CRISPR/Cas9 Induced Somatic Recombination at the CRTISO Locus in Tomato
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Plasmids Construction
2.3. Plant Transformation and Genotyping
2.4. NHEJ Analysis Using Illumina HTS
2.5. HR Events Analysis in the CRTISO Region
2.6. CO Events Analysis Using Illumina Whole Genome Sequencing (WGS)
3. Results
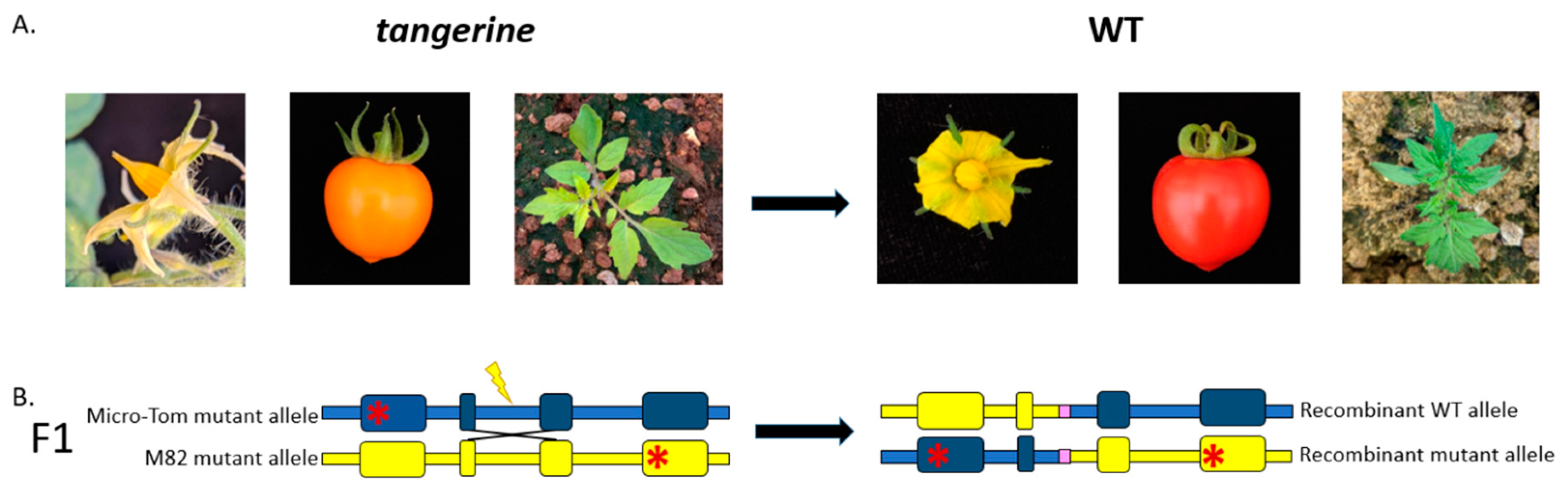
3.1. Gain-of-Function Phenotypic Assay Design for HR Events Identification
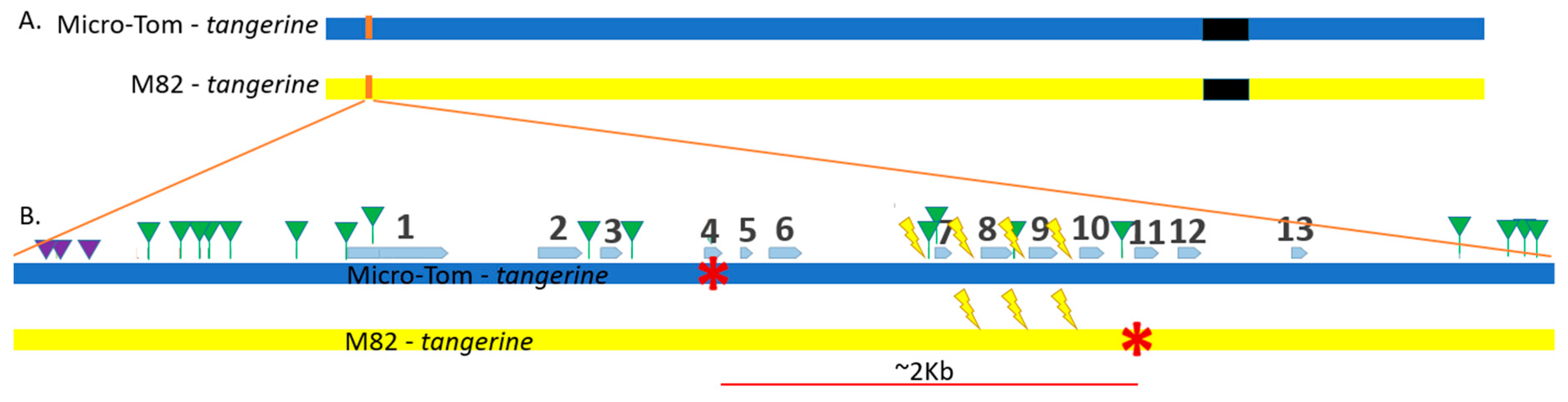
3.2. Three out of Four CRTISO Targets Induce Efficient NHEJ DSB Repair
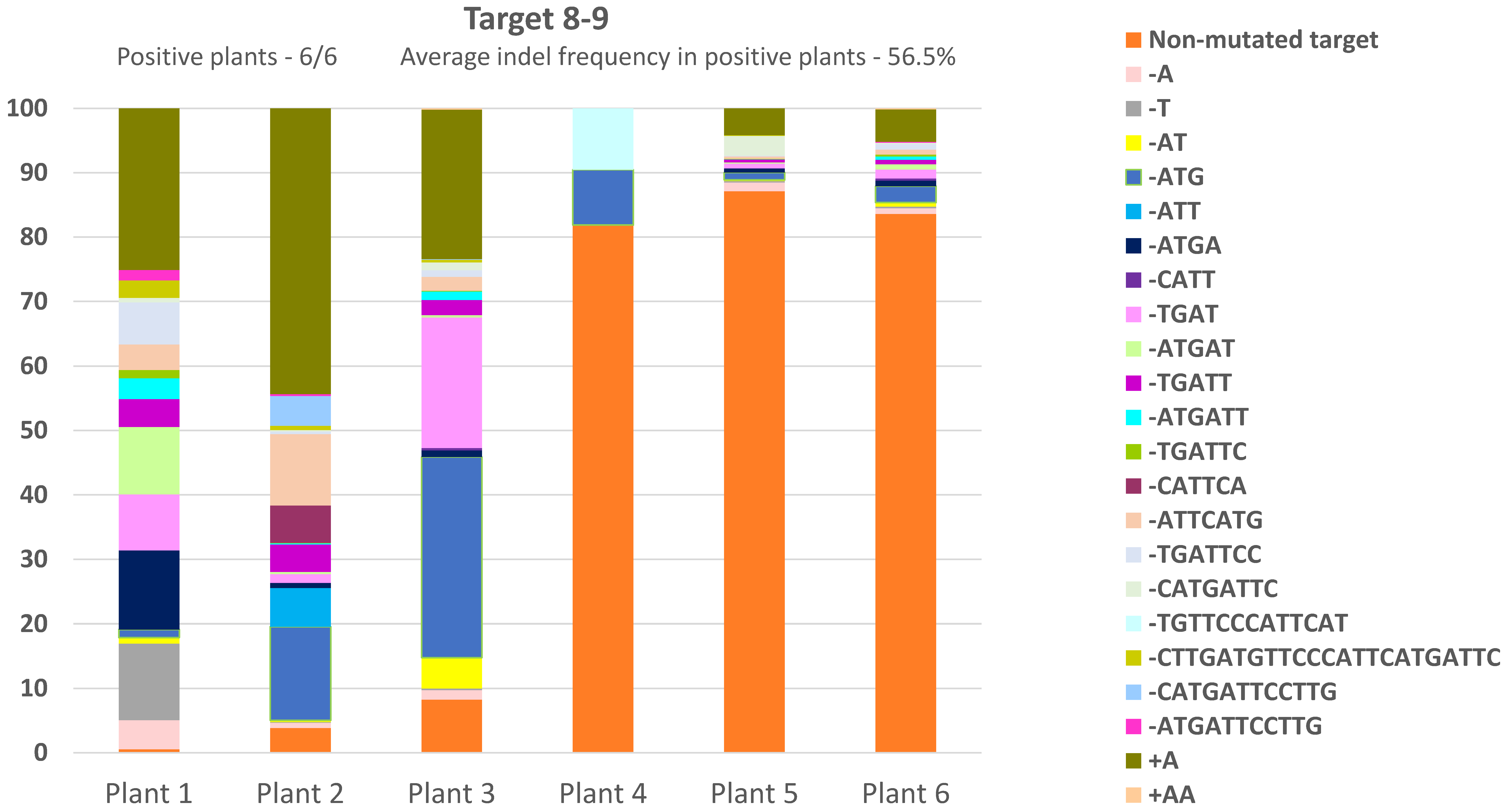
3.3. CRTISO Target 8–9 Gives Germinal Transmission of Somatic DSB IHSR Events
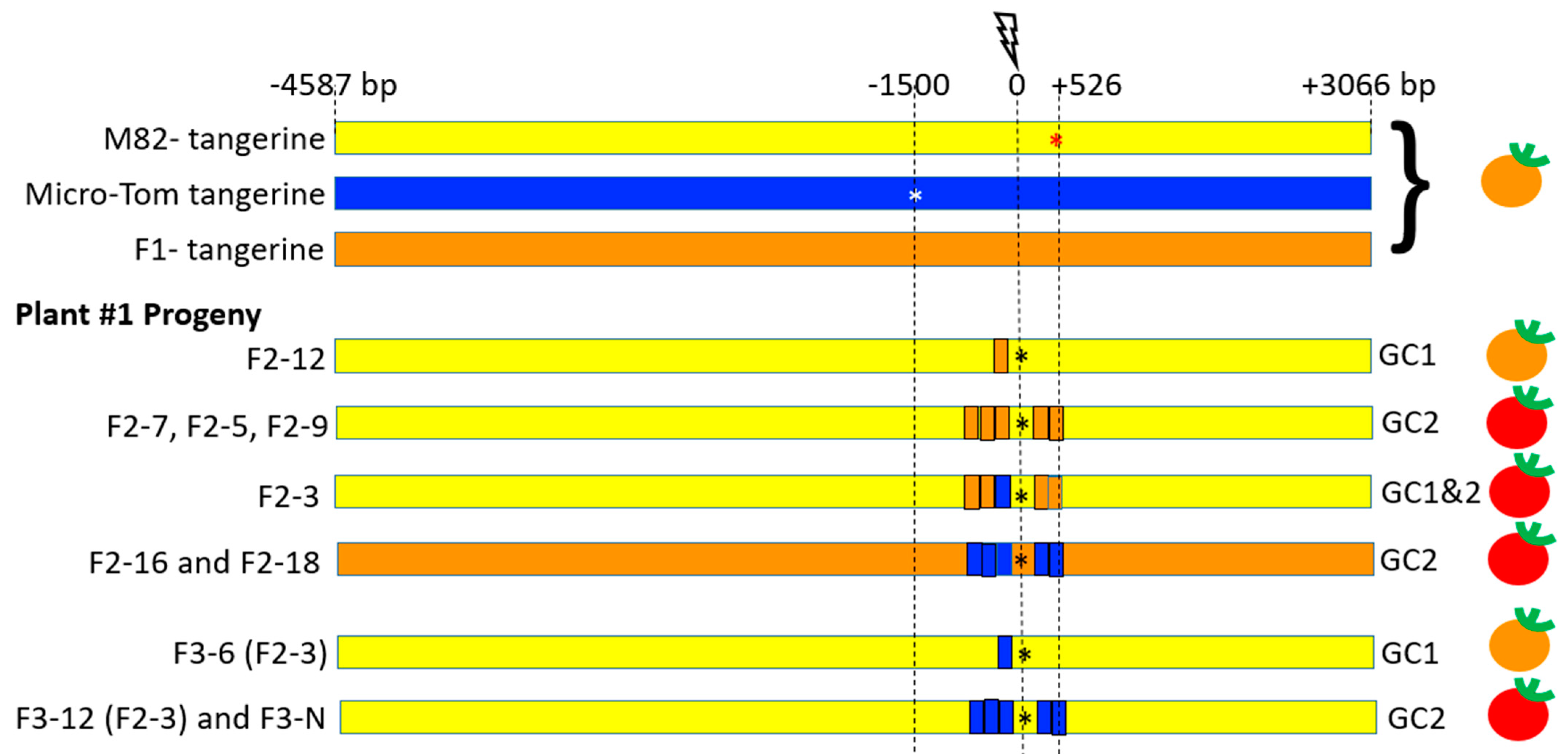
3.4. CRTISO Target 8–9 Activity in Somatic Tissue Leads to Germinal Transmission of GC and CO Events
