Abstract
Gene duplication is an important evolutionary mechanism allowing to provide new genetic material and thus opportunities to acquire new gene functions for an organism, with major implications such as speciation events. Various processes are known to allow a gene to be duplicated and different models explain how duplicated genes can be maintained in genomes. Due to their particular importance, the identification of duplicated genes is essential when studying genome evolution but it can still be a challenge due to the various fates duplicated genes can encounter. In this review, we first describe the evolutionary processes allowing the formation of duplicated genes but also describe the various bioinformatic approaches that can be used to identify them in genome sequences. Indeed, these bioinformatic approaches differ according to the underlying duplication mechanism. Hence, understanding the specificity of the duplicated genes of interest is a great asset for tool selection and should be taken into account when exploring a biological question.
1. Introduction
The eukaryotic genome organization is complex and contains different types of sequences with much of them being non-coding sequences that may have an important impact on genome functioning and regulation. Moreover, genomes are highly dynamic with several ongoing processes allowing the creation of genetic novelty necessary for species to evolve and adapt to changing environments. Among the different possibilities, gene duplication is a very important mechanism providing new genetic material and opportunities to acquire new functions [1].
In particular, numerous examples have described the role of duplication in some cases of adaptation to environmental conditions [2]. For example, gene duplication has played a role in nutrient transport under stress conditions, in protection against heat, cold, or salty environments, in the resistance to drugs and pesticides, but also in the adaptation to domestication. Gene duplication can also be involved in speciation, especially via whole genome duplication (WGD) as it is suspected in plants, where a correlation has been observed between WGD and increased rates of speciation or divergence [3]. In particular, this mechanism is thought to have generated the new flowering plant Mimulus peregrinus within the last 140 years [4]. Although less numerous than in plants, some examples also exist in animals such as in Drosophila where the hybrid-male sterility gene Odysseus was formed by gene duplication [5]. On the other hand, duplication may also have important deleterious effects in humans and can be associated with some diseases [6]. For example, the analysis of human genes linked to diseases made it possible to show that 80% of them have been duplicated in their evolutionary history, the disease-associated mutation being associated with only one of the duplicated copies [7]. Recently, the analysis of the evolution of cancer suppression in mammals revealed that species known to be resistant to cancer contain the most cancer gene copies [8].
Duplicated genes are also called paralogs in contrast to orthologs, to refer to their homologous relationship, i.e., the fact that they descend from a common ancestor via a duplication event rather than a speciation event. The terminology concerning duplicated genes can be complex and depends upon different factors (for a review, see [9]). In particular, it may be difficult to assess precisely the evolutionary relationships between duplicated genes since duplication is often followed by speciation and gene loss. Several definitions have been proposed to integrate more or less precise ideas concerning the mechanism of formation and the evolutionary relationship among paralogs. For example, ohnologs correspond to paralogs that have been created by WGD [10]. Three new definitions, pro-ortholog, semi-ortholog, and trans-homolog, were proposed to account for situations in which one or both lineages that lead to two present-day genes involve gene duplication [11]. In that respect, a pro-ortholog is a gene that is orthologous to the ancestor of a set of paralogs of the gene under consideration whereas a semi-ortholog is one of the descendants of an ortholog of the gene under consideration, after that gene has duplicated. Trans-homologs can be defined as genes related to each other via two independent duplication events from the same ancestral gene. Moreover, it is also possible to link paralogous relationships to speciation events with the definition of in-paralogs and out-paralogs [12]. When paralogs from a given lineage have evolved by gene duplication that happened after a speciation event, they can be referred to as in-paralogs. On the opposite, paralogous genes which have evolved by duplication events happening before a speciation event, can be referred to as out-paralogs. Many other terms, although less used, have been proposed to take into account chromosomal position retention, the combination of vertical and horizontal transmissions or to highlight paralogous genes appearing to be orthologs due to differential gene loss [9].
In a genome, duplicated genes can thus be formed by various mechanisms and may have different ages and fates. This makes their bioinformatic identification all the more difficult since according to the methods used, different duplicated gene datasets will be identified inside the same organism. In this review, we thus aim at describing the evolutionary processes implicated in the formation and the fate of duplicated genes as well as the different bioinformatic approaches that can be used to identify them in genome sequences. The question of deciphering the evolutionary relationships among duplicated genes will not be discussed in detail, for reviews on the subject see [13,14].
2. Evolutionary Processes Leading to the Formation and the Fate of Duplicated Genes
2.1. How to Make New from Old: Duplication Mechanisms
Duplicated genes can appear under various forms which are the consequences of the mechanisms that generated them. Some of the mechanisms can be particularly well documented but it is not always the case, at least for some organisms. According to the mechanism, the results concerning the gene content can be different since it can either involve individual genes or all genes on entire chromosomes (Figure 1).
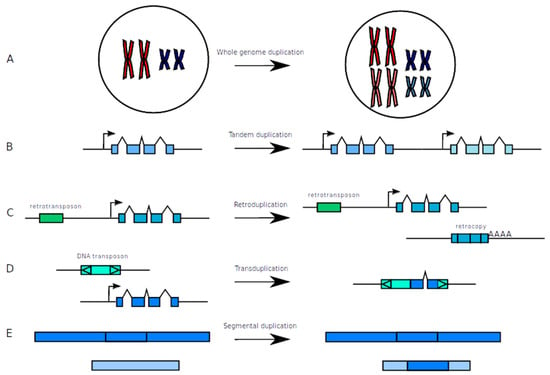
Figure 1.
The different types of duplications. (A) Whole genome duplication which implies complete chromosome duplication. (B) Tandem duplications which produce identical adjacent sequences. (C) Retroduplication, which produces a retrocopy of a gene devoid of introns and with a polyA tail. (D) Transduplication in which a DNA transposon acquires fragments of genes. (E) Segmental duplications which correspond to long stretches of duplicated sequences with high identity.
2.1.1. Whole Genome Duplication (WGD)
In the first mechanism, duplicated genes arise from the duplication of complete chromosomes, which correspond to what is called whole genome duplication (WGD) (Figure 1A). In that case, all chromosomes from a genome will be duplicated, leading each gene from the genome to exist in two copies. This type of duplication has been well documented in plants and is defined as polyploidization, for which it is possible to distinguish the mechanism of hybridization between different species (allopolyploidization) or inside a given species (autopolyploidization) [15]. Different mechanisms have been shown to produce this outcome such as polyspermy, non-reduced gametes or incomplete mitosis during the early stage of embryo development [16]. Gene duplication, independently of the mechanism of formation, is largely present in plant genomes since on average 64.5% of genes have been recognized as duplicated in an analysis that considered 41 genomes and used the same methodology to build gene families, with a range going from 45.5% in a moss to 84.4% in the apple tree [17]. It is possible to estimate that several WGD events took place during the evolution of plant species, the most ancient happening in the ancestor of all seed plants about 319 million years ago and another more recent before the diversification of angiosperms 192 million years ago [18]. A large number of WGD are also consecutive to recent events. For example, the wheat group has evolved through different complex hybridizations among species from the plant genera Aegilops and Triticum followed by genome doubling. The most recent event giving birth to allotetraploid wheat (two different diploid parental species) has been proposed to occur about 300,000 to 500,000 years ago, while an allohexaploid wheat (three different diploid parental species) was formed only about 10,000 years ago [19]. Another domesticated plant, the Oilseed crop (Brassica napus L.) originated between around 6700 to 51,000 years ago by hybridization between two species, which were themselves polyploids [20,21,22,23]. The consequences of the different types of hybridization, and thus WGD, are that many plants arising from these processes have very large genomes. On the contrary, in other organisms, there are still some debates concerning the occurrence of WGD versus several more local duplications. This is the case in vertebrates in which the “2R hypothesis”, originally proposed by Susumu Ohno [1], assumes the existence of two rounds of WGD in their early evolution. The “2R hypothesis” has been the subject of numerous studies to prove this theory. This has led to numerous works published during the last twenty years either in favor of the “2R hypothesis”, or in favor of only one round of WGD, or rejecting any idea of WGD (see for a review [24]). The main reason explaining the difficulty to determine whether two rounds of WGD happened or not very anciently comes from two phenomena which could blur the signal. Both phenomena make it harder to detect ancient WGD either through the loss of signal (fractionation) or increased complexity (diploidization). The fractionation is characterized by a heavy loss of duplicated genes following WGD [25]. The diploidization refers to the chromosomal rearrangements and segment loss often observed after WGD when the genome goes back to a diploid state [26]. Indeed, a return to diploidization involves the transition to disomic inheritance as it has been proposed in Salmonid species, for example [27]. In a recent work, 61 animal genomes were used to reconstruct the gene order of the ancestral Amniota genome, to identify duplicated genes produced by the 2R in this genome, and to reconstruct the timeline of events conducting a pre-vertebrate genome going from 17 chromosomes to 54 after the occurrence of two successive WGD [28]. Although a lot of arguments seem now to be more in favor of the “2R hypothesis”, the question is still not completely resolved. Very recently, an investigation using phylogenetic approaches and tree topology comparisons of gene families containing at least three members and located on several human chromosomes led to the conclusion that small-scale duplication (SSD) events scattered on all the animal history were more likely to be involved in vertebrate genome evolution rather than WGD [29].
