 201, 34–41. [Google Scholar] [CrossRef]
- Ma, Q.Y.; Sun, T.L.; Li, S.S.; Wen, J.; Zhu, L.; Yin, T.M.; Yan, K.Y.; Xu, X.; Li, S.X.; Mao, J.F.; et al. The Acer truncatum genome provides insights into nervonic acid biosynthesis. Plant J. 2020, 104, 662–678. [Google Scholar] [CrossRef]
- Li, X.X.; Guo, C.; Ahmad, S.; Wang, Q.; Yu, J.; Liu, C.; Guo, Y.F. Systematic analysis of MYB family genes in potato and their multiple roles in development and stress responses. Biomolecules 2019, 9, 317. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative Toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Chen, C.H.; Chen, X.Q.; Han, J.; Lu, W.L.; Ren, Z.H. Genome-wide analysis of the WRKY gene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses. BMC Plant Biol. 2020, 20, 443. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.J.; Zhang, J.S.; Cheng, H.; Aslam, M.; Lv, H.W.; Dong, W.; Hu, A.Q.; Guo, M.L.; Liu, Q.; Qin, Y. Identification and evolutionary analysis of FAD2 gene family in green plants. Trop. Plant Biol. 2021, 14, 239–250. [Google Scholar] [CrossRef]
- Gao, S.Q.; Li, L.Z.; Han, X.L.; Liu, T.T.; Jin, P.; Cai, L.; Xu, M.; Zhang, T.Y.; Zhang, F.; Chen, J.P.; et al. Genome-wide identification of the histone acetyltransferase gene family in Triticum aestivum. BMC Genom. 2021, 22, 49. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.Y.; Wang, X.; Xu, Y.T.; Deng, X.X.; Xu, Q. Genome-wide analysis of the R2R3-MYB transcription factor gene family in sweet orange (Citrus sinensis). Mol. Biol. Rep. 2014, 41, 6769–6785. [Google Scholar] [CrossRef]
- Cao, Z.H.; Zhang, S.Z.; Wang, R.K.; Zhang, R.F.; Hao, Y.J. Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene confering abiotic stress tolerance in plants. PLoS ONE 2018, 8, e69955. [Google Scholar] [CrossRef]
- Zhang, H.; Yu, Z.J.; Yao, X.D.; Chen, J.L.; Chen, X.; Zhou, H.W.; Lou, Y.X.; Ming, F.; Jin, Y. Genome-wide identification and characterization of small auxin-up RNA (SAUR) gene family in plants: Evolution and expression profiles during normal growth and stress response. BMC Plant Biol. 2021, 21, 4. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Cai, K.W.; Pei, X.N.; Li, Y.; Hu, Y.B.; Meng, F.J.; Song, X.S.; Tigabu, M.; Ding, C.J.; Zhao, X.Y. Genome-wide identification of NAC transcription factor family in Juglans mandshurica and their expression analysis during the fruit development and ripening. Int. J. Mol. Sci. 2021, 22, 12414. [Google Scholar] [CrossRef]
- Liu, Y.S.; Wang, Y.Z.; Pei, J.B.; Li, Y.D.; Sun, H.Y. Genome-wide identification and characterization of COMT gene family during the development of blueberry fruit. BMC Plant Biol. 2021, 21, 5. [Google Scholar] [CrossRef]
- Zuo, X.Y.; Wang, S.X.; Xiang, W.; Yang, H.R.; Tahir, M.M.; Zheng, S.G.; An, N.; Han, M.Y.; Zhao, C.P.; Zhang, D. Genome-wide identification of the 14-3-3 gene family and its participation in floral transition by interacting with TFL1/FT in apple. BMC Genom. 2021, 22, 41. [Google Scholar] [CrossRef]
- Chen, Z.; Shen, Z.J.; Zhao, D.; Xu, L.; Zhang, L.J.; Zou, Q. Genome-wide analysis of LysM-containing gene family in wheat: Structural and phylogenetic analysis during development and defense. Genes 2020, 12, 31. [Google Scholar] [CrossRef]
- Cai, K.W.; Liu, H.X.; Chen, S.; Liu, Y.; Zhao, X.Y.; Chen, S. Genome-wide identification and analysis of class III peroxidases in Betula pendula. BMC Genom. 2021, 22, 314. [Google Scholar] [CrossRef] [PubMed]
- Ylli, D. The response to DNA damage at telomeric repeats and its consequences for telomere function. Genes 2019, 10, 318. [Google Scholar]
- Fan, Y.X.; Lin, F.K.; Zhang, R.F.; Wang, M.M.; Gu, R.H.; Long, C.L. Acer truncatum Bunge: A comprehensive review on ethnobotany, phytochemistry and pharmacology. J. Ethnopharmacol. 2021, 282, 114572. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.X.; Yang, Y.; Liu, C.; Hou, Y.X.; Yang, S.Z.; Wang, L.S.; Zhang, X.Q. Potential suitable habitat of two economically important forest trees (Acer truncatum and Xanthoceras sorbifolium) in east Asia under current and future climate scenarios. Forests 2021, 12, 1263. [Google Scholar] [CrossRef]
- Geethalakshmi, S.; Barathkumar, S.; Prabu, G. The MYB transcription factor family genes in Sugarcane (Saccharum sp.). Plant Mol. Biol. Rep. 2015, 33, 512–531. [Google Scholar] [CrossRef]
- Sun, W.J.; Ma, Z.T.; Chen, H.; Liu, M.Y. MYB gene family in Potato (Solanum tuberosum L.): Genome-wide identification of hormone-responsive reveals their potential functions in growth and development. Int. J. Mol. Sci. 2019, 20, 4847. [Google Scholar] [CrossRef] [Green Version]
- El-Esawi, M.; Alayafi, A. Overexpression of StDREB2 transcription factor enhances drought stress tolerance in Cotton (Gossypium barbadense L.). Genes 2019, 10, 142. [Google Scholar] [CrossRef] [Green Version]
- Kanofsky, K.; Riggers, J.; Staar, M.; Strauch, C.J.; Arndt, L.C.; Hehl, R. A strong NF-κB p65 responsive cis -regulatory sequence from Arabidopsis thaliana interacts with WRKY40. Plant Cell Rep. 2019, 38, 1139–1150. [Google Scholar] [CrossRef]
- Cui, X.X.; Yan, Q.; Gan, S.P.; Xue, D.; Wang, H.T.; Xing, H.; Zhao, J.M.; Guo, N. GmWRKY40, a member of the WRKY transcription factor genes identified from Glycine max L., enhanced the resistance to phytophthora sojae. BMC Plant Biol. 2019, 19, 598. [Google Scholar] [CrossRef] [Green Version]
- Huangfu, J.Y.; Li, J.C.; Li, R.; Ye, M.; Kuai, P.; Zhang, T.F.; Lou, Y.G. The transcription factor OsWRKY45 negatively modulates the resistance of rice to the brown planthopper nilaparvata lugens. Int. J. Mol. Sci. 2016, 17, 697. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.F.; Li, M.Z.; Xiong, Y.P.; Wu, J.M.; Silva, J.A.T.D.; Ma, G.H. Genome-wide characterization, expression profile analysis of WRKY family genes in Santalum album and functional identification of their role in abiotic stress. Int. J. Mol. Sci. 2019, 20, 5676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Wang, D.D.; Yang, C.H.; Kong, N.N.; Shi, Z.; Zhao, P.; Nan, Y.Y.; Nie, T.K.; Wang, R.Q.; Ma, H.L.; et al. Genome-wide identification of the potato WRKY transcription factor family. PLoS ONE 2017, 12, e0181573. [Google Scholar] [CrossRef] [Green Version]
- Li, D.H.; Liu, P.; Yu, J.Y.; Wang, L.H.; Dossa, K.; Zhang, Y.X.; Zhou, R.; Wei, X.; Zhang, X.R. Genome-wide analysis of WRKY gene family in the sesame genome and identification of the WRKY genes involved in responses to abiotic stresses. BMC Plant Biol. 2017, 17, 152. [Google Scholar] [CrossRef]
- Yue, H.; Wang, M.; Liu, S.Y.; Du, X.H.; Song, W.N.; Nie, X.J. Transcriptome-wide identification and expression profiles of the WRKY transcription factor family in Broomcorn millet (Panicum miliaceum L.). BMC Genom. 2016, 17, 343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fei, X.T.; Hou, L.X.; Shi, J.W.; Yang, T.X.; Liu, Y.L.; Wei, A.Z. Patterns of drought response of 38 WRKY transcription factors of Zanthoxylum bungeanum Maxim. Int. J. Mol. Sci. 2018, 20, 68. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Liu, Q.; Liu, T.Y.; Cui, X.; Ning, W. Expression of putative luteolin biosynthesis genes and WRKY transcription factors in Taraxacum antungense kitag. Plant Cell Tissue Organ Cult. 2021, 145, 649–665. [Google Scholar] [CrossRef]
- Liu, Z.; Leng, S.; Chang, Q.Y.; Cheng, C.W.; Zheng, Z.M.; Yu, S. Identification of yellowhorn (Xanthoceras sorbifolium) WRKY transcription factor family and analysis of abiotic stress response model. J. For. Res. 2020, 32, 987–1004. [Google Scholar] [CrossRef]
- Guo, C.L.; Guo, R.R.; Xu, X.Z.; Gao, M.; Li, X.Q.; Song, J.Y.; Zheng, Y.; Wang, X.P. Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J. Exp. Bot. 2014, 65, 1513–1528. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, J.; Zhou, X.H.; Liu, S.Y.; Zhuang, Y. Identification of WRKY gene family and characterization of cold stress-responsive WRKY genes in eggplant. PeerJ 2020, 8, e8777. [Google Scholar] [CrossRef]
- He, G.; Guan, C.N.; Chen, Q.X.; Gou, X.J.; Liu, W.; Zeng, Q.Y.; Lan, T. Genome-wide analysis of the glutathione s-transferase gene family in Capsella rubella: Identification, expression, and biochemical functions. Front. Plant Sci. 2016, 7, 1325. [Google Scholar] [CrossRef] [Green Version]
- Hao, F.; Yang, G.; Zhou, H.J.; Yao, J.J.; Liu, D.R.L.; Zhao, P.; Zhang, S.X. Genome-wide identification and transcriptional expression profiles of transcription factor WRKY in common Walnut (Juglans regia L.). Genes 2021, 12, 1444. [Google Scholar] [CrossRef]
- Hu, W.J.; Ren, Q.Y.; Chen, Y.L.; Xu, G.L.; Qian, Y.X. Genome-wide identification and analysis of WRKY gene family in maize provide insights into regulatory network in response to abiotic stresses. BMC Plant Biol. 2021, 21, 427. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.R.; Li, Y.Y.; Sun, Y.D.; Lu, L.; Zhao, Z.X.; Zhou, J.G.; Li, X.Z. Comparative RNA-seq analysis reveals candidate genes associated with fruit set in pumpkin. Sci. Hortic. 2021, 288, 110255. [Google Scholar] [CrossRef]
- Malwina, P.; Hamilton, R.S.; Hong, W.Y.; Sharkey, A.M.; Paul, R.; Erlyani, A.H.N.; Eric, J.; Stephen, C.D.; Burton, G.J.; Tereza, C. RNA-Seq reveals changes in human placental metabolism, transport and endocrinology across the first-second trimester transition. Biol. Open 2021, 10, bio058222. [Google Scholar]
- Zhang, H.W.; Li, M.; Kong, M.; Dunwell, J.M.; Zhang, Y.Y.; Yue, C.; Wu, J.Y.; Zhang, S.L. Study on the differences of gene expression between pear and apple wild cultivation materials based on RNA-seq technique. BMC Plant Biol. 2021, 21, 256–269. [Google Scholar] [CrossRef] [PubMed]
- Zuzic, M.; Arias, J.E.R.; Wohl, S.G.; Busskamp, V. Retinal miRNA functions in health and disease. Genes 2019, 10, 377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, J.; Wang, F.; Hou, X.L.; Wang, Z.; Huang, Z.N. Genome-wide fractionation and identification of WRKY transcription factors in Chinese Cabbage (Brassica rapa ssp. pekinensis) reveals collinearity and their expression patterns under abiotic and biotic stresses. Plant Mol. Biol. Rep. 2014, 32, 781–795. [Google Scholar] [CrossRef]
- Wang, L.N.; Zhu, W.; Fang, L.C.; Sun, X.M.; Su, L.Y.; Liang, Z.C.; Wang, N.; Londo, J.P.; Li, S.H.; Xin, H.P. Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol. 2014, 14, 103. [Google Scholar] [CrossRef] [Green Version]
- Ling, J.; Jiang, W.J.; Zhang, Y.; Yu, H.J.; Mao, Z.C.; Gu, X.F.; Huang, S.W.; Xie, B.Y. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom. 2011, 12, 471. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.R.; Wang, R.; Zheng, M.; Liu, X.B.; Meng, F.; Wu, H.L.; Yao, Y.Y.; Xin, M.M.; Peng, H.R.; Ni, Z.F.; et al. Ta WRKY 51 promotes lateral root formation through negative regulation of ethylene biosynthesis in wheat (Triticum aestivum L.). Plant J. 2018, 96, 372–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nevaiah, B.N.; Karthikeyan, A.S.; Raghothama, K.G. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007, 143, 1789–1801. [Google Scholar]
- Zhang, J.; Peng, Y.L.; Guo, Z.J. Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res. 2008, 18, 508–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.X.; Yarra, R.; Jin, L.F.; Cao, H.X. Genome-wide identification and expression analysis of MYB gene family in oil palm (Elaeis guineensis Jacq.) under abiotic stress conditions. Environ. Exp. Bot. 2020, 180, 104245. [Google Scholar] [CrossRef]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.P.; Zhao, Y.; Xu, S.J.; Zhang, Z.Y.; Xu, Y.Y.; Zhang, J.Y.; Chong, K. OsMADS57 together with OsTB1 coordinates transcription of its target OsWRKY94 and D14 to switch its organogenesis to defense for cold adaptation in rice. New Phytol. 2018, 218, 219–231. [Google Scholar] [CrossRef] [Green Version]
- Zou, C.S.; Jiang, W.B.; Yu, D.Q. Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. J. Exp. Bot. 2010, 61, 3901–3914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.J.; Mao, S.S.; Gao, Y.L.; Zhu, L.Y.; Wu, D.M.; Cui, Y.X.; Li, J.N.; Qian, W. Genome-wide identification and expression analysis of WRKY transcription factors under multiple stresses in Brassica napus. PLoS ONE 2016, 11, e0157558. [Google Scholar] [CrossRef] [Green Version]







| Gene Name | Gene ID | SL 1 | CDS (Length) | AA 2 | PI 3 | MW (D) 4 | Group |
|---|---|---|---|---|---|---|---|
| AtruWRKY1 | Atru.chr5.112 | cytosol | 693 | 230 | 7.31 | 25,946.49 | Ⅱ b |
| AtruWRKY2 | Atru.chr5.580 | nucleus | 1122 | 373 | 6.13 | 40,527.27 | Ⅲ c |
| AtruWRKY3 | Atru.chr5.640 | nucleus | 1107 | 368 | 5.35 | 40,969.65 | Ⅲ a |
| AtruWRKY4 | Atru.chr5.983 | nucleus | 798 | 265 | 5.24 | 30,044.15 | Ⅲ c |
| AtruWRKY5 | Atru.chr5.585 | cytosol | 930 | 309 | 6.32 | 33,994.84 | Ⅰ a |
| AtruWRKY6 | Atru.chr5.856 | nucleus | 1164 | 387 | 8.50 | 40,844.83 | Ⅲ e |
| AtruWRKY7 | Atru.chr5.965 | nucleus | 945 | 314 | 6.50 | 34,739.92 | Ⅱ b |
| AtruWRKY8 | Atru.chr5.562 | nucleus | 1122 | 373 | 6.13 | 40,527.27 | Ⅲ c |
| AtruWRKY9 | Atru.chr5.1597 | nucleus | 1044 | 347 | 9.67 | 38,515.61 | Ⅲ b |
| AtruWRKY10 | Atru.chr11.1895 | nucleus | 1878 | 625 | 6.66 | 68,360.37 | Ⅲ e |
| AtruWRKY11 | Atru.chr11.1706 | nucleus | 1551 | 516 | 8.85 | 56,006.74 | Ⅰ b |
| AtruWRKY12 | Atru.chr11.1674 | nucleus | 807 | 268 | 5.73 | 29,060.95 | Ⅱ b |
| AtruWRKY13 | Atru.chr11.1669 | nucleus | 915 | 304 | 6.12 | 32,948.26 | Ⅱ b |
| AtruWRKY14 | Atru.chr7.2541 | nucleus | 1029 | 342 | 7.62 | 38,513.83 | Ⅲ d |
| AtruWRKY15 | Atru.chr7.1919 | nucleus | 492 | 163 | 5.63 | 18,559.46 | Ⅱ a |
| AtruWRKY16 | Atru.chr7.53 | nucleus | 1482 | 493 | 5.62 | 53,646.88 | Ⅰ b |
| AtruWRKY17 | Atru.chr7.2542 | nucleus | 819 | 272 | 8.70 | 30,682.41 | Ⅲ d |
| AtruWRKY18 | Atru.chr7.2657 | nucleus | 1020 | 339 | 9.33 | 36,585.20 | Ⅲ b |
| AtruWRKY19 | Atru.chr13.744 | nucleus | 1140 | 379 | 9.45 | 41,219.55 | Ⅲ b |
| AtruWRKY20 | Atru.chr2.3906 | nucleus | 2241 | 746 | 5.78 | 80,810.51 | Ⅰ b |
| AtruWRKY21 | Atru.chr2.3616 | nucleus | 1725 | 574 | 6.88 | 61,806.15 | Ⅰ b |
| AtruWRKY22 | Atru.chr2.840 | nucleus | 1086 | 361 | 9.84 | 40,045.18 | Ⅲ b |
| AtruWRKY23 | Atru.chr2.3488 | nucleus | 1656 | 551 | 7.84 | 60,264.59 | Ⅲ e |
| AtruWRKY24 | Atru.chr2.3873 | cytosol | 216 | 71 | 8.95 | 8012.95 | Ⅱ a |
| AtruWRKY25 | Atru.chr10.684 | nucleus | 1548 | 515 | 6.05 | 55,775.07 | Ⅲ c |
| AtruWRKY26 | Atru.chr10.1574 | nucleus | 693 | 230 | 5.56 | 25,497.48 | Ⅲ c |
| AtruWRKY27 | Atru.chr10.1366 | nucleus | 984 | 327 | 6.46 | 36,108.74 | Ⅱ b |
| AtruWRKY28 | Atru.chr10.2262 | nucleus | 1845 | 614 | 6.07 | 65,386.01 | Ⅲ e |
| AtruWRKY29 | Atru.chr10.2411 | nucleus | 1038 | 345 | 8.57 | 38,343.75 | Ⅲ d |
| AtruWRKY30 | Atru.chr8.2579 | nucleus | 1740 | 579 | 5.90 | 62,385.94 | Ⅲ e |
| AtruWRKY31 | Atru.chr8.2526 | nucleus | 831 | 276 | 9.56 | 29,968.99 | Ⅰ b |
| AtruWRKY32 | Atru.chr8.2350 | nucleus | 1692 | 563 | 6.10 | 61,892.15 | Ⅰ b |
| AtruWRKY33 | Atru.chr8.95 | nucleus | 1050 | 349 | 5.50 | 39,460.85 | Ⅲ a |
| AtruWRKY34 | Atru.chr8.344 | nucleus | 1458 | 485 | 6.84 | 53,270.12 | Ⅰ b |
| AtruWRKY35 | Atru.chr8.1995 | nucleus | 981 | 326 | 4.82 | 36,337.09 | Ⅲ a |
| AtruWRKY36 | Atru.chr1.2580 | nucleus | 1788 | 595 | 7.73 | 65,397.05 | Ⅰ b |
| AtruWRKY37 | Atru.chr1.252 | nucleus | 828 | 275 | 8.93 | 30,322.86 | Ⅲ a |
| AtruWRKY38 | Atru.chr9.2113 | nucleus | 1011 | 336 | 5.41 | 37,384.57 | Ⅲ c |
| AtruWRKY39 | Atru.chr9.2017 | nucleus | 1089 | 362 | 5.51 | 40,377.47 | Ⅲ a |
| AtruWRKY40 | Atru.chr9.2304 | nucleus | 1662 | 553 | 7.22 | 59,525.46 | Ⅲ e |
| AtruWRKY41 | Atru.chr4.410 | nucleus | 621 | 206 | 6.59 | 22,872.99 | Ⅱ a |
| AtruWRKY42 | Atru.chr4.480 | nucleus | 699 | 232 | 8.78 | 26,451.89 | Ⅱ b |
| AtruWRKY43 | Atru.chr4.3047 | nucleus | 1296 | 431 | 5.80 | 46,645.38 | Ⅱ b |
| AtruWRKY44 | Atru.chr6.3354 | nucleus | 1689 | 562 | 6.07 | 61,083.23 | Ⅰ b |
| AtruWRKY45 | Atru.chr6.3220 | nucleus | 1911 | 636 | 6.25 | 69,112.06 | Ⅲ e |
| AtruWRKY46 | Atru.chr6.1828 | nucleus | 558 | 185 | 9.41 | 21,199.56 | Ⅰ a |
| AtruWRKY47 | Atru.chr6.2590 | nucleus | 612 | 203 | 9.32 | 22,806.53 | Ⅰ a |
| AtruWRKY48 | Atru.chr6.2076 | nucleus | 1035 | 344 | 6.73 | 37,984.86 | Ⅱ b |
| AtruWRKY49 | Atru.chr6.1003 | nucleus | 1224 | 407 | 6.18 | 45,486.52 | Ⅲ e |
| AtruWRKY50 | Atru.chr3.267 | nucleus | 1047 | 348 | 9.74 | 37,932.62 | Ⅲ b |
| AtruWRKY51 | Atru.chr3.2384 | nucleus | 435 | 144 | 6.43 | 16,327.83 | Ⅰ b |
| AtruWRKY52 | Atru.chr12.203 | nucleus | 1113 | 370 | 5.87 | 39,919.36 | Ⅲ a |
| AtruWRKY53 | Atru.chr12.1782 | nucleus | 2307 | 768 | 5.31 | 83,984.96 | Ⅰ b |
| AtruWRKY54 | Atru.chr12.201_Atru.chr12.202 | nucleus | 1017 | 338 | 5.78 | 37,079.06 | Ⅲ a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Li, X.; Wei, J.; Cai, K.; Zhang, H.; Ge, L.; Ren, Z.; Zhao, C.; Zhao, X. Genome-Wide Identification and Analysis of the WRKY Gene Family and Cold Stress Response in Acer truncatum. Genes 2021, 12, 1867. https://doi.org/10.3390/genes12121867
Li Y, Li X, Wei J, Cai K, Zhang H, Ge L, Ren Z, Zhao C, Zhao X. Genome-Wide Identification and Analysis of the WRKY Gene Family and Cold Stress Response in Acer truncatum. Genes. 2021; 12(12):1867. https://doi.org/10.3390/genes12121867
Chicago/Turabian StyleLi, Yan, Xiang Li, Jiatong Wei, Kewei Cai, Hongzhi Zhang, Lili Ge, Zengjun Ren, Chunli Zhao, and Xiyang Zhao. 2021. "Genome-Wide Identification and Analysis of the WRKY Gene Family and Cold Stress Response in Acer truncatum" Genes 12, no. 12: 1867. https://doi.org/10.3390/genes12121867
APA StyleLi, Y., Li, X., Wei, J., Cai, K., Zhang, H., Ge, L., Ren, Z., Zhao, C., & Zhao, X. (2021). Genome-Wide Identification and Analysis of the WRKY Gene Family and Cold Stress Response in Acer truncatum. Genes, 12(12), 1867. https://doi.org/10.3390/genes12121867