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Page, S.L.; Hawley, R.S. Chromosome choreography: The meiotic ballet. Science 2003, 301, 785–789. [Google Scholar] [CrossRef]
- Choi, K.; Zhao, X.; Tock, A.J.; Lambing, C.; Underwood, C.J.; Hardcastle, T.J.; Serra, H.; Kim, J.; Cho, H.S.; Ziolkowski, P.A.; et al. Nucleosomes and DNA methylation shape meiotic DSB frequency in Arabidopsis thaliana transposons and gene regulatory regions. Genome Res. 2018, 28, 532–546. [Google Scholar] [CrossRef]
- He, Y.; Wang, M.; Dukowic-Schulze, S.; Zhou, A.; Tiang, C.L.; Shilo, S.; Sidhu, G.K.; Eichten, S.; Bradbury, P.; Springer, N.M.; et al. Genomic features shaping the landscape of meiotic double-strand-break hotspots in maize. Proc. Natl. Acad. Sci. USA 2017, 114, 12231–12236. [Google Scholar] [CrossRef]
- International Wheat Genome Sequencing Consortium (IWGSC). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014, 345, 1251788. [Google Scholar] [CrossRef]
- Shilo, S.; Melamed-Bessudo, C.; Dorone, Y.; Barkai, N.; Levy, A.A. DNA crossover motifs associated with epigenetic modifications delineate open chromatin regions in arabidopsis. Plant Cell 2015, 27, 2427–2436. [Google Scholar] [CrossRef]
- Mercier, R.; Mezard, C.; Jenczewski, E.; Macaisne, N.; Grelon, M. The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 2015, 66, 297–327. [Google Scholar] [CrossRef] [PubMed]
- Traas, J. Organogenesis at the Shoot Apical Meristem. Plants 2018, 8, 6. [Google Scholar] [CrossRef]
- Evans, H.J. Chromatid aberrations induced by γ irradiation. I. The structure and frequency of chromatid interchanges in diploid and tetraploid cells of Vicia faba. Genetics 1961, 46, 257–275. [Google Scholar] [PubMed]
- Gorbunova, V.; Avivi-Ragolski, N.; Shalev, G.; Kovalchuk, I.; Abbo, S.; Hohn, B.; Levy, A.A. A new hyperrecombinogenic mutant of Nicotiana tabacum. Plant J. 2000, 24, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Filler-Hayut, S.; Melamed-Bessudo, C.; Levy, A.A. Targeted recombination between homologous chromosomes for precise breeding in tomato. Nat Commun. 2017, 8, 15605. [Google Scholar] [CrossRef] [PubMed]
- Isaacson, T.; Ronen, G.; Zamir, D.; Hirschberg, J. Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of β-carotene and xanthophylls in plants. Plant Cell 2002, 2, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Dahan-Meir, T.; Filler-Hayut, S.; Melamed-Bessudo, C.; Bocobza, S.; Czosnek, H.; Aharoni, A.; Levy, A.A. Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 2018, 95, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Menda, N.; Semel, Y.; Peled, D.; Eshed, Y.; Zamir, D. In silico screening of a saturated mutation library of tomato. Plant J. 2004, 38, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Ensembl Plants. Available online: http://plants.ensembl.org (accessed on 1 December 2020).
- Sarrion-Perdigones, A.; Palaci, J.; Granell, A.; Orzaez, D. Design and construction of multigenic constructs for plant biotechnology using the Golden Braid cloning strategy. Methods Mol. Biol. 2014, 1116, 133–151. [Google Scholar] [CrossRef] [PubMed]
- Fauser, F.; Schiml, S.; Puchta, H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 2014, 79, 348–359. [Google Scholar] [CrossRef]
- Park, J.; Lim, K.; Kim, J.S.; Bae, S. Cas-analyzer: An online tool for assessing genome editing results using NGS data. Bioinformatics 2017, 33, 286–288. [Google Scholar] [CrossRef] [PubMed]
- Blecher-Gonen, R.; Barnett-Itzhaki, Z.; Jaitin, D.; Amann-Zalcenstein, D.; Lara-Astiaso, D.; Amit, I. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat. Protoc. 2013, 8, 539–554. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Picard Tool. Available online: https://broadinstitute.github.io/picard/ (accessed on 1 December 2020).
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Demirci, S.; van Dijk, A.D.; Sanchez Perez, G.; Aflitos, S.A.; de Ridder, D.; Peters, S.A. Distribution, position and genomic characteristics of crossovers in tomato recombinant inbred lines derived from an interspecific cross between Solanum lycopersicum and Solanum pimpinellifolium. Plant J. 2017, 89, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Tsaponina, O.; Haber, J.E. Frequent Interchromosomal Template Switches during Gene Conversion in S. cerevisiae. Mol. Cell. 2014, 55, 615–625. [Google Scholar] [CrossRef] [PubMed]
- Wright, W.D.; Shah, S.S.; Heyer, W.D. Homologous recombination and the repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10524–10535. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ben Shlush, I.; Samach, A.; Melamed-Bessudo, C.; Ben-Tov, D.; Dahan-Meir, T.; Filler-Hayut, S.; Levy, A.A. CRISPR/Cas9 Induced Somatic Recombination at the CRTISO Locus in Tomato. Genes 2021, 12, 59. https://doi.org/10.3390/genes12010059
Ben Shlush I, Samach A, Melamed-Bessudo C, Ben-Tov D, Dahan-Meir T, Filler-Hayut S, Levy AA. CRISPR/Cas9 Induced Somatic Recombination at the CRTISO Locus in Tomato. Genes. 2021; 12(1):59. https://doi.org/10.3390/genes12010059
Chicago/Turabian StyleBen Shlush, Ilan, Aviva Samach, Cathy Melamed-Bessudo, Daniela Ben-Tov, Tal Dahan-Meir, Shdema Filler-Hayut, and Avraham A. Levy. 2021. "CRISPR/Cas9 Induced Somatic Recombination at the CRTISO Locus in Tomato" Genes 12, no. 1: 59. https://doi.org/10.3390/genes12010059
APA StyleBen Shlush, I., Samach, A., Melamed-Bessudo, C., Ben-Tov, D., Dahan-Meir, T., Filler-Hayut, S., & Levy, A. A. (2021). CRISPR/Cas9 Induced Somatic Recombination at the CRTISO Locus in Tomato. Genes, 12(1), 59. https://doi.org/10.3390/genes12010059