2.1.2. Tandem Duplications
At smaller scales, local events called tandem duplication, create a novel copy of a gene next to it producing tandemly arrayed genes (TAGs) (Figure 1B). The molecular mechanism involved consists in unequal crossing overs, which can produce regions containing one or several genes, depending on the position of the breakage on the chromosomes [30]. These unequal crossing overs are either the result of homologous recombination between sequences (on homologous chromosomes or on sister chromatids) or of non-homologous recombination by replication-dependent chromosome breakages [31]. When multiple occurrences of unequal crossovers happen, it might lead to increasing or decreasing copy numbers in gene families. The molecular mechanism allowing the recombination depends on the sequences that promote the exchange between chromosomes or chromatids, which can be long direct repeats (>100 bp) and short ones (>12 bp) [32]. When repeats are long, the tandem duplication can arise via the homologous recombination whereas when they are short, duplication arises by single-strand annealing, template switching, or non-homologous end joining. This type of duplication leads to the formation of clusters of duplicated genes sometimes representing specific gene families. For example, this mechanism has been shown to confer soybean resistance against cyst nematode (Heterodera glycines) at Rhg1, a quantitative trait locus on chromosome 18, by changing the copy number variation that increases the gene expression [33]. In maize, thousands of tandem gene duplicates were identified that correspond to about 10% of the annotated genes [34]. Some of them may contribute to a phenotypic variation such as the White Cap locus, which provided the possibility to select white-grain color [35].
2.1.3. Duplications Via the Action of Transposable Elements
Duplicated sequences can also be formed by the action of transposable elements (TEs) according to different ways. TEs are repeated sequences with the ability to move from one position to another along and across chromosomes and which may represent a very large proportion in genomes, going from about 3% in yeast to more than 80% in maize [36,37]. When they are mobilized, some of them can drag host sequences with them or can target the gene transcript, all of these having the consequence to duplicate the host sequences. There are two mechanisms by which TEs can promote duplication of complete genes or part of genes as a direct consequence of their transposition: The retroposition and the transduplication (Figure 1C,D). The retroposition mechanism consists of the reverse transcription of a messenger RNA from a host gene into a cDNA then inserted in another location of the genome by the action of the enzymes of a retrotransposon [38]. Genes submitted to this mechanism are located in the 3′ side of retrotransposons and benefit from a transcription read-through initiated inside the TE [39,40]. This new gene, that is called a retrocopy, has particular features such as the presence of a polyA tail in its 3′ end, the loss of introns, and the presence of target site duplication at both extremities which are the signature of its insertion. Retrocopies have been discovered in different organisms such as in mammals, and especially in the human genome where thousands of them have been identified [41,42]. Although less numerous, retrocopies have also been identified in insects such as in Drosophila melanogaster [43,44], or in the mosquito Anopheles gambiae [45]. Interestingly, it has been observed a bias in the location in the genome of these retrocopieswhich move from the X chromosome toward the autosomes in the insects [43,45]. In mammals, X chromosomes seem to have generated and recruited more retrocopies than the other chromosomes [46]. This type of duplicated gene is also found in plant genomes. For example, in the rice genome (Oryza sativa), between 491 and 1235 retrocopies were identified according to the methodology [47,48]. In Arabidopsis thaliana, 271 retrocopies were identified [48]. The other mechanism that involves TEs, the transduplication, happens when DNA transposons incorporate unspliced fragments of different genes, although the true mechanism is still unknown [49]. The gene fragments may still contain introns. First discovered in maize, this mechanism has then been documented only in plants such as A. thaliana, Japanese morning glory, soybean, and rice [49,50,51,52,53,54]. In rice especially, a particular type of DNA transposons called Pack-MULE, which represent about 3000 insertions in the genome, has been shown to contain sequence fragments derived from more than a thousand genes [54].
2.1.4. Segmental Duplications
At a larger scale, segmental duplications, also called “low copy repeats”, correspond to very long stretches of duplicated sequences that can span between 1 to 200 kb and that share a sequence identity higher than 95% (Figure 1E; [55]). They have been first observed in several eukaryotic organisms such as the yeast [56] and humans [57]. These duplications are formed from the replicative transpositions of small portions of chromosomes. However, the exact mechanism is unclear and the fact that these duplications do not generate tandem repeats and that no short direct repeats at junction have been found suggests that neither unequal crossing-overs nor double-stranded breakages followed by repair are involved [55]. It has been proposed that in yeast, the segmental duplications could result from replication accidents [58] and that most of these sequences present a certain level of instability that can be rescued when translocation within another chromosome happens [59]. In Drosophila, high enrichment in TEs at segmental duplication extremities have been observed, indicating their possible implication in the duplication formation by homologous repair ends [60]. Similarly in mammals, particular types of TEs were found to be enriched at the junction of segmental duplications [61,62]. In the human genome, the sequence divergence of the duplicated segments has been used to estimate their evolutionary age which corresponds to the divergence between the New and Old World monkeys, 35 million years ago [63]. Segmental duplications account for an average of 13.7% of the total human genome, located in pericentromeric and subtelomeric regions [64]. Moreover, some chromosomes seem to be enriched in duplicated segments of this type such as the Y chromosome where they represent 50.4% of this chromosome [64]. In other mammals such as rat, mouse, or dog, this type of duplication is less abundant [64]. The comparative analysis of several genomes of Lepidoptera species made it possible to determine a large variation in the content of segmental duplications, going from 1.2% in the silkworm (Bombyx mori) to 15.2% in the postman butterfly (Heliconius melpomene) [65].
2.1.5. Differences among Duplication Types
Notable differences depending on the formation mechanisms in terms of function, expression, evolutionary constraints, and protein interactions have been reported. For example, in yeast duplicated genes issued from WGD are associated with different sets of functions when compared to duplicated genes generated by SSDs [66,67]. This has also been shown in plants [68,69,70,71,72]. In Arabidopsis and rice, for example, TAGs were found to be enriched with genes that encode membrane proteins and with functions in “abiotic and biotic stress” when compared to other duplicated genes. TAGs were also underrepresented in genes involved in transcription and DNA or RNA binding functions compared to non-TAG duplicated genes [73]. More recently, Acharya et al. [74] reported a higher multifunctionality, estimated by the number of GO and Pfam annotations, for WGD duplicated genes compared to SSD genes in humans. They also observed a significantly higher proportion of essential genes among the WGD genes relative to SSD genes.
It has also been observed that duplicated genes differ in divergence of expression according to the mode of duplication. In Arabidopsis and in poplar, for example, WGD genes were found to display a lower divergence of expression than other duplicated genes [71,75]. In a study deciphering more deeply the different types of duplicated genes, Wang et al. [48] observed that in Arabidopsis and rice, WGD genes and TAGs displayed a lower divergence of expression than proximal, retrotransposed dispersed, and DNA based transposed duplicated genes.
In a recent study in Angiosperms, WGD duplicated genes were shown to be under stronger constraints to diverge at the sequence and expression level relative to SSDs [76]. It has also been observed that among WGD genes, those that are also involved in local duplications showed higher non synonymous substitution rates (Ka) and selection rates (Ka/Ks) than nonlocally duplicated WGD genes indicating that they evolve faster [77].
When considering protein-protein interactions (PPI) networks, it has been observed that the fraction of shared PPI between paralogous genes was higher when the genes shared the same function and showed a higher co-expression [78]. Among duplicated genes, WGD gene pairs displayed a higher fraction of shared PPIs than other duplicated gene pairs [78]. Arsovski et al. [79] examined the density of Arabidopsis DNaseI footprints, which are locations of protein binding sites, in the 1000 bp flanking upstream and downstream sequences of duplicated genes. They found that WGD duplicated genes had more footprints than TAGs. Moreover, WGD duplicated genes formed denser and more complex regulatory networks than TAGs when genome-wide regulatory networks were analyzed.
In summary, mechanisms that can lead to the formation of duplicated genes are various. The fates encountered by the new duplicated genes are also distinct and may depend on several factors.
2.2. Evolutionary Fates of Duplicated Genes
2.2.1. Pseudogenization and Neo-Functionalization
After their formation, duplicated genes can encounter various fates (for a complete review on this matter, see [80]). The most likely is the pseudogenization or the complete loss of one copy since only one gene copy will continue to be under purifying selective constraints for its current function, leaving the other one free to accumulate deleterious mutations. These pseudogenes can be conserved in the genome. For example, A. thaliana and the rice contain thousands of pseudogenes in their genomes [81]. In humans, the olfactory receptor gene families have been shown to be composed of between 60–70% pseudogenes whereas in dogs pseudogenes represent less than 20% in those gene families, explaining the reduced sense of smell in humans [82,83]. Sometimes, however, the process of mutation accumulation can drive to a completely different outcome. Different models of population genetics have been proposed to highlight evolutionary mechanisms explaining the different fates of duplicated genes allowing them to be maintained in organisms (for specific reviews on this subject, see [84,85]). It has been proposed that three main steps are needed for duplicated genes to be maintained: Phase 1 consists of the origin of a genetic change through mutation, phase 2 corresponds to the fixation period when the mutation segregates in the population, and phase 3 corresponds to the preservation period where the duplication is conserved. Although infrequent, a mutation can provide a new allele giving rise to a new function for the gene copy. If this function is advantageous, it will be subjected to distinct selective constraints leading to its fixation in the population, in a process called neo-functionalization. There are two models to explain this mechanism. The Dyhkhuzen-Hartl model proposes that the mutations at the duplicated gene are fixed by drift and later, during a change in the environment, the new gene will become advantageous for the organism [86], whereas the “Adaptation model” proposes that an adaptive mutation is fixed at one of the duplicated locus because it is immediately advantageous [85]. Various examples of neo-functionalization have been described. The analysis of the copper transporter gene family, which contains the two genes Ctr1 and Ctr2, suggested that the metazoan Ctr2 arose several hundred million years ago via a duplication event of the Ctr1 genomic locus. The resulting Ctr2 then lost the ability to transport copper but gained the ability to regulate Ctr1 cleavage [87]. In mammals, the family of retinoic acid receptors (RARs), which play a role in the embryonic development, contains three duplicated genes, RARα, β, and γ, with RARβ having kept the ancestral RAR role, while the two others have diverged both in ligand-binding capacity and in expression patterns suggesting that neo-functionalization occurred at both the expression and the functional levels for these genes [88]. A wide transcriptomic analysis in maize made it possible to determine that 13% of all gene pairs generated by WGD have been submitted to regulatory neo-functionalization in leaves [89]. The analysis of a gene family containing three members in the D. melanogaster genome made it possible to show that the family was created by two rounds of tandem gene duplication in the last five million years and that the two new duplicated copies have diverged in function from the parental copy [90].
2.2.2. Sub-Functionalization and Functional Redundancy
Alternatively to the possibilities of pseudogenization and neo-functionalization, the duplicated genes can be submitted to sub-functionalization. In this process, accumulation of mutations drives the subdivision of the ancestral gene function among the duplicated genes. The complementarity can come from a change in the regulatory sequences, leading the two copies to have different expression patterns that will recapitulate the ancestral one when taken together, for example [91]. Several models have been proposed to explain this mechanism. In the first model called duplication–degeneration–complementation (DDC) the two gene copies will acquire complementary functions through independent mutations, which will lead to the need of the two copies to fulfill the original function by drift rather than by selective constraints [91]. Another possibility is described by the “gene sharing” model in which the acquisition of two expression domains could predate the duplication, with each copy losing one of the two afterward [92,93]. A close model corresponds to the “specialization” model [94] which proposes that an ancestral function is split among paralogs that will be expressed in different tissues or developmental stages. These two last models predict that the duplication will be followed by advantageous mutations in all duplicated genes with positive selection patterns detectable in their sequences. Moreover, it is supposed that the ancestral gene is able to fulfill the function of all duplicated genes but not so well. Numerous examples of sub-functionalization have been identified in eukaryotes. For example, in mammals, the Agouti-melanocortin system is represented by the Agouti protein (ASIP) and the Agouti-related protein (AgRP) whose expression patterns with distinct physiologic functions were acquired through sub-functionalization such that the current expression pattern and function of each protein correspond to a subset of the ancestral gene [95]. In tomato, two members of the gene family encoding phytochromes, which are light receptors playing a role in plant development, exhibit both common and non-redundant functions suggesting that they have sub-functionalized since their duplication [96]. Finally, it is also possible for the two copies of a gene to be both maintained in the genome by dosage subfunctionalization, each expressing the ancestral function, leading to a functional redundancy [97,98]. A model proposed to explain this possibility stipulates that expression reduction could help the retention of duplicates and the conservation of their ancestral function [99]. Several cases have been identified such as, for example, two members of the mammalian HOX gene complex, Hoxa3 and Hoxd3, implicated in the embryonic development, that have been shown to display a similar function in mice [100]. In the yeast S. cerevisiae, duplicated genes were shown to maintain functional redundancy for several million years [101].
2.2.3. The Fates of Duplicated Genes Depend on Different Factors
These different fates can be conditioned by the mechanism that led to the formation of the duplicated genes. Indeed, it was suggested that tandem duplication could more often produce duplicated genes having differential partitioning of regulatory sequences which implies that both genes would be necessary to recapitulate the ancestral expression pattern [102]. In A. thaliana, it was proposed that pseudogenes are more often derived from tandem duplications although this could be a bias due to the higher proportion of this type of mechanism compared to others in this organism [70]. The fate of retrocopies is often to become pseudogenes because of the lack of regulatory sequences [38]. However, it is sometimes possible for retrocopies to recruit other regulatory sequences allowing them to develop a new function. The structure of these retrogenes is usually chimeric with coding or regulatory features not present in the original genes [43,103,104,105,106]. Moreover, it has been observed in mammals that retrocopies located on the same chromosome than their parental gene have more chance to remain active indicating a role for the genomic context to maintain their expression [107]. In plants, a positive correlation has been observed between the size of gene family and the number of pseudogenes, with large families being more subjected to gene loss [81]. However, the gene function is also an important factor in the fate of duplicated genes. Indeed, in A. thaliana, pseudogenes tend to have functional counterparts in disease resistance, specialized metabolism cell wall modification, and protein degradation, whereas transcription factor and receptor-like kinase gene families are devoid of pseudogenes [70,81]. Other factors may also influence the fate of duplicated genes such as the number of protein interactions [76,108] as well as particular structural features [109]. According to the organisms, the outcome and formation mechanism of duplicated genes can also be different. In human and mouse, for example, the relative contributions of two types of duplication mechanisms made it possible to show that tandem duplications contributed more to duplications in the entire genome than retroposition, except for the two-copy gene families, and generated duplicated genes with more chance to be retained [110]. At another scale in primates, recent duplicated genes originated more often from segmental duplication than in other mammals in which the main mechanism to generate them rather corresponds to tandem duplication [111]. WGD in humans was proposed to have generated duplicated genes functionally more divergent but with a higher proportion of essential genes, which is the opposite trend to what was observed in yeast [74]. In Drosophila, young duplicated genes were shown to be preferentially subjected to neo-functionalization, implying the retention of almost two-thirds of these duplicated genes [112]. In plants, where most duplicated genes are derived from WGD and tandem duplication, a functional bias can be observed in genes according to their mechanism of formation [70]. Thus, genes involved in responses to environmental stimuli and upregulated in stress conditions are rather generated by tandem duplication, which implies that this mechanism is important for adaptive evolution in changing environments. Recently, a model was proposed to explain the gene retention after WGD in Paramecium species by dosage constraints, i.e., the majority of duplicated genes keep their ancestral function and are retained to produce the requested amount of proteins to perform this ancestral function [98].
In the next section, we will present in detail some of the current bioinformatic methodologies available to identify and analyze duplication in genomes with the goal to emphasize their advantages and weaknesses according to the situation.
3. Bioinformatic Approaches to Identify Duplications in Genomes
The identification of duplication within or between genomes is a complex process. Many algorithms have been developed for this purpose and different approaches can be used that have different aims and computation costs. Moreover, some of them are more suitable to search for a particular duplication event, are more optimized for large genomes, can deal with multiple genomes, or can handle genomes that have undergone multiple duplication and rearrangement events. In addition, there may be difficulties in the installation, the configuration, the launch, and the parsing of the results. This means for the user that programming skills may be required to use some of these softwares. There are also variations in the input data and the pre-processing requirements, the computing time or the associated visualizations, all of this making the choice of a tool not easy. Moreover, these tools do not all identify the same type of duplication and may therefore, be more or less adapted according to the biological question investigated. In summary, there is no stand-alone software that can solve all these problems and the choice of the tool will depend on computer skills but also on the genomes being compared and the biological questions being asked. In the following sections, we will present the different types of algorithms highlighting their specificities, advantages and weaknesses, with a focus on some tools that will be presented in more detail.
3.1. Paralog Detection
As said before, homologs are genes that share a common ancestry and are divided between orthologs (derived by speciation) and paralogs (derived by duplication). Based on this definition, the search for duplicated genes can be done through the identification of paralogous relationships. Therefore, it can be conducted by either identifying homologous genes in a given genome, which by definition can only be paralogous, or between multiple genomes before distinguishing orthologs from paralogs. Several approaches exist to this aim that we will present below.
3.1.1. Homology Assessment
Homology, even if defined by a few words, is a challenging concept to be detected through bioinformatic tools (for a broad overview, see [13,113]). The only material given to us to infer common ancestry that may have started millions of years ago is the sequences of contemporary organisms. A notable exception to this limit came with the rise of paleogenomics which aims at sequencing genomes of extinct species through preserved elements such as ancient seeds or fossilized body parts [114]. However, even if paleogenomics provides useful information, the amount of material is scarce compared to the number of contemporary species. Two methods are typically used to assess the homology between genes: The sequence similarity and the gene structure. Both methods rely on the idea that common ancestry (i.e., homology) is the most likely explanation when two genes share a strong similarity and/or structure. The limitations of these methods account for the aforementioned problem of inferring history through present traces: Divergence becomes difficult to detect when the distance between species increases. Hence, when two genes share sequence similarity or structure, it is a strong indication of homology, but when two genes do not share those, it hardly says anything about their homology.
The sequence similarity can be tested with a sequence alignment algorithm. The most popular ones such as BLAST [115], Psi-BLAST [116], and HMMER3 [117] are heuristic methods. Thus, they might not give the best results, but they drastically save computational time compared to a classical method such as the Smith and Waterman algorithm [118] even though some implementations have tried to make it faster as PARALIGN [119] or SWIMM [120]. In the case of homology, the alignment is generally performed on protein sequence instead of the gene. This allows a greater sensitivity since amino acid substitutions occur less frequently than nucleotide substitutions allowing silent mutations and because introns generate a lot of noise [121]. With these methods, the homology is tested against a cutoff on three different metrics: E-value, bit-score, or percent identity. The e-value is a statistic representing the expected number of times a given alignment score would occur by chance given the length and number of sequences being aligned. It is the most widely used metric as a first step to assess homology. Since the e-value is dependent on the database size, a potential caveat when setting a cutoff is to apprehend how the results might change for different databases. The bit-score is another metric measuring the sequence similarity given the raw score and the score system used but independent of the length or the number of sequences being aligned. The bit-score might be preferred in the case of a comparison between alignments since it relies only on the two sequences being aligned. Finally, percent identity is a straightforward metric giving how many amino acids are identical in the local alignment. When assessing homology on a genome-wide scale, the difficulty resides in setting the right cutoff for these metrics. For instance, to capture duplicates that diverged in function, the threshold needs to be relaxed, but with the risk of increasing the number of false positives. Based on empirical results, a 30% identity is generally accepted as a significant cutoff for protein homology [122]. However, countless identified homologs have an identity percentage lower than 30%. The same problem arises when using only e-value or bit-score. To allow better identification, more complex similarity-based metrics were developed. For example, Rost [123] proposed a formula based on the homology-derived secondary structure of proteins (HSSP) curve defined by Sander and Schneider [122] and considering the number L of aligned residues between two proteins to define a curve to separate true and false positives. Two proteins are then considered homologous if the proportion p of identical residues over L aligned residues is higher than the cutoff point defined by the formula. Li et al. [124] proposed a rewording of Rost’s formula to define different sets of duplicated genes with different stringencies in human. Since a gold standard cutoff is impossible to determine, a variety of values are used, sometimes combining different metrics leading to different results (Table 1).

Table 1.
Estimation of the amount of duplicated genes in different species.
When not working on a genome-scale but on specific sequences, homology imputation can be reinforced by looking at the gene structure. Domains shared by proteins are strong indicators of homology. Conserved domains can be found in databases such as Pfam [132] or InterPro [133] and searched against sequences of interest. This method is also a great tool to unravel complex evolution such as gene splitting and fusion for multi-domain proteins but require a time consuming manual expertise.
When searching for duplicated genes within a genome, assessing homology inside this genome is enough. However, when comparing multiple genomes, a link needs to be made between homologs of the different genomes. This raises the issue of resolving ortholog and paralog relationships. For this, a different kind of method needs to be applied. At first, methods to identify orthologous genes were only constructing orthologous groups because they focused on one-to-one ortholog relationships across multiple species. However, with the addition of one-to-many and many-to-many relationships, paralogs were included. Therefore, it could be argued that these methods are eligible to detect duplicated genes across multiple genomes. They are generally split into two categories: The graph-based methods and the tree-based methods [14,134]. Generally, graph-based methods construct a homology graph then build clusters of genes based on the types of inferred relationships. On the contrary, tree-based methods identify clusters of genes before constructing a tree along which the types of relationships are inferred.
3.1.2. Multispecies Graph-Based Methods
In graph-based methods, each gene is a vertex and a homology relationship is depicted by an edge. These edges are first drawn by assessing sequence similarity in the various forms described before. At this step, edges only correspond to potential homology relationships which can be orthology, paralogy, or noise. The noise can be removed by the clustering step. Depending on the clustering method, some paralogous relationships can also be removed. It is important to note that for the resolution of ortholog and paralog relationships, all these methods consider that for a given speciation event, in-paralogs are less diverged than orthologs that are less diverged than out-paralogs.
One of the first proposed clustering methods was the identification of triangle patterns inside a graph where at least three genomes are used [135]. It relies on the idea that two similar genes from two genomes, which are also similar to a third gene from another genome are highly susceptible to be orthologs. Then, triangles sharing similar edges are added to the same group until no other can be added. These groups, called clusters of orthologous groups (COGs) can therefore contain paralogs. However, the nature of the paralogs included in a group is hard to control. Hence, another way to detect paralogs based on graph exploration was proposed with InParanoid [136]. Here, two genes from different species with a best reciprocal hit are defined as orthologs and will be used as a seed for the group. Any gene having a better score with the seed gene of the same species than with the seed gene from the other species is included inside the group as an in-paralog relative to the speciation event. Thus, only in-paralogs in regard to the speciation event considered should be added, allowing a better control over the group formation. The method Hieranoid expanded this idea with the use of a guiding species tree for a better scalability when using many species [137]. The algorithm enlarges groups by exploring the guiding tree. It first runs InParanoid between two closely related species. Then, it creates a pseudo-species where each identified homologous group is represented by either a consensus sequence or a Hidden Markovian Model profile, depending on the number of sequences. InParanoid is then used again between the pseudo-species and the next closest neighbor. The process is repeated until all species are included in the analysis. By keeping track of groups formed at each step, it is possible to identify the speciation event encompassing any in-paralog pairs. Acting as a synthesis between InParanoid and COGs, both eggNOG [138] and OrthoDB [139] start by identifying groups of in-paralogs for each species then link them between species using triangulation.
Considering that the InParanoid method was reliable to detect “ancient” paralogs but not “recent” ones, Li et al. added steps and proposed another method, OrthoMCL [140]. It begins by the same ortholog seed approach but with the constraint that in-paralogs must have a better score with the seed genes from their respective species than with any other sequences from any species. In addition, a Markovian Cluster algorithm is run to simulate a random walk on the graph with each edge having a transition probability depending on the similarity score. This makes it possible to identify robust subgraphs and notably separate diverged paralogs. Using also a similar approach than InParanoid, the method OrthoInspector starts by constructing species-wise in-paralogous groups. Inside a species, an in-paralogous group is inferred for each protein [141]. Inside a species, a group of potential in-paralogs is inferred for each protein. When two proteins are potential in-paralogs, only the intersection of their respective potential groups is conserved as the final in-paralogous group. Therefore, if we have three proteins A, B, and C, they will belong to the definitive in-paralogous group (A, B, C) if and only if all three potential in-paralogous groups constructed for each protein give (A, B, C). This stringent method creates groups of lowly diverged in-paralogs. In-paralogous groups or single proteins are then grouped between species based on best-reciprocal hits.
Finally, two other methods add an interesting consideration regarding homologs. Aiming to tackle a well-known problem of sequence alignment, OrthoFinder [142] allows a reliable incorporation of short sequences. Indeed, alignment score is correlated with the sequence length, which is a problem for short sequences giving high scores even when not related. OrthoFinder proposes a normalization of the alignment score after a grouping according to the sequence lengths into equally sized bins. This normalization makes the score for short and really long sequences less dependent on the sequence size. Another interesting method, OMA [143], proposes to detect falsely imputed orthology inferences due to paralogs with differential gene loss. The detection is performed by using a third species containing both paralogs which acts as an evidence of non-orthology. OMA is also more permissive in the grouping of paralogs because it takes into account that paralogs may evolve faster than orthologs [144].
When studying genes, especially across species, representing their evolutionary relationships as a tree is easier to analyze. However, constructing such a tree is done at the cost of computational time. In addition, different strategies can be adopted for the tree reconstruction.
3.1.3. Multispecies Tree-Based Methods
In tree-based methods, homology is assessed according to the various forms described before, then groups of homologs are constructed across species. Genes from these groups are aligned to build gene trees. Paralog and ortholog relationships are then resolved by the reconciliation of the gene trees and the associated species tree. Therefore, in these methods, the detection of duplicated genes is only performed at the first step. The tree construction only influences the evolutionary history used to explain the appearance of such duplications.
In regards to the homolog grouping strategy, tree-based methods are generally more inclusive than graph-based methods. Indeed, after the group construction, they use all the sequences from all the species to infer paralog and ortholog relationships. Therefore, they can extract more information and are less restricted by false homology prediction and thus are able to capture more diverged homologs. Most of them construct homologous groups by clustering all genes that have a significant alignment score, defined differently according to the method used such as TreeFam [145], BranchClust [146], HOGENOM [147], or PhylomeDB [148]. Some tree-based methods use pre-processed homologous groups and are only used to reconcile the gene and species trees such as Orthostrapper [149], Softparsmap [150], or LOFT [151]. Therefore, graph-based methods can be used as an entry-point to combine the power of both methods.
When reconciling the gene and species trees, all these methods use the Maximum Parsimony principle [152]. This is translated by minimizing the number of duplication events, which are assumed to be rare events. A notable exception is PrIME-GSR [153] that tries to take into account the duplication and loss of genes through a probabilistic model. Apart from this exception, tree-based methods differ according to the type of species tree they accept, how they root the gene trees, and how tree uncertainty is assessed. Since it does not affect duplication detection, they are not as thoroughly explored as the graph-based method (for a complete review, see [14]).
3.2. Detection of Syntenic Blocks (WGD-Segmental Duplications)
A syntenic block can be defined as a region of the genome spanning a number of genes that are orthologous and co-arranged compared to another genome [154]. Two regions of a genome with a number of homologous genes co-arranged with each other can also be defined as a syntenic block. Here, we focus on this second definition because pairs of homologous genes between these pairs of regions correspond to duplicated genes.
It can be interesting to access different databases storing pre-calculated syntenic blocks shared between different species. This makes it possible for an easy and direct access to reliable information without any computation. Nevertheless, these databases cannot include every contemporary species nor information about recently released genomes. This implies that depending on the organism being studied it can be necessary to manually identify syntenic blocks using different tools. To accurately detect homologous chromosomal segments within a genome or between different ones, many approaches and tools are available. The choice of the tool depends on various parameters.
A first important parameter is the degree of preservation of duplicates in the compared genomes. This will influence the level at which the study should be conducted, and thus will impact the choice of the tool since each of them works at a particular level. For closely related genomes, synteny can be studied at the DNA level using tools such as Satsuma [155] or SyMap 3.4 [156]. In the case of more distant genomes, the DNA level cannot be used because the sequences will be too divergent. A solution is to perform analysis at the protein level because coding genes may retain for a longer time enough amino-acid sequence similarity and a similar relative order along chromosomes. Tools such as MCScanX [157], i-ADHoRe [158], CYNTENATOR [159], or SynChro [160] search for syntenic blocks using protein sequences and can therefore be adapted to this type of genome comparison. Finally, in the case of more distant genomes, it is more appealing to use tools based on analyses at the protein level and on the construction of profiles, graphs, or statistical models to help manage the evolutionary distance.
Four types of approaches can be applied to search for syntenic blocks. The first one is based on the construction of a sparse matrix of homologous genes. The matrix is investigated to look for dense diagonals which correspond to the syntenic blocks. Tools such as i-ADHoRe 3.0 [158], DiagHunter [161], FISH [162], SyMAP [163], or Cinteny [164] implement this type of approach. The second approach corresponds to different greedy algorithms that will be optimized by dynamic programming at the benefit of computational costs. This type of algorithm operates by constructing chains of collinear gene pairs, called anchoring genes. It is implemented in tools such as DAGchainer [165], MCScanX [157], or LineUp [166]. An important sub-category of this approach consists of algorithms based on aligning sequences using a modified Smith-Waterman algorithm as in ColinearScan [167] or CYNTENATOR [159]. To continue, the graph approach aims at building graphs allowing the identification of the syntenic blocks. To do this, local collinear alignments are constructed between the input genomes. By combining the local alignments with the blocks, a graph can be constructed which allows, after different analyses, the reconstruction of the syntenic blocks. This approach can be found in tools such as DRIMM-Synteny [168] or Enredo [169]. Finally, another approach aims at inferring syntenic blocks based on genomic rearrangements. This type of approach can be useful in the reconstruction of ancestral genomes and has been implemented in different tools such as GASTS [170] or PMAG++ [171,172]. This approach is not covered in the present review but has already been discussed in another recent review article [173].
All these tools are able to answer different questions and their use depends on the number of studied genomes as well as the level of divergence among them. Most of them have many critical parameters, sometimes with important pre-processing requirements, which need to be mastered before obtaining reliable results. Most of the tools are presented in a comprehensive format in Table 2. Therefore, the purpose of this section is to examine in detail a representative sample of tools illustrative of each approach.

Table 2.
Summary of the characteristics of different existing tools for identifying syntenic blocks.
3.2.1. Approaches Based on the Construction of Homologous Gene Matrices
These approaches correspond to tools based on the search for syntenies using clustering of neighboring matching gene pairs. The basic concept is to consider the homology in or between genomes as a sparse matrix. In summary, homologous gene pairs are considered as 1, other cases are encoded as 0. The goal is to detect syntenic regions by searching for dense diagonals of 1 in the matrix. Tandem duplication can also be accounted for by detecting horizontal or vertical lines of 1.
The main advantage of this approach is that it is designed around a formal definition of the syntenic blocks. Moreover, statistical validation can be performed on putative syntenic blocks to filter out false positives. However, several weaknesses exist for this approach. To begin with, the important impact of the parameters requires a good knowledge of the biological question asked. With this type of approach different parameters are critical and need to be finely tuned. An example being the size of the gap allowed between genes considered as belonging to the same block. This parameter can deeply affect the results, and needs to be configured according to the specificity of the study. The size of the gap depends mainly on the density of the matrix, i.e., the density of the pairs of homologous genes between the segments constituting the matrix. On one hand, a small gap value results in many small syntenic blocks that are more difficult to analyze. On the other hand, a high gap value produces long blocks that are easier to analyze but allow for more false positives. Moreover, the metrics used to estimate the distance between genes in a matrix are also an important setting. Two types of metrics are often proposed: The Diagonal Pseudo Distance (used by i-ADHoRe and DiagHunter), and the Manhattan Distance (used by FISH, SynMAP, or Cinteny). The Diagonal Pseudo Distance promotes genes near the diagonal axis and therefore, the distance inflates rapidly the further away genes are from this diagonal. In contrast, the Manhattan Distance tends to give smaller distances between aligned genes on the vertical or horizontal axis. Other types of distances have been implemented in tools such as PhylDiag [174] that uses the Euclidean Distance or the Chebyshev Distance in addition to the others mentioned above. Thus, the choice of the distance is not easy and will impact the results as surely as a wrongly set gap value. A benchmark analysis suggested that the Manhattan Distance gives the best results among these four distances [174]. The importance of the configuration is really to be taken into account when using this approach in an optimal way and makes these algorithms difficult to use without a minimum of expertise on both the tool and the biological question. Moreover, statistical tests to evaluate homologous regions are based on the assumption that the rate of gene loss is balanced between homologous regions. This is not the case for many species. Furthermore, some differences in terms of genome structure, especially the gene density and repetition in chromosomal regions, both locally and at the genome level, are difficult to account for with this approach. Finally, matrices can only compare genomes by pair, which implies that benefits of comparing multiple genomes at once, including WGD studies or diverged synteny blocks, cannot be done. Moreover, this approach cannot resolve multiple relationships between genes, detect inversions, nor non-overlapping syntenic blocks. To finish, not all of these implementations can detect tandem duplication. In the already cited tools, only i-ADHoRe and FISH can handle them. We will present these tools in more detail below.
i-ADHoRe (Iterative Automatic Detection of Homologous Regions) 3.0
i-ADHoRe [158] is one of the most used programs to find syntenic blocks and can be considered as a state-of-the-art algorithm. In its latest version, i-ADHoRe enables the detection of genomic homology through the identification of gene collinearity. This version is well optimized to handle a large number of genomes, taking advantage of parallel computing.
The algorithm begins by assimilating tandem duplicated genes as a single representative. Then, for each pair of genes, a sparse gene homology matrix is constructed. In this matrix, homologous genes are considered as dots, making collinearity zones seen as dense diagonals. Gene clusters with at least three homologous gene pairs are included in diagonals after a statistical validation taking into account the overall background density of the matrix. In the case of multiple clusters found, a correction for multiple testing is performed using the Bonferroni or False Discovery Rate (FDR) method. This part corresponds to the traditional homology matrix approach. The next part of the algorithm is an optimization by dynamic programming.
Significant collinear regions found during this initial detection are aligned using the progressive Needleman–Wunsch (pNW) algorithm or a greedy graph-based algorithm [178]. The results of this alignment are stored into a profile, which contains the combined content of the two collinear regions and constitutes a more sensitive probe to find new homologous regions including more degenerated ones. Using this newly constructed profile, a search is performed in an iterative way. Thus, this profile is used to search for new sequences that can be aligned with it. If possible, the new matches are added to the profile. As long as new collinear regions can be added to a profile, these two steps are repeated.
The results are provided as text files and two associated plots: A dot plot and a set of graphs representing each final aligned profile.
This tool has many key parameters that directly influence the quality of the results:
- prob_cutoff, is used to store the maximum probability for a cluster to be generated by chance. The default value is 0.001.
- gap_size, indicating the maximum distance between genes in a cluster. The default value is 15.
- cluster_gap, indicating the maximum distance between individual base clusters in a cluster. The default value is 20.
- q_value, storing the minimum r2-value which measures the quality of the linear regression prediction.
- anchor_points, the minimum number of anchor points which is comprised between 3 and 6.
The main advantage of this tool is to allow the computation of multiple genomes thanks to different optimizations including the use of parallel computation, an efficient statistical model to estimate p-values of diagonals before including them, the use of greedy graph-based alignment algorithms, and the use of ordered gene lists instead of genome sequence. This level of abstraction allows a more efficient detection of collinearity and thus the divergent intergenic sequences will have less impact on the algorithm.
OrthoCluster
In this category, OrthoCluster [177] appears as particular. It is not based on the classical approach of homology matrix construction although it is using the same philosophy by identifying syntenies via the clustering of neighboring matching gene pairs. This program is based on an algorithm implementing a strategy of tree enumeration to detect orthologous gene clusters. This tool can handle many genomes and makes it possible to overcome some of the weaknesses of the other classical approaches. Indeed, it can detect four types of genome rearrangements including insertions, transpositions, insertions/deletions, and reciprocal translocations via different algorithms. To detect reciprocal translocations (exchange of DNA parts by recombination), OrthoCluster merges syntenic blocks to build the longest possible blocks, identifying blocks not broken by duplications, inversions or transpositions. To detect transpositions (regions moved from a chromosome and inserted into a non-homologous chromosome), OrthoCluster searches in each adjacent syntenic block for a region between their homologous syntenic block in the other genome. If a fragment of less than 50 genes is found between them, a transposition is identified. Then, the detection of inverted segments in the genome is performed by checking if the order of the genes is the same in each syntenic block. If the gene order is inverted between the two, this region constitutes an inversion. Finally, in order to detect insertion or deletion of genes, OrthoCluster compares the pairs of adjacent syntenic blocks in the reference genome. Genes identified between these blocks are considered as insertions/deletions and reported. It can also detect segmental duplications and resolve one-to-many relationships. Moreover, the orientation and the order of genes are taken in to account. Nevertheless, this tool is limited to the orthology detection and can therefore only be applied on closely related organisms.
The fine-tuned configuration of this tool is crucial to obtain reliable results. Eight parameters can be defined by the user to set up the algorithm according to the needs:
- l max defining the upper bound on the number of genes in each cluster.
- l min defining the lower bound on the number of genes in each cluster.
- op maximal percentage of out-map genes allowed.
- ip defining the maximal percentage of mismatched in-map genes allowed.
- op and ip can control the number of genes involved in transpositions in synteny block.
- i maximal number of mismatched in-map genes allowed.
- o maximal number of out-map genes allowed.
- r to find order-preserving clusters.
- s to find strandedness-preserving clusters.
3.2.2. Algorithms Using Dynamic Programming Optimizations
This type of approach generally implements algorithms more costly in computation than the homology matrix approaches. Some methods benefit from dynamic programming to build a chain of collinear pairwised genes. In these methods, the dynamic programming algorithms are implemented in the search for collinear genes, allowing an exhaustive and fast search. A scoring system is set up allowing to build pairs of adjacent collinear genes, which constitute anchoring genes, and to penalize the distance between them. The main advantage of a multi-alignment of collinear chromosomal regions is its ability to reveal past WGD events and complex chromosomal rearrangement relationships. In this type of approach, the syntenic blocks are composed of anchoring genes that are located at collinear positions and between them non-anchoring genes that are assumed to have undergone mutations. Nevertheless, the user needs to already know what to look for and the characteristics of the genomes and syntenic blocks being studied.
MCScanX and MultiSyn
MCScanX [154] is one of the most used tools aiming at searching for syntenic blocks and is implemented in the webtool MultiSyn [176], allowing biologists with no informatic skills to use this approach. Moreover, this tool produces additional visualizations allowing a simplified analysis.
The MCScanX algorithm takes place in three steps. The first step uses the results of an all-against-all comparison using BLASTP [179] to find collinear blocks. BLASTP matches are sorted according to their genomic positions. To handle tandem regions, all consecutive genes with a BLASTP match that are separated by less than five genes, are collapsed into a single representative. Then, the highest scoring chains of collinear gene pairs are searched for using dynamic programming. Non-overlapping chains involving at least five collinear gene pairs are saved. In a pair of collinear blocks, two distinct genomic locations with aligned collinear genes are assigned as anchors.
The second step makes it possible to assign each syntenic block to a gene class. To do that, all genes are first assigned to the singleton class. Genes with BLASTP hits to other genes are assigned to the class “dispersed duplicates”. If the hits are close enough, they are assigned to the class “proximal duplicates”. If the hits are neighboring, they are assigned to the class “tandem duplicates”. To finish, anchored genes are assigned to the WGD/segmental class.
In the last step, twelve downstream analyses can be performed using different scripts and correspond to the computation of the nonsynonymous and synonymous rates (Ka and Ks), the generation of various plots, the construction of gene families with associated analyses, the detection of collinear tandem arrays, the computation of the number of intra- and inter-species collinear blocks at each locus of reference genomes, and the display of statistics on gene numbers at different duplication depths.
To be functional this tool needs to be configured using at least six parameters:
- match_score, defines a threshold used to validate a synteny block. Default value is 50.
- gap_penalty, defines the penalty added when opening a gap. Default value is 21.
- match_size, defines the number of genes required to consider it as a collinear block. Default value is 5.
- e_value, defines the statistical significance of the synteny block alignment. Default value is 1e-10.
- max_gaps, maximum number of gaps allowed. Default value is 25.
- overlap_window stores the maximum number of genes to collapse BLAST matches. Default value is 5.
The special feature and strength of MCScanX is that each chromosome is used as a reference. Thus, all collinear segments in pairs are mapped. This is followed by a multiple alignment procedure of homologous genes, described as “transitive homology” [180]. This approach allows MCScanX to match regions that were not initially detected based on their collinearity with the reference.
To conclude, this tool is powerful and allows performing many analyses, if the user has the ability to install and configure it properly. MultiSyn eases the configuration step, the initial formatting of the data and the analyses using a graphical interface. As a final advantage, this tool can be deployed locally. As for i-ADHoRe, the use of ordered gene lists instead of a genome sequence allows getting more reliable results at lower computational costs.
SynChro
SynChro [160] is based on Reciprocal Best-Hits (RBH) to construct syntenic blocks. This algorithm is faster and easy to use thanks to its unique parameter (Δ) which represents syntenic block stringency. To go into more details, this algorithm is composed of three simple steps. In the first step, RBH are identified using Opscan (http://wwwabi.snv.jussieu.fr/public/opscan/), a tool based on the FASTA algorithm [181]. RBH can be defined as two genes whose best hit is mutual. In the second step, the RBH makes it possible to define syntenic blocks using co-localizing RBH (defined by Δ) along the chromosomes of two genomes as anchors. In the third step, syntenic blocks are completed by non-RBH homologs. Genes are defined as non-RBH if they share 30% of similarity and if the ratio of the length of the match between the two sequences and the length of the smallest sequence is greater than 0.5.
This tool provides various graphical outputs including dotplots, chromosome painting, and synteny maps, as well as text results. Therefore, it makes it possible to obtain in a limited computational time very good quality results with a fast handling in an “all in one” manner allowing to easily visualize the results.
CYNTENATOR
CYNTENATOR [159] is a tool aiming at identifying conserved syntenic regions between distant genomes. This tool is based on a progressive multiple gene order alignment. The main advantage of this tool is its scalability allowing it to work on more than 10 genomes contrary to many other approaches. Moreover, it makes it possible to get rid of heavy preprocessing steps due to its high flexibility.
To begin, a progressive pairwise alignment between genomes is performed. These alignments are based on a user-defined phylogenetic tree that directs the order in which the genomes will be compared. Only valid alignments gathering homologous regions of all species are retained for collinearity search in the next genome. This filtering step helps lower the computational costs and allows determining the gene order conservation between distant genomes. This step is followed by a pairwise alignment using a Smith-Waterman local alignment weighted by the phylogenetic distance. The results of these alignments constitute the syntenic blocks. The use of a progressive alignment algorithm makes it possible to conduct studies on several large genomes while taking into account the phylogeny of the studied species. The absence of a heavy pre-processing on the input data, except an all against all homology score, allows to avoid bias.
SyMap
SyMap [163] is a tool based on DAGChainer [165]. The advantage of this software is that its interface via a web application allows the user to be free from any configuration and data preparation via the code. In addition, this tool allows retrieving various additional information and in particular the Ka/Ks ratio using PAML [182]. The intermediate results can be retrieved and the final results can be visualized in an interactive dot-plot. Once the genomes have been added to the database and the parameters have been defined, the computations are launched. The SyMap algorithm works as follows. First, the genomes are aligned using an alignment software. Different tools can be used for this step, including BLAST. Then, different filters are applied and in particular the condensation of tandemly duplicated genes into a single occurrence and filtering out of repeated sequences. The syntenic blocks within this homologous matrix are then searched for using DAGChainer. Finally, different visualizations are constructed. The main advantages of this tool are its speed, the ease of use, and the visualizations. However, some parameters are not configurable and it does not allow the study of more than two genomes at the same time.
3.2.3. Approaches Based on Graphs
The principle of this type of algorithm is to construct a graph gathering all the pairs of homologous genes shared by the compared genomes. These approaches aim at solving many problems raised by the methods presented before, in particular the possibility of studying several large genomes and to detect non-overlapping syntenic blocks. The previous approaches have difficulties decoding more complex genomic architectures that have undergone phases of significant duplication followed by reploidization. The search for non-overlapping syntenic blocks is of great interest because it makes it possible to focus on rearrangements that happened after the duplication events. However, the search for non-overlapping syntenic blocks is not just about simply decomposing overlapping blocks. Different algorithms have been proposed to meet these needs. The first algorithms as GRIMM-Synteny [183] or MAGIC [184] were suitable for small sets of genomes, but were not able to handle genomes with large duplications and deletions, and were not able to find non-overlapping blocks. Later, Enredo [169] was written to solve this problem. One last problem with the algorithms from the two previous approaches is that as more and more genomes are integrated into comparative studies, the number of genes shared between genomes decreases. This has a strong impact on the algorithms with the risk of rejecting the blocks because they are statistically nonsignificant, as it happens with tools such as GRIMM-Synteny.
Typically, the algorithms used in the graph-based approaches follow different steps. First, input genomes are locally aligned and the resulting alignments are used to construct a graph. Then, the different sub-structures (depending on the initial graph structure) are searched for to find the different segmentations of the genomes. The type of graph structure has a major influence on the results. Indeed, some of them handle these problems with more or less success and can therefore not find similar results. Four graph structures are predominant to analyze syntenic blocks.
The first structure corresponds to an alignment graph. The graph contains a vertex referring to each character in the sequence and edges referring to aligned characters. It is then possible to obtain collinear or noncollinear alignments by solving the maximum weight trace problem. Duplicated regions are more easily visible in an alignment graph structure. Nevertheless, this structure does not allow the user to get inversion information.
The second structure corresponds to A-Bruijn graphs that can be found in DRIMM-Synteny [168]. The main idea behind this graph is to merge aligned vertices. Thus, A-Bruijn graphs have one vertex for each aligned sequence. The links represent only the sequence. The main problem with this approach is represented by the short cycles, which tend to make local alignments hide a local collinearity. As an alignment graph structure, it does not allow access to inversion information, meaning that it is not possible to differentiate between the tandem repeats and palindromes.
The third structure, known as the Enredo graphs, can be found in Enredo [169]. It aims at managing genomes partitioned into segments. The nucleotide alignments are then made. Thus, Enredo graphs have two vertices per set of aligned segments, a head vertex and a tail vertex. It is then possible to eliminate various substructures from the Enredo graph in order to obtain the final segmentation of the genome. An Enredo structure can help find non-overlapping blocks and is suitable to consider non-overlapping inversions.
To finish, the Cactus graphs [185,186] have also been proposed. They are structures with vertices for adjacencies and undirected edges for genome segments. This type of graph is Eulerian meaning that there is a path that crosses all the nodes only once. This graph is also subdivided into independent units where each edge is part at most of one cycle. The cactus structure is a unique sub structure that allows an easy detection of short cycles.
These different graph structures allow the study of some particular sub-structures to identify syntenic blocks. One of the most important corresponds to the collinear paths. These sub-structures are a set of blocks that appear in genomes consecutively, without breaks and with the same orientation thus representing syntenic blocks. A second sub-structure corresponds to the presence of microblocks within larger regions that tend to introduce breaks into syntenic blocks. A third sub-structure corresponds to the short cycles. They are the mark of rearrangement. Indeed, similarities between sequences make them appear and thus break the collinearity. An important number of short cycles is problematic because they can aggregate into complex networks and hide true collinear blocks. We will detail a little bit more on one algorithm implementing the graph based approach below.
DRIMM-Synteny
This algorithm is the update of GRIMM-Synteny and aims at solving various problems from this previous version. In particular, the suppression of blocks due to their statistical non-significance in the case of the study of several distant genomes. The syntenic blocks that do not overlap are identified, which allows, in a second step, to bypass the threading problem based on the use of an A-Bruijn graph structure. This type of graph is an Eulerian and undirected multigraph. Edges are weighted by the number of times a gene pair is consecutive in the analyzed genomes.
In DRIMM-Synteny, an A-Bruijn graph is constructed by collapsing together identically labeled vertices from all genomes. From this graph, syntenic blocks can be found. In fact, a perfectly repeated block corresponds to a path in the graph. Perfectly repeated regions that do not share genes with other regions in the set of genomes being studied will appear as unconnected paths. These are referred to as the maximum paths in the graph, satisfying the condition that all of their internal vertices have only two neighboring vertices. This algorithm solves some existent problems known for this type of approach using different subroutines. There may be small differences between the different syntenic genes, which leads to short cycles. DRIMM-Synteny is able to detect them by computing a shaft at maximum range. A heuristic then allows detecting the links that create them in order to remove them. In addition, the presence of syntenic microblocks separate the long unbranched paths into several subpaths, thus complicating the detection of the blocks. Finally, the short palindromic regions that can be found within syntenic blocks form thornes that have the same effect as the microblocks.
3.3. Detection of Tandemly Arrayed Genes (TAGs)
Specific methods have been developed to handle specifically tandem duplication detection. TAGs are gene family members that are tightly clustered on a chromosome [73]. The vast majority of the methods are home-made pipelines available from the authors and may require programming skills. A few tools, particularly those related to the identification of syntenic blocks, are able to help in the identification of TAGs because they are generally summarized in a single occurrence of the dataset to lower the statistical noise. In general, they are not dedicated methods but more trivial algorithms. However, they have the advantage of being simpler to use. Most of these algorithms rely on protein comparisons, making them dependent on genome annotation. However, there exists very few methods that can deal with genomic sequences to search for long DNA tandem repeats. The advantage of these latest methods is that they can detect pseudogenes that originated from duplication or short ORFs generally missed by automatic genome annotation. We will first describe TAG detection in the genome at protein level, then at DNA level.
3.3.1. Detection at Protein Level
These methods begin with the identification of homologous gene pairs. This can be done using different algorithms, in most cases an all-by-all BLASTP comparison of the proteome against itself or between the proteomes of two species, followed by a filtering using a threshold to retain only homologous pairs. The difference between these approaches lies in the homology assessment and the degree of sophistication to filter out false positives.
The most straightforward, but trivial, way is used in the first step of WGD detection algorithms such as MCScanX or i-ADHoRe [157,158]. These algorithms take as input homologous gene pairs, the preferred format being the BLASTP output. Then, the program classifies homologous pairs according to their rank along the chromosome. If consecutive BLASTP matches have a common gene and its paired genes are separated by fewer than five genes, these matches (forming a TAG) are collapsed using a representative pair with the smallest BLASTP e-value. The advantage of this approach is its speed but the drawback is that it can miss divergent homologous genes. Moreover, even if few programming skills are required, a parsing step is still necessary to obtain the list of identified TAGs.
To alleviate some problems related to the input (an all-against-all BLASTP), it is possible to use gene families as input. They can be constructed by different algorithms summarized in Table 1. Then, a TAG is defined as a block of adjacent genes belonging to the same family and separated by spacers that are generally genes not belonging to the homologous family. Several definitions can be used for the allowed number of spacers, mostly 0 or 1 but also ranging from 0 to 10 spacers [73,127,129,187]. The construction of gene families allows incorporating more distantly related homologs than the previous approach. The definition of homologous genes can be improved by merging all non-overlapping HSP of one hit [73]. The most widely used clustering algorithms are the single linkage algorithm, and more and more Markov clustering (MCL) and its variants. It is an efficient approach but adjusting the inflation and expansion parameters of MCL is not easy. The inflation parameter controls the flux between groups of classification (i.e., the number of steps in the random walk along the similarity graph). The expansion parameter controls the strength of links by strengthening them inside the clusters and weakening them between clusters.
3.3.2. Detection at DNA Level
The vast majority of Tandem Repeat detection methods at DNA level deal with the identification of short highly repeated sequences. They are used to mask sequences corresponding to TEs or/and segments of low complexity before genome annotation or to explore the amplification of short duplications associated with human diseases for example, or copy number variation (CNV) between genotypes. These types of DNA duplication are not the focus of this review and will not be treated in detail. Here, we give a list of some famous short DNA Tandem Repeat detection tools able to deal with large datasets: DUSTMASKER [188], SEGMASKER [189], Tandem Repeat Finder (TRF) [190], TANTAN [191] and more recently ULTRA [192], TARDIS [193], and dot2dot [194].
We will now focus on long tandem duplication detection because all studies on TAGs based on protein similarity are biased by the quality of the available genome annotation. They exclude RNA genes or degenerated copies [195]. However, duplicated pseudogenes are an important evolutionary residue of a genome past activity [196]. A genome-wide approach has been proposed to take into account pseudogenes in TAG detection [197]. It scans, using TBLASTN, each protein against its chromosomal regions (the surrounding DNA sequences is three times longer than the CDS) and filter hits according to a refined bit-score, called the BTF score, that takes into account all non-overlapping HSP of less than 20% on the same strand. Then, it looks at CDSs in the ascending order of their chromosomal positions to extract TAGs. This mixed approach (at DNA and protein levels) is implemented in Python 2.4. The scripts are available from the authors and need a step of manual curation to eliminate false positive TAGs, due to the presence of minisatellites.
This previous approach is based on proteins and therefore depends on genome annotation. It has mainly been used on compact genomes [195]. ReD Tandem is an alternative method that circumvents this limitation [195]. Indeed, the main problem of detecting TAGs at genomic level is that large duplications despite being close, are far from being contiguous. The authors thus proposed to define tandemly duplicated segments as paralogous segments of size l with adjacent copies separated by a maximum distance T (in A. thaliana, the parameter values are l = 500 bp, T = 150 kb). The algorithm begins with anchors (paralogous segments of size l) and chains then using DAGchainer or OSfinder [175] into longer duplicated regions (called tandem units). Such alignable units are anchors of length l and separated by less than L bases (L = 40 kb for A. thaliana). Then, the tandem units are assembled into TAGs (i.e., tandem units separated by less than T bases, with T = 150 kb for A. thaliana). The C++ scripts are available but need some computational skills to be installed. Nevertheless, this elegant approach has allowed the authors to identify in A. thaliana several types of TAGs previously undetectable for genome-wide approaches. In decreasing order of importance, these new TAGs correspond to trans-elements genes, pseudogenes, pre-tRNAs, other RNAs, miRNAs, snoRNAs, and unknown genes [195].
3.4. Databases Storing Syntenic Block or Homology Information
3.4.1. Syntenic Information
These databases have the advantage to not require computations and therefore no programming skills for the user. Some of them also offer visualizations and search tools. The main disadvantage is that they do not contain information from all organisms. Each of these databases provide particular features but some elements are common. In some cases, it is possible to access all the syntenic blocks between two organisms. The list of organisms is more or less extended depending on the database. Some of them propose to visualize these blocks using various representations such as circular visualizations, chromosome painting, or dot-plots. Some databases allow manually importing genomes to identify blocks of synteny. In this case, different tools may be implemented for the identification and are more or less easy to configure. For example, Ensembl [198] stores different information including syntenic blocks generated by Pecan [169] as a multi-alignment algorithm and Enredo to detect syntenic blocks. Synteny portal [199] and Genomicus [200] provide also syntenic blocks generated by inferCars [201] for different species but also multiple visualizations. Finally, other databases exist including ECRbase [202] with syntenic blocks generated from the DNA level. OrthoClusterDb [203] is a good example of what can be found inside these databases. Two main possibilities are available. First, it allows online access to the OrthoCluster tool [177] and to carry out identification of syntenic blocks on a remote server using a graphical interface facilitating the configuration and the retrieval of the results. Another possibility is to access different pre-computed syntenic blocks by OrthoCluster for different species. Pre-computed species belong to different groups (Mammals, Pseudomonaceae, Drosophila, Plasmodium, and Caenorhabditis) with 54 species available. The syntenic blocks can be visualized on a figure called genome painting which allows visualizing the chromosomes of the compared species with a system of colored segments highlighting the syntenic blocks. It is also possible to retrieve raw output files or to access to syntenic blocks using an online genome browser.
3.4.2. Homology Relationships Databases
Dataset of duplicated genes without specification of the underlying mechanism of duplication can be retrieved from public databases. These databases can be associated or not with a specific methodology with available tools for a local use. The INPARANOID 8 database for example, provides the InParanoid tool and proposes orthology analysis between 273 proteomes, mostly eukaryotic. The dataset of orthologous and paralogous relationships between genes can be downloaded by pairs of species [204]. In HOGENOM, gene families are built from complete genomes from all three domains of life [147]. Its clustering pipeline is based on the SiLiX clustering method [205]. Even if this database is regularly updated, users can only retrieve families one by one according to keywords. The total amount of paralogous genes in a species is only available upon request directly to the authors.
Ensembl Compara is a specific section of the Ensembl database providing cross-species resources and analyses, at both the sequence and the gene levels. The main Ensembl database is dedicated to chordate genomes and displays now counterparts for several groups of organisms (Ensembl Genomes, Ensembl Bacteria, Ensembl Protists, Ensembl Fungi, Ensembl Plants, and Ensembl Metazoa). All these databases are associated with the Ensembl Compara system. This system provides access to protein gene families via a Perl API [198]. These families are built using all proteins from Ensembl through a classical process using BLASTP for similarity searches and a MCL clustering with scores as weight for edges in the initial graph. A final step aligns all sequences from a family using MAFFT [206]. It is to note that the Ensembl Compara protein families correspond to the most similar proteins compared to its gene tree section, where paralogous relationships are also available but in a tree format. Many other repositories are available but our goal is not to be exhaustive. Among the most generalist, we can cite PhylomeDB [148], OMA [207], OrthