X Chromosome Inactivation during Grasshopper Spermatogenesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Squashing and Spreading of Seminiferous Tubules
2.2. Immunofluorescence Microscopy
2.3. Feulgen–Rossenbeck Reaction
2.4. Histological Sections
2.5. Image Acquisition and Processing
2.6. Fluorescence Quantification
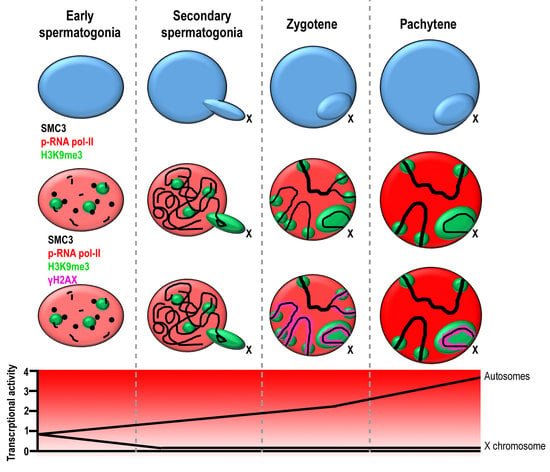
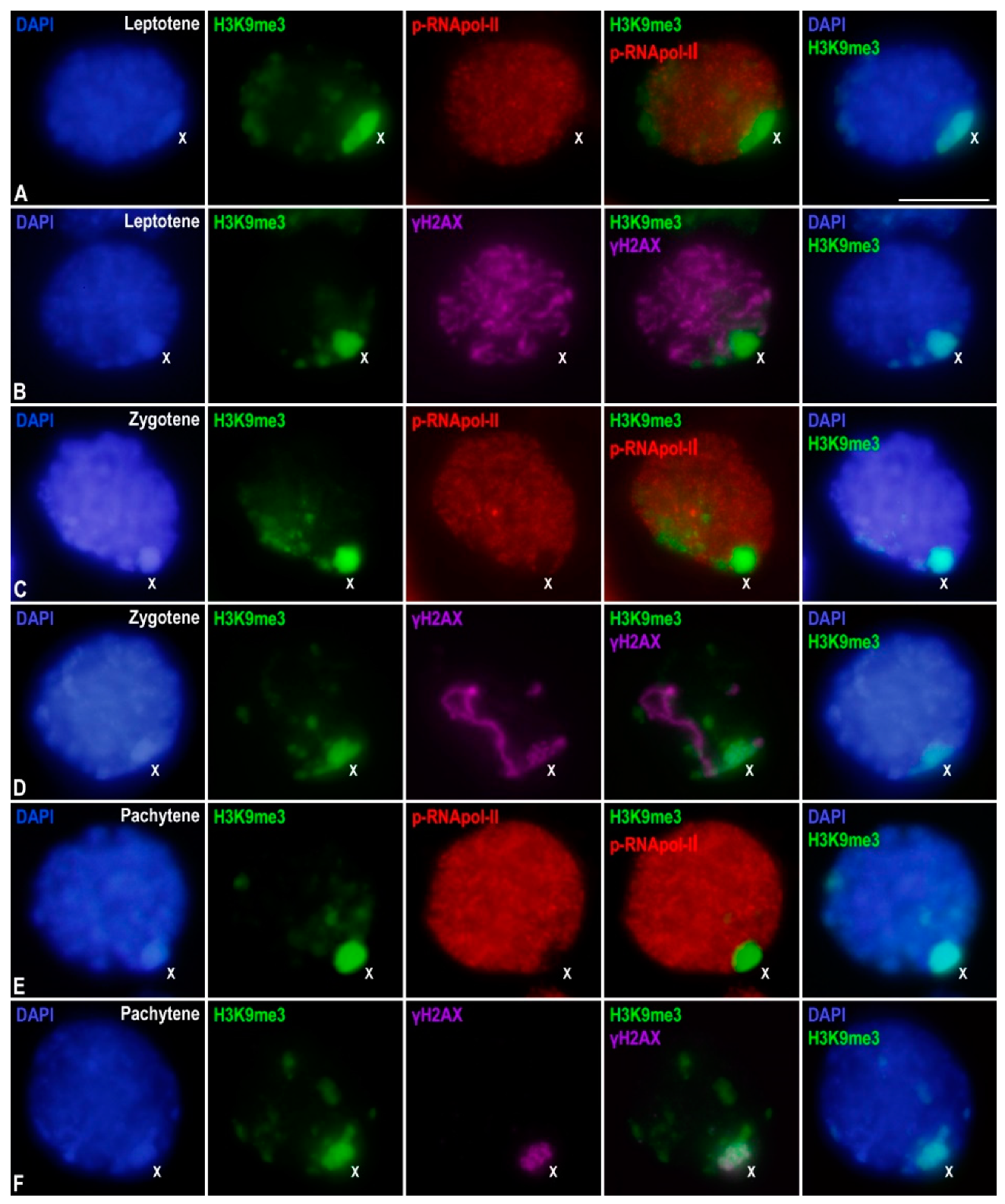
3. Results
3.1. Transcriptional Activity in E. plorans Spermatocytes and Spermatids
3.2. Epigenetic Marks of Transcriptional Inactivation in E. plorans Spermatocytes
3.3. The X Chromosome Inactivation Occurs in Premeiotic Cells
3.4. Transcription Is Maintained in the Transition from Premeiotic Cells to Spermatocytes
4. Discussion
4.1. The Pattern of Meiotic Transcription in Spermatocytes of E. plorans Differs from That Displayed by Eutherian Mammals
4.2. X Chromosome Heterochromatinization and Inactivation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zickler, D.; Kleckner, N. Recombination, pairing, and pynapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 2015, 7, a016626. [Google Scholar] [CrossRef] [Green Version]
- Baudat, F.; Manova, K.; Yuen, J.P.; Jasin, M.; Keeney, S. Chromosome synapsis defects and sexually dimorphic mei-otic progression in mice lacking Spo11. Mol. Cell 2000, 6, 989–998. [Google Scholar] [CrossRef]
- Grelon, M.; Vezon, D.; Gendrot, G.; Pelletier, G. AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 2001, 20, 589–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keeney, S.; Giroux, C.N.; Kleckner, N. Meiosis-Specific DNA Double-Strand Breaks Are Catalyzed by Spo11, a Member of a Widely Conserved Protein Family. Cell 1997, 88, 375–384. [Google Scholar] [CrossRef] [Green Version]
- Peoples, T.L.; Dean, E.; Gonzalez, O.; Lambourne, L.; Burgess, S.M. Close, stable homolog juxtaposition during meio-sis in budding yeast is dependent on meiotic recombination, occurs independently of synapsis, and is distinct from DSB-independent pairing contacts. Genes Dev. 2002, 16, 1682–1695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romanienko, P.J.; Camerini-Otero, R. The Mouse Spo11 Gene Is Required for Meiotic Chromosome Synapsis. Mol. Cell 2000, 6, 975–987. [Google Scholar] [CrossRef]
- Madigan, J.P.; Chotkowski, H.L.; Glaser, R.L. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res. 2002, 30, 3698–3705. [Google Scholar] [CrossRef]
- Redon, C.; Pilch, D.; Rogakou, E.; Sedelnikova, O.; Newrock, K.; Bonner, W. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 2002, 12, 162–169. [Google Scholar] [CrossRef]
- Fernandez-Capetillo, O.; Allis, C.D.; Nussenzweig, A. Phosphorylation of Histone H2B at DNA Double-Strand Breaks. J. Exp. Med. 2004, 199, 1671–1677. [Google Scholar] [CrossRef] [Green Version]
