Relationship of Cultivated Grain Amaranth Species and Wild Relative Accessions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA Extraction of In Vitro Grown Seedlings
2.3. SNP Marker Analysis
2.4. SNP Calling and Data Analysis
3. Results
3.1. Characterization of SNP Markers
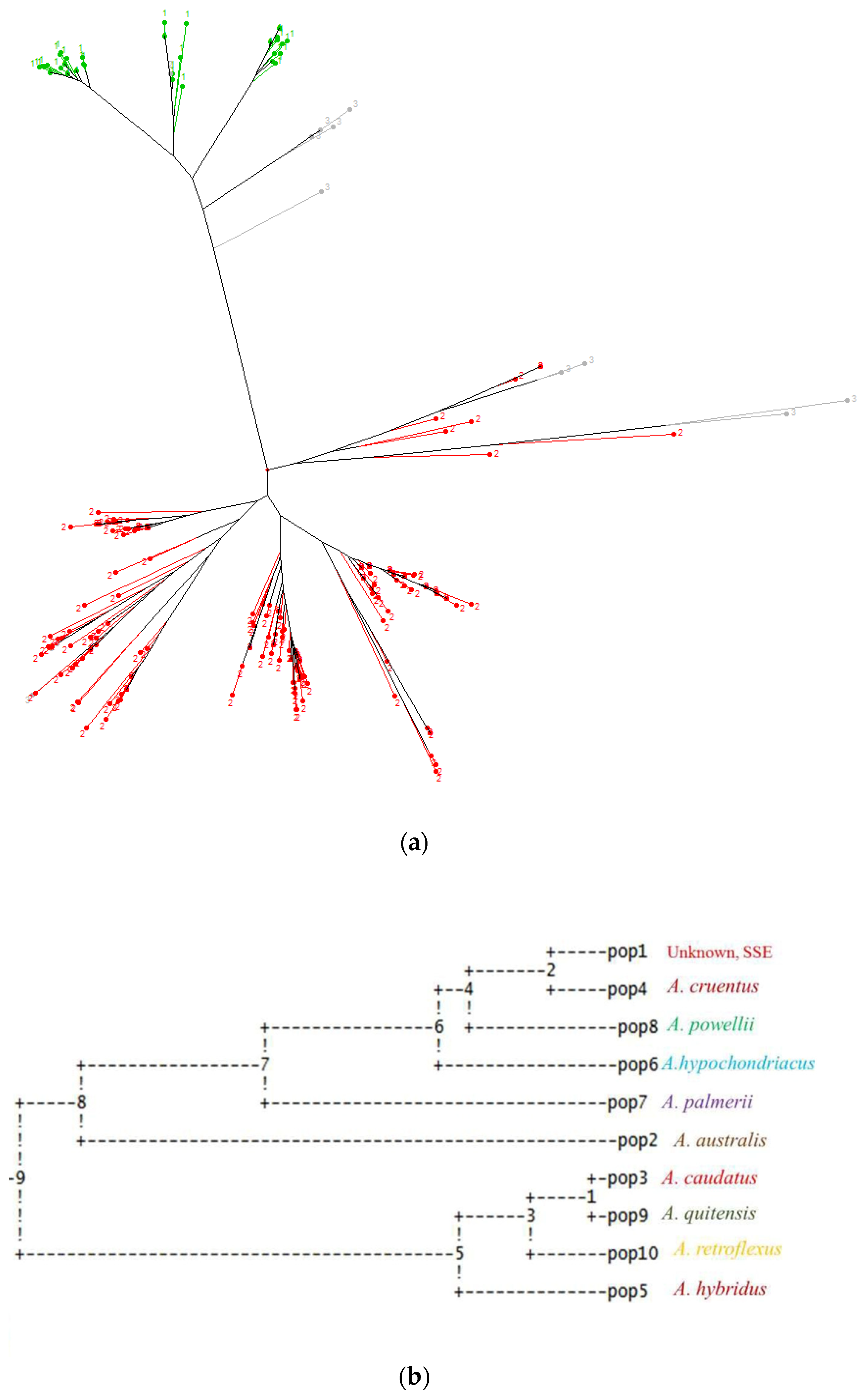
3.2. Relationships between Grain Amaranth Accessions
3.3. Cluster Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Suresh, S.; Chung, J.W.; Cho, G.T.; Sung, J.S.; Park, J.H.; Gwag, J.G.; Baek, H.J. Analysis of molecular genetic diversity and population structure in Amaranthus germplasm using SSR markers. Plant. Biosyst. 2014, 148, 635–644. [Google Scholar] [CrossRef]
- Das, S. Taxonomy and phylogeny of grain amaranths. In Amaranthus: A Promising Crop of Future; Springer: Singapore, 2016; pp. 57–94. [Google Scholar]
- Sauer, J.D. The grain amaranths and their relatives: A revised taxonomic and geographic survey. Ann. Missouri Bot. Gard. 1967, 54, 103–137. [Google Scholar] [CrossRef]
- Brenner, D.M.; Baltensperger, D.D.; Kulakow, P.A.; Lehmann, J.W.; Myers, R.L.; Slabbert, M.M.; Sleugh, B.B. Genetic resources and breeding of Amaranthus. Plant Breed. Rev. 2010, 19, 227–285. [Google Scholar]
- He, Q.; Park, Y.J. Evaluation of genetic structure of amaranth accessions from the United States. Weed Turfgrass Sci. 2013, 2, 230–235. [Google Scholar] [CrossRef] [Green Version]
- Aguilar, E.G.; Albarracín, G.; Uñates, M.A.; Piola, H.D.; Camiña, J.M.; Escudero, N.L. Evaluation of the nutritional quality of the grain protein of new amaranths varieties. Pl. Food Hum. Nutr. 2015, 70, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Akin-Idowu, P.E.; Odunola, O.A.; Gbadegesin, M.A.; Ademoyegun, O.T.; Aduloju, A.O.; Olagunju, Y.O. Nutritional evaluation of five species of grain amaranth–an under-utilized crop. Intern. J. Sci. 2017, 6, 18–27. [Google Scholar]
- Tovar-Pérez, E.G.; Lugo-Radillo, A.; Aguilera-Aguirre, S. Amaranth grain as a potential source of biologically active peptides: A review of their identification, production, bioactivity, and characterization. Food Rev. Intern. 2019, 35, 221–245. [Google Scholar] [CrossRef]
- Amare, E.; Mouquet-Rivier, C.; Servent, A.; Morel, G.; Adish, A.; Haki, G.D. Protein quality of amaranth grains cultivated in Ethiopia as affected by popping and fermentation. Food Nutr. Sci. 2015, 6, 38. [Google Scholar] [CrossRef] [Green Version]
- Akin-Idowu, P.E.; Ademoyegun, O.T.; Olagunju, Y.O.; Aduloju, A.O.; Adebo, U.G. Phytochemical content and antioxidant activity of five grain amaranth species. Am. J. Food Sci. Tech. 2017, 5, 249–255. [Google Scholar]
- Bhat, A.; Satpathy, G.; Gupta, R.K. Evaluation of Nutraceutical properties of Amaranthus hypochondriacus L. grains and formulation of value-added cookies. J. Pharmacogn. Phytochem. 2015, 3, 51–54. [Google Scholar]
- Gresta, F.; Guerrini, A.; Sacchetti, G.; Tacchini, M.; Sortino, O.; Ceravolo, G.; Onofri, A. Agronomic, chemical, and antioxidant characterization of grain amaranths grown in a Mediterranean environment. Crop Sci. 2017, 57, 2688–2698. [Google Scholar] [CrossRef]
- Karamać, M.; Gai, F.; Longato, E.; Meineri, G.; Janiak, M.A.; Amarowicz, R.; Peiretti, P.G. Antioxidant activity and phenolic composition of amaranth (Amaranthus caudatus) during plant growth. Antioxidants 2019, 8, 173. [Google Scholar] [CrossRef] [Green Version]
- Temesgen, A.; Bultosa, G. Physicochemical characteristics and nutrient composition of three grain amaranth species grown in Hirna, Eastern Ethiopia. East. Afr. J. Sci. 2017, 11, 17–26. [Google Scholar]
- Wu, H.; Sun, M.; Yue, S.; Sun, H.; Cai, Y.; Huang, R.; Brenner, D.; Corke, H. Field evaluation of an Amaranthus genetic resource collection in China. Genet. Res. Crop. Evol. 2000, 47, 43–53. [Google Scholar] [CrossRef]
- Hejazi, S.N.; Orsat, V.; Azadi, B.; Kubow, S. Improvement of the in vitro protein digestibility of amaranth grain through optimization of the malting process. J. Cereal Sci. 2016, 68, 59–65. [Google Scholar] [CrossRef]
- Shukla, S.; Bhargava, A.; Chatterjee, A.; Pandey, A.C.; Mishra, B.K. Diversity in phenotypic and nutritional traits in vegetable amaranth (Amaranthus tricolor), a nutritionally underutilized crop. J. Sci. Food Agr. 2010, 90, 139–144. [Google Scholar] [CrossRef]
- Brown, A.H.D. Core collections: A practical approach to genetic resources management. Genome 1989, 31, 818–824. [Google Scholar] [CrossRef]
- Espitia, E. Amaranth germplasm development and agronomic studies in Mexico. Food Rev. Int. 1992, 8, 71–86. [Google Scholar] [CrossRef]
- Chan, K.F.; Sun, M. Genetic diversity and relationships detected by isozyme and RAPD analysis of crop and wild species of Amaranthus. Theor. Appl. Genetic. 1997, 95, 865–873. [Google Scholar] [CrossRef]
- Yudina, R.S.; Zheleznova, N.B.; Zakharova, O.V.; Zheleznov, A.V.; Shumny, V.K. Isozyme analysis in a genetic collection of amaranths (Amaranthus, L.). Russ. J. Genet. 2005, 41, 1395–1400. [Google Scholar] [CrossRef]
- Sammour, R.H.; Radwan, S.A.; Mira, M. Genetic diversity in genus Amaranthus: From morphology to genomic DNA. Res. Rev. Biosci. 2012, 6, 351–360. [Google Scholar]
- Park, Y.J.; Nishikawa, T.; Matsushima, K.; Minami, M.; Nemoto, K. A rapid and reliable PCR-restriction fragment length polymorphism (RFLP) marker for the identification of Amaranthus cruentus species. Breed. Sci. 2014, 64, 422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mallory, M.A.; Hall, R.V.; McNabb, A.R.; Pratt, D.B.; Jellen, E.N.; Maughan, P.J. Development and characterization of microsatellite markers for the grain amaranths. Crop. Sci. 2008, 48, 1098–1106. [Google Scholar] [CrossRef]
- Oo, W.H.; Park, Y.J. Analysis of the genetic diversity and population structure of amaranth accessions from South America using 14 SSR Markers. Kor. J. Crop Sci. 2013, 58, 336–346. [Google Scholar] [CrossRef] [Green Version]
- Maughan, P.J.; Yourstone, S.M.; Jellen, E.N.; Udall, J.A. SNP discovery via genomic reduction, barcoding, and 454-pyrosequencing in amaranth. Plant Genome 2009, 2, 260–270. [Google Scholar] [CrossRef] [Green Version]
- Maughan, P.J.; Smith, S.M.; Fairbanks, D.J.; Jellen, E.N. Development, characterization, and linkage mapping of single nucleotide polymorphisms in the grain amaranths (Amaranthus sp.). Plant Genome 2011, 4, 92–101. [Google Scholar] [CrossRef] [Green Version]
- Faseela, K.V.; Joseph, S. Molecular characterization of amaranth landraces and assessment of interspecific relationships among Amaranthus spp. (L.) using RAPD markers. Indian J. Genet. Plant Breed. 2007, 67, 12–17. [Google Scholar]
- Transue, D.K.; Fairbanks, D.J.; Robison, L.R.; Andersen, W.R. Species identification by RAPD analysis of grain amaranth genetic resources. Crop. Sci. 1994, 34, 1385–1389. [Google Scholar] [CrossRef]
- Mandal, N.; Das, P.K. Intra-and interspecific genetic diversity in grain Amaranthus using random amplified polymorphic DNA markers. Plant Tissue Cult. 2002, 12, 49–56. [Google Scholar]
- Xu, F.; Sun, M. Comparative analysis of phylogenetic relationships of grain amaranths and their wild relatives (Amaranthus; Amaranthaceae) using internal transcribed spacer, amplified fragment length polymorphism, and double-primer fluorescent intersimple sequence repeat markers. Mol. Phylogenetics Evol. 2001, 21, 372–387. [Google Scholar]
- He, C.; Holme, J.; Anthony, J. SNP genotyping: The KASP assay. In Crop Breeding: Methods and Protocols; Humana Press: New York, NY, USA, 2014; pp. 75–86. [Google Scholar]
- Mammadov, J.; Aggarwal, R.; Buyyarapu, R.; Kumpatla, S. SNP markers and their impact on plant breeding. Internat. J. Pl. Genom. 2012, 2012, 728398. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Blair, M.W. Genotyping by sequencing (GBS) polymorphism diversity in grain amaranths and relatives. Front. Plant Sci. 2017, 8, 1960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stetter, M.G.; Müller, T.; Schmid, K.J. Genomic and phenotypic evidence for an incomplete domestication of South American grain amaranth (Amaranthus caudatus). Mol. Ecol. 2017, 26, 871–886. [Google Scholar] [CrossRef]
- Steemers, F.J.; Gunderson, K.L. Whole genome genotyping technologies on the BeadArray™ platform. Biotechnol. J. 2007, 2, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, F.R.; Maughan, P.J.; Alvarez, A.; Kietlinski, K.D.; Smith, S.M.; Pratt, D.B.; Elzinga, D.B.; Jellen, E.N. Assessment of genetic diversity in Peruvian amaranth (Amaranthus caudatus and A. hybridus) germplasm using single nucleotide polymorphism markers. Crop. Sci. 2013, 53, 532. [Google Scholar] [CrossRef]
- Thapa, R.; Blair, M.W. Morphological assessment of cultivated and wild progenitor grain amaranth diversity. Agronomy 2018, 8, 272. [Google Scholar] [CrossRef] [Green Version]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Anderson, J.A.; Churchill, G.A.; Sutrique, J.E.; Tanksley, S.D.; Sorrells, M.E. Optimizing parental selection for genetic linkage maps. Genome 1993, 36, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Molecular Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Earl, D.A. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Res. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maughan, P.J.; Yourstone, S.M.; Byers, R.L.; Smith, S.M.; Udall, J.A. Single-nucleotide polymorphism genotyping in mapping populations via genomic reduction and next-generation sequencing: Proof of concept. Plant Genome 2010, 3, 166–178. [Google Scholar] [CrossRef] [Green Version]
- Hauptli, H.; Jain, S. Allozyme variation and evolutionary relationships of grain amaranths (Amaranthus spp.). Theor. Appl. Genet. 1984, 69, 153–165. [Google Scholar] [CrossRef]
- Stetter, M.G.; Zeitler, L.; Steinhaus, A.; Kroener, K.; Biljecki, M.; Schmid, K.J. Crossing methods and cultivation conditions for rapid production of segregating populations in three grain amaranth species. Front. Plant Sci. 2016, 7, 816. [Google Scholar] [CrossRef] [Green Version]
- Lightfoot, D.J.; Jarvis, D.E.; Ramaraj, T.; Lee, R.; Jellen, E.N.; Maughan, P.J. Single-molecule sequencing and Hi-C-based proximity-guided assembly of amaranth (Amaranthus hypochondriacus) chromosomes provide in- sights into genome evolution. BMC Biol. 2017, 15, 74. [Google Scholar] [CrossRef] [Green Version]

| SNP Name | SNP Type | MAF | GD | H | PIC |
|---|---|---|---|---|---|
| AM17977 | C/T | 0.5888 | 0.5208 | 0.4094 | 0.4283 |
| AM18039 | A/G | 0.7156 | 0.4280 | 0.0109 | 0.3664 |
| AM19011 | C/T | 0.6920 | 0.4348 | 0.0000 | 0.3520 |
| AM19559 | C/T | 0.8533 | 0.2534 | 0.2717 | 0.2263 |
| AM19643 | G/T | 0.6703 | 0.4949 | 0.1957 | 0.4433 |
| AM19746 | A/G | 0.4891 | 0.5277 | 0.1812 | 0.4158 |
| AM20177 | A/G | 0.8007 | 0.3268 | 0.1014 | 0.2858 |
| AM21724 | A/G | 0.7772 | 0.3576 | 0.1196 | 0.3111 |
| AM21859 | A/T | 0.4783 | 0.6347 | 0.0072 | 0.5631 |
| AM22137 | A/C | 0.7373 | 0.3892 | 0.0036 | 0.3163 |
| AM22892 | A/G | 0.8116 | 0.3072 | 0.1159 | 0.2622 |
| AM23006 | A/C | 0.6775 | 0.4437 | 0.0145 | 0.3545 |
| AM23128 | C/T | 0.7428 | 0.3875 | 0.0145 | 0.3204 |
| AM23196 | C/T | 0.8188 | 0.3004 | 0.0290 | 0.2613 |
| AM23262 | G/T | 0.5417 | 0.5891 | 0.0109 | 0.5159 |
| AM24029 | A/G | 0.7373 | 0.3946 | 0.0036 | 0.3273 |
| AM24210 | C/T | 0.3877 | 0.6581 | 0.0000 | 0.5837 |
| AM24401 | C/G | 0.7391 | 0.3911 | 0.0072 | 0.3226 |
| AM27616 | A/G | 0.7482 | 0.3820 | 0.0036 | 0.3169 |
| AM27642 | A/G | 0.4275 | 0.6534 | 0.0000 | 0.5800 |
| AM17870 | A/G | 0.6232 | 0.4826 | 0.2681 | 0.3824 |
| AM18185 | C/T | 0.7591 | 0.3675 | 0.0326 | 0.3026 |
| AM19378 | G/T | 0.5960 | 0.4873 | 0.6268 | 0.3754 |
| AM19426 | A/T | 0.6178 | 0.4951 | 0.6993 | 0.4007 |
| AM19707 | C/G | 0.6268 | 0.5285 | 0.0000 | 0.4649 |
| AM19842 | A/G | 0.7301 | 0.4081 | 0.0616 | 0.3452 |
| AM19855 | C/T | 0.4493 | 0.6213 | 0.0000 | 0.5419 |
| AM20180 | A/G | 0.7138 | 0.4183 | 0.0072 | 0.3447 |
| AM20403 | C/T | 0.4982 | 0.5337 | 0.0833 | 0.4249 |
| AM20533 | C/T | 0.8714 | 0.2251 | 0.2500 | 0.2013 |
| AM21310 | A/T | 0.7681 | 0.3562 | 0.0000 | 0.2928 |
| AM21336 | C/G | 0.8406 | 0.2691 | 0.3043 | 0.2348 |
| AM21842 | A/G | 0.5942 | 0.5286 | 0.1377 | 0.4440 |
| AM22476 | A/G | 0.7409 | 0.3910 | 0.0036 | 0.3251 |
| AM22487 | C/G | 0.6757 | 0.4472 | 0.2210 | 0.3594 |
| AM22649 | C/T | 0.6141 | 0.4821 | 0.7500 | 0.3759 |
| AM23703 | A/G | 0.5688 | 0.4997 | 0.0072 | 0.3852 |
| AM24078 | A/G | 0.7917 | 0.3328 | 0.0036 | 0.2820 |
| AM24266 | C/G | 0.8478 | 0.2591 | 0.0072 | 0.2274 |
| AM24531 | C/T | 0.6884 | 0.4472 | 0.0580 | 0.3723 |
| AM24819 | A/G | 0.6558 | 0.4718 | 0.2609 | 0.3871 |
| AM26171 | C/T | 0.6993 | 0.4308 | 0.0072 | 0.3523 |
| AM27610 | A/T | 0.7120 | 0.4162 | 0.0109 | 0.3382 |
| AM27626 | A/C | 0.6975 | 0.4242 | 0.0471 | 0.3372 |
| Mean | 0.6776 | 0.4363 | 0.1215 | 0.3648 |
| Source of Variation | Df | SS | MS | % | Type | Fixation Index | p-Value |
|---|---|---|---|---|---|---|---|
| Among species | 6 | 2095.326 | 349.221 | 52% | FST | 0.520 | 0.001 |
| Among individuals within species | 240 | 1967.552 | 8.198 | 24% | FIS | 0.498 | 0.001 |
| Within individuals | 247 | 678.000 | 2.745 | 24% | FIT | 0.759 | 0.001 |
| Total | 493 | 4740.879 | 100% |
| Species Name | A. caudatus | A. cruentus | A. hybridus | A. hypo. | A. powellii | A. quitensis | A. retroflexus |
|---|---|---|---|---|---|---|---|
| A. caudatus | 0.000 | ||||||
| A. cruentus | 0.643 | 0.000 | |||||
| A. hybridus | 0.291 | 0.345 | 0.000 | ||||
| A. hypochondriacus | 0.703 | 0.410 | 0.398 | 0.000 | |||
| A. powellii | 0.703 | 0.377 | 0.334 | 0.424 | 0.000 | ||
| A. quitensis | 0.062 | 0.635 | 0.306 | 0.702 | 0.714 | 0.000 | |
| A. retroflexus | 0.717 | 0.510 | 0.307 | 0.539 | 0.346 | 0.752 | 0.000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thapa, R.; Edwards, M.; Blair, M.W. Relationship of Cultivated Grain Amaranth Species and Wild Relative Accessions. Genes 2021, 12, 1849. https://doi.org/10.3390/genes12121849
Thapa R, Edwards M, Blair MW. Relationship of Cultivated Grain Amaranth Species and Wild Relative Accessions. Genes. 2021; 12(12):1849. https://doi.org/10.3390/genes12121849
Chicago/Turabian StyleThapa, Ranjita, Matthew Edwards, and Matthew W. Blair. 2021. "Relationship of Cultivated Grain Amaranth Species and Wild Relative Accessions" Genes 12, no. 12: 1849. https://doi.org/10.3390/genes12121849
APA StyleThapa, R., Edwards, M., & Blair, M. W. (2021). Relationship of Cultivated Grain Amaranth Species and Wild Relative Accessions. Genes, 12(12), 1849. https://doi.org/10.3390/genes12121849