- Celeste, A.; Fernandez-Capetillo, O.; Kruhlak, M.J.; Pilch, D.R.; Staudt, D.; Lee, A.; Bonner, R.F.; Bonner, W.M.; Nussenzweig, A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 2003, 5, 675–679. [Google Scholar] [CrossRef]
- Grey, C.; de Massy, B. Chromosome Organization in Early Meiotic Prophase. Front. Cell Dev. Biol. 2021, 9, 688878. [Google Scholar] [CrossRef] [PubMed]
- Bishop, D.K.; Park, D.; Xu, L.; Kleckner, N. DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 1992, 69, 439–456. [Google Scholar] [CrossRef]
- Brown, M.S.; Bishop, D.K. DNA Strand Exchange and RecA Homologs in Meiosis. Cold Spring Harb. Perspect. Biol. 2014, 7, a016659. [Google Scholar] [CrossRef] [Green Version]
- Shinohara, A.; Ogawa, H.; Ogawa, T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 1992, 69, 457–470. [Google Scholar] [CrossRef]
- Ashley, T.; Plug, A.W.; Xu, J.; Solari, A.J.; Reddy, G.; Golub, E.I.; Ward, D.C. Dynamic changes in Rad51 distribution on chromatin during meiosis in male and female vertebrates. Chromosoma 1995, 104, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Barlow, A.L.; Benson, F.E.; West, S.; Hultén, M.A. Distribution of the Rad51 recombinase in human and mouse spermatocytes. EMBO J. 1997, 16, 5207–5215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franklin, A.E.; McElver, J.; Sunjevaric, I.; Rothstein, R.; Bowen, B.; Cande, W.Z. Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell. 1999, 11, 809–824. [Google Scholar] [CrossRef] [Green Version]
- Moens, P.B.; Chen, D.J.; Shen, Z.; Kolas, N.; Tarsounas, M.; Heng, H.H.Q.; Spyropoulos, B. Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma 1997, 106, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Moens, P.B.; Kolas, N.K.; Tarsounas, M.; Marcon, E.; Cohen, P.E.; Spyropoulos, B. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J. Cell Sci. 2002, 115, 1611–1622. [Google Scholar] [CrossRef]
- Pawlowski, W.P.; Golubovskaya, I.N.; Cande, W.Z. Altered Nuclear Distribution of Recombination Protein RAD51 in Maize Mutants Suggests the Involvement of RAD51 in Meiotic Homology Recognition. Plant Cell 2003, 15, 1807–1816. [Google Scholar] [CrossRef]
- Rockmill, B.; Sym, M.; Scherthan, H.; Roeder, G.S. Roles for two RecA homologs in promoting meiotic chromosome synapsis. Genes Dev. 1995, 9, 2684–2695. [Google Scholar] [CrossRef] [Green Version]
- Tsubouchi, H.; Roeder, G. The Importance of Genetic Recombination for Fidelity of Chromosome Pairing in Meiosis. Dev. Cell 2003, 5, 915–925. [Google Scholar] [CrossRef] [Green Version]
- Viera, A.; Santos, J.L.; Page, J.; Parra, M.T.; Calvente, A.; Cifuentes, M.; Gómez, R.; Lira, R.; Suja, J.A.; Rufas, J.S. DNA double-strand breaks, recombination and synapsis: The timing of meiosis differs in grasshoppers and flies. EMBO Rep. 2004, 5, 385–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, L.; Kang, R.; Rosenberg, S.C.; Qiu, Y.; Raviram, R.; Chee, S.; Hu, R.; Ren, B.; Cole, F.; Corbett, K.D. Dynamic reorganization of the genome shapes the recombination landscape in meiotic prophase. Nat. Struct. Mol. Biol. 2019, 26, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Vara, C.; Paytuvi-Gallart, A.; Cuartero, Y.; Le Dily, F.; Garcia, F.; Salva-Castro, J.; Gomez, H.L.; Julia, E.; Moutinho, C.; Cigliano, R.A.; et al. Three-Dimensional Genomic Structure and Cohesin Occupancy Correlate with Transcriptional Activity during Spermato-genesis. Cell Rep. 2019, 28, 352–367.e359. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente, R.; Pratto, F.; Hernández-Hernández, A.; Manterola, M.; López-Jiménez, P.; Gómez, R.; Viera, A.; Parra, M.T.; Kouznetsova, A.; Camerini-Otero, R.D.; et al. Epigenetic Dysregulation of Mammalian Male Meiosis Caused by In-terference of Recombination and Synapsis. Cells 2021, 10, 2311. [Google Scholar] [CrossRef]
- Kota, S.K.; Feil, R. Epigenetic Transitions in Germ Cell Development and Meiosis. Dev. Cell 2010, 19, 675–686. [Google Scholar] [CrossRef] [Green Version]
- Lam, K.-W.G.; Brick, K.; Cheng, G.; Pratto, F.; Camerini-Otero, R.D. Cell-type-specific genomics reveals histone modification dynamics in mammalian meiosis. Nat. Commun. 2019, 10, 3821. [Google Scholar] [CrossRef] [Green Version]
- van der Heijden, G.; Derijck, A.A.H.A.; Pósfai, E.; Giele, M.; Pelczar, P.; Ramos, L.; Wansink, D.G.; Van der Vlag, J.; Peters, A.H.F.M.; De Boer, P. Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nat. Genet. 2007, 39, 251–258. [Google Scholar] [CrossRef]
- Almstrup, K.; Nielsen, J.E.; Hansen, M.A.; Tanaka, M.; Skakkebæk, N.E.; Leffers, H. Analysis of Cell-Type-Specific Gene Expression During Mouse Spermatogenesis1. Biol. Reprod. 2004, 70, 1751–1761. [Google Scholar] [CrossRef]
- da Cruz, I.; Rodríguez-Casuriaga, R.; Santiñaque, F.F.; Farías, J.; Curti, G.; Capoano, C.A.; Folle, G.A.; Benavente, R.; Sote-lo-Silveira, J.R.; Geisinger, A. Transcriptome analysis of highly purified mouse spermatogenic cell populations: Gene ex-pression signatures switch from meiotic-to postmeiotic-related processes at pachytene stage. BMC Genom. 2016, 17, 294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monesi, V. Synthetic activities during spermatogenesis in the mouse: RNA and protein. Exp. Cell Res. 1965, 39, 197–224. [Google Scholar] [CrossRef]
- Page, J.; de la Fuente, R.; Manterola, M.; Parra, M.T.; Viera, A.; Berrios, S.; Fernandez-Donoso, R.; Rufas, J.S. Inactiva-tion or non-reactivation: What accounts better for the silence of sex chromosomes during mammalian male meiosis? Chromosoma 2012, 121, 307–326. [Google Scholar] [CrossRef] [PubMed]
- Shima, J.E.; McLean, D.J.; McCarrey, J.R.; Griswold, M.D. The Murine Testicular Transcriptome: Characterizing Gene Expression in the Testis during the Progression of Spermatogenesis1. Biol. Reprod. 2004, 71, 319–330. [Google Scholar] [CrossRef] [Green Version]
- Baarends, W.M.; Wassenaar, E.; van der Laan, R.; Hoogerbrugge, J.; Sleddens-Linkels, E.; Hoeijmakers, J.H.J.; de Boer, P.; Grootegoed, J.A. Silencing of Unpaired Chromatin and Histone H2A Ubiquitination in Mammalian Meiosis. Mol. Cell. Biol. 2005, 25, 1041–1053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manterola, M.; Page, J.; Vasco, C.; Berrios, S.; Parra, M.T.; Viera, A.; Rufas, J.S.; Zuccotti, M.; Garagna, S.; Fernan-dez-Donoso, R. A high incidence of meiotic silencing of unsynapsed chromatin is not associated with substantial pachy-tene loss in heterozygous male mice carrying multiple simple robertsonian translocations. PLoS Genet. 2009, 5, e1000625. [Google Scholar] [CrossRef] [Green Version]
- Schimenti, J. Synapsis or silence. Nat. Genet. 2005, 37, 11–13. [Google Scholar] [CrossRef]
- Turner, J.M.; Mahadevaiah, S.K.; Fernandez-Capetillo, O.; Nussenzweig, A.; Xu, X.; Deng, C.X.; Burgoyne, P.S. Si-lencing of unsynapsed meiotic chromosomes in the mouse. Nat. Genet. 2005, 37, 41–47. [Google Scholar] [CrossRef]
- Mahadevaiah, S.K.; Turner, J.M.; Baudat, F.; Rogakou, E.P.; De Boer, P.; Blanco-Rodríguez, J.; Jasin, M.; Keeney, S.; Bonner, W.M.; Burgoyne, P.S. Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 2001, 27, 271–276. [Google Scholar] [CrossRef]
- Turner, J.M.A. Meiotic sex chromosome inactivation. Development 2007, 134, 1823–1831. [Google Scholar] [CrossRef] [Green Version]
- Turner, J.M.; Aprelikova, O.; Xu, X.; Wang, R.; Kim, S.; Chandramouli, G.V.; Barrett, J.; Burgoyne, P.S.; Deng, C.-X. BRCA1, Histone H2AX Phosphorylation, and Male Meiotic Sex Chromosome Inactivation. Curr. Biol. 2004, 14, 2135–2142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, D.S.; Shakes, D.C. Spermatogenesis. Adv. Exp. Med. Biol. 2013, 757, 171–203. [Google Scholar] [CrossRef] [PubMed]
- Das, N.K.; Siegel, E.P.; Alfert, M. Synthetic activities during spermatogenesis in the locust. J. Cell Biol. 1965, 25, 387–395. [Google Scholar] [CrossRef]
- de Almeida, B.R.R.; Noronha, R.C.R.; da Costa, M.J.R.; Nagamachi, C.Y.; Pieczarka, J.C. Meiosis in the scorpion Tityus silvestris: New insights into achiasmatic chromosomes. Biol. Open 2019, 8, bio040352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, S.A. Differential Ribonucleic Acid Synthesis of X and Autosomes during Meiosis. Nature 1963, 200, 1235. [Google Scholar] [CrossRef] [PubMed]
- Henderson, S.A. RNA synthesis during male meiosis and spermiogenesis. Chromosoma 1964, 15, 345–366. [Google Scholar] [CrossRef]
- Hennig, W.; Weyrich, A. Histone modifications in the male germ line of Drosophila. BMC Dev. Biol. 2013, 13, 7. [Google Scholar] [CrossRef] [Green Version]
- Palacios-Gimenez, O.M.; Martí, D.A.; Cabral-De-Mello, D.C. Neo-sex chromosomes of Ronderosia bergi: Insight into the evolution of sex chromosomes in grasshoppers. Chromosoma 2015, 124, 353–365. [Google Scholar] [CrossRef]
- Sotero-Caio, C.G.; de Souza, M.J.; Cabral-de-Mello, D.C.; Brasileiro-Vidal, A.C.; Guerra, M. Phosphorylation of Histone H3S10 in Animal Chromosomes: Is There a Uniform Pattern? Cytogenet. Genome Res. 2011, 135, 111–117. [Google Scholar] [CrossRef]
- Traut, W.; Schubert, V.; Daliková, M.; Marec, F.; Sahara, K. Activity and inactivity of moth sex chromosomes in somatic and meiotic cells. Chromosoma 2019, 128, 533–545. [Google Scholar] [CrossRef]
- Viera, A.; Parra, M.T.; Rufas, J.S.; Page, J. Transcription reactivation during the first meiotic prophase in bugs is not dependent on synapsis. Chromosoma 2016, 126, 179–194. [Google Scholar] [CrossRef] [Green Version]
- Calvente, A.; Viera, A.; Page, J.; Parra, M.T.; Gómez, R.; Suja, J.A.; Rufas, J.S.; Santos, J.L. DNA double-strand breaks and homology search: Inferences from a species with incomplete pairing and synapsis. J. Cell Sci. 2005, 118, 2957–2963. [Google Scholar] [CrossRef] [Green Version]
- Viera, A.; Calvente, A.; Page, J.; Parra, M.; Gómez, R.; Suja, J.; Rufas, J.; Santos, J. X and B chromosomes display similar meiotic characteristics in male grasshoppers. Cytogenet. Genome Res. 2004, 106, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Viera, A.; Santos, J.L.; Parra, M.T.; Calvente, A.; Gomez, R.; de la Fuente, R.; Suja, J.A.; Page, J.; de la Vega, C.G.; Rufas, J.S. Incomplete synapsis and chiasma localization: The chicken or the egg? Cytogenet. Genome Res. 2010, 128, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Viera, A.; Santos, J.L.; Rufas, J.S. Relationship between incomplete synapsis and chiasma localization. Chromosoma 2009, 118, 377–389. [Google Scholar] [CrossRef]
- Page, J.; Suja, J.A.; Santos, J.L.; Rufas, J.S. Squash Procedure for Protein Immunolocalization in Meiotic Cells. Chromosom. Res. 1998, 6, 639–642. [Google Scholar] [CrossRef]
- Parra, M.T.; Yen, T.J.; He, D.; Rufas, J.S.; Suja, J.A.; Page, J.; Valdeolmillos, A. Expression and behaviour of CENP-E at kinetochores during mouse spermatogenesis. Chromosoma 2002, 111, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Peters, A.H.; Plug, A.W.; van Vugt, M.J.; de Boer, P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 1997, 5, 66–68. [Google Scholar] [CrossRef]
- Page, J.; Berríos, S.; Rufas, J.S.; Parra, M.T.; Suja, J.A.; Heyting, C.; Fernández-Donoso, R. The pairing of X and Y chromosomes during meiotic prophase in the marsupial species Thylamys elegans is maintained by a dense plate developed from their axial elements. J. Cell Sci. 2003, 116, 551–560. [Google Scholar] [CrossRef] [Green Version]
- Viera, A.; Parra, M.T.; Page, J.; Santos, J.L.; Rufas, J.S.; Suja, J.A. Dynamic relocation of telomere complexes in mouse meiotic chromosomes. Chromosom. Res. 2003, 11, 797–807. [Google Scholar] [CrossRef]
- Darlington, D.C.; LaCour, L.F. The Handling of Chromosomes; G. Allen & Unwin: London, UK, 1969; 272p. [Google Scholar]
- Viera, A.; Page, J.; Rufas, J. Inverted Meiosis: The True Bugs as a Model to Study. Meiosis 2008, 5, 137–156. [Google Scholar] [CrossRef]
- Church, K. The grasshopper X chromosome. I. States of condensation and the nuclear envelope at G1, S and G2 of premeiotic interphase and at early meiotic prophase. Chromosoma 1979, 71, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Geisinger, A.; Rodríguez-Casuriaga, R.; Benavente, R. Transcriptomics of Meiosis in the Male Mouse. Front. Cell Dev. Biol. 2021, 9, 626020. [Google Scholar] [CrossRef] [PubMed]
- Kotaja, N. MicroRNAs and spermatogenesis. Fertil. Steril. 2014, 101, 1552–1562. [Google Scholar] [CrossRef]
- Shiu, P.K.; Raju, N.B.; Zickler, D.; Metzenberg, R.L. Meiotic Silencing by Unpaired DNA. Cell 2001, 107, 905–916. [Google Scholar] [CrossRef] [Green Version]
- Turner, J.M. Meiotic Silencing in Mammals. Annu. Rev. Genet. 2015, 49, 395–412. [Google Scholar] [CrossRef]
- Cabrero, J.; Teruel, M.; Carmona, F.D.; Jiménez, R.; Camacho, J. Histone H3 lysine 9 acetylation pattern suggests that X and B chromosomes are silenced during entire male meiosis in a grasshopper. Cytogenet. Genome Res. 2007, 119, 135–142. [Google Scholar] [CrossRef]
- Inagaki, A.; Schoenmakers, S.; Baarends, W.M. DNA double strand break repair, chromosome synapsis and tran-scriptional silencing in meiosis. Epigenetics 2010, 5, 255–266. [Google Scholar] [CrossRef] [Green Version]
- Cowell, I.G.; Aucott, R.; Mahadevaiah, S.K.; Burgoyne, P.S.; Huskisson, N.; Bongiorni, S.; Prantera, G.; Fanti, L.; Pimpinelli, S.; Wu, R.; et al. Heterochromatin, HP1 and methyla-tion at lysine 9 of histone H3 in animals. Chromosoma 2002, 111, 22–36. [Google Scholar] [CrossRef]
- Iannelli, F.; Galbiati, A.; Capozzo, I.; Nguyen, Q.; Magnuson, B.; Michelini, F.; D’Alessandro, G.; Cabrini, M.; Roncador, M.; Francia, S.; et al. A damaged genome’s transcriptional landscape through multilayered expression profiling around in situ-mapped DNA double-strand breaks. Nat. Commun. 2017, 8, 15656. [Google Scholar] [CrossRef]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA Double-stranded Breaks Induce Histone H2AX Phosphorylation on Serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Federici, F.; Mulugeta, E.; Schoenmakers, S.; Wassenaar, E.; Hoogerbrugge, J.W.; van der Heijden, G.W.; van Cappellen, W.A.; Slotman, J.A.; van IJcken, I.W.F.; Laven, J.S.; et al. Incomplete meiotic sex chromosome inacti-vation in the domestic dog. BMC Genom. 2016, 16, 291. [Google Scholar]
- Daish, T.J.; Casey, A.E.; Grützner, F. Lack of sex chromosome specific meiotic silencing in platypus reveals origin of MSCI in therian mammals. BMC Biol. 2015, 13, 106. [Google Scholar] [CrossRef] [Green Version]
- Guioli, S.; Lovell-Badge, R.; Turner, J.M.A. Error-Prone ZW Pairing and No Evidence for Meiotic Sex Chromosome Inactivation in the Chicken Germ Line. PLoS Genet. 2012, 8, e1002560. [Google Scholar] [CrossRef] [Green Version]
- Checchi, P.M.; Engebrecht, J. Caenorhabditis elegans histone methyltransferase MET-2 shields the male X chromosome from checkpoint machinery and mediates meiotic sex chromosome inactivation. PLoS Genet. 2011, 7, e1002267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larson, B.J.; Van, M.V.; Nakayama, T.; Engebrecht, J. Plasticity in the Meiotic Epigenetic Landscape of Sex Chromosomes in Caenorhabditis Species. Genetics 2016, 203, 1641–1658. [Google Scholar] [CrossRef] [Green Version]
- Rappaport, Y.; Achache, H.; Falk, R.; Murik, O.; Ram, O.; Tzur, Y.B. Bisection of the X chromosome disrupts the initiation of chromosome silencing during meiosis in Caenorhabditis elegans. Nat. Commun. 2021, 12, 4802. [Google Scholar] [CrossRef] [PubMed]
- Vibranovski, M. Meiotic Sex Chromosome Inactivation in Drosophila. J. Genom. 2014, 2, 104–117. [Google Scholar] [CrossRef] [Green Version]
- Mahadevaraju, S.; Fear, J.M.; Akeju, M.; Galletta, B.J.; Pinheiro, M.M.L.S.; Avelino, C.C.; Cabral-De-Mello, D.C.; Conlon, K.; Dell’Orso, S.; Demere, Z.; et al. Dynamic sex chromosome expression in Drosophila male germ cells. Nat. Commun. 2021, 12, 892. [Google Scholar] [CrossRef]
- Namekawa, S.H.; Park, P.J.; Zhang, L.F.; Shima, J.E.; McCarrey, J.R.; Griswold, M.D.; Lee, J.T. Postmeiotic sex chro-matin in the male germline of mice. Curr. Biol. 2006, 16, 660–667. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.; Raman, M.J. Dynamics of radiation induced γH2AX foci in chromatin subcompartments of mouse pachytene spermatocytes and round spermatids. Mol. Reprod. Dev. 2014, 81, 484–496. [Google Scholar] [CrossRef] [PubMed]
- Checchi, P.M.; Lawrence, K.S.; Van, M.; Larson, B.J.; Engebrecht, J. Pseudosynapsis and Decreased Stringency of Meiotic Repair Pathway Choice on the Hemizygous Sex Chromosome of Caenorhabditis elegans Males. Genetics 2014, 197, 543–560. [Google Scholar] [CrossRef] [Green Version]
- Gil-Fernández, A.; Ribagorda, M.; Martín-Ruiz, M.; López-Jiménez, P.; Laguna, T.; Gómez, R.; Parra, M.T.; Viera, A.; Veyru-nes, F.; Page, J. Meiotic Behavior of Achiasmate Sex Chromosomes in the African Pygmy Mouse Mus mattheyi Offers New Insights into the Evolution of Sex Chromosome Pairing and Segregation in Mammals. Genes 2021, 12, 1434. [Google Scholar] [CrossRef] [PubMed]
- Costa, Y.; Speed, R.M.; Gautier, P.; Semple, C.A.; Maratou, K.; Turner, J.M.; Cooke, H.J. Mouse MAELSTROM: The link between meiotic silencing of unsynapsed chromatin and microRNA pathway? Hum. Mol. Genet. 2006, 15, 2324–2334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Handel, M.A. The XY body: A specialized meiotic chromatin domain. Exp. Cell Res. 2004, 296, 57–63. [Google Scholar] [CrossRef]
- Hoyer-Fender, S. Molecular aspects of XY body formation. Cytogenet. Genome Res. 2003, 103, 245–255. [Google Scholar]
- Barchi, M.; Mahadevaiah, S.; di Giacomo, M.; Baudat, F.; de Rooij, D.G.; Burgoyne, P.S.; Jasin, M.; Keeney, S. Surveil-lance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Mol. Cell. Biol. 2005, 25, 7203–7215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgoyne, P.S.; Mahadevaiah, S.K.; Turner, J.M.A. The consequences of asynapsis for mammalian meiosis. Nat. Rev. Genet. 2009, 10, 207–216. [Google Scholar] [CrossRef]
- Royo, H.; Polikiewicz, G.; Mahadevaiah, S.K.; Prosser, H.; Mitchell, M.; Bradley, A.; de Rooij, D.; Burgoyne, P.S.; Turner, J.M. Evidence that Meiotic Sex Chromosome Inactivation Is Essential for Male Fertility. Curr. Biol. 2010, 20, 2117–2123. [Google Scholar] [CrossRef] [Green Version]
- Waters, P.D.; Ruiz-Herrera, A. Meiotic Executioner Genes Protect the Y from Extinction. Trends Genet. 2020, 36, 728–738. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Viera, A.; Parra, M.T.; Arévalo, S.; García de la Vega, C.; Santos, J.L.; Page, J. X Chromosome Inactivation during Grasshopper Spermatogenesis. Genes 2021, 12, 1844. https://doi.org/10.3390/genes12121844
Viera A, Parra MT, Arévalo S, García de la Vega C, Santos JL, Page J. X Chromosome Inactivation during Grasshopper Spermatogenesis. Genes. 2021; 12(12):1844. https://doi.org/10.3390/genes12121844
Chicago/Turabian StyleViera, Alberto, María Teresa Parra, Sara Arévalo, Carlos García de la Vega, Juan Luis Santos, and Jesús Page. 2021. "X Chromosome Inactivation during Grasshopper Spermatogenesis" Genes 12, no. 12: 1844. https://doi.org/10.3390/genes12121844
APA StyleViera, A., Parra, M. T., Arévalo, S., García de la Vega, C., Santos, J. L., & Page, J. (2021). X Chromosome Inactivation during Grasshopper Spermatogenesis. Genes, 12(12), 1844. https://doi.org/10.3390/genes12121844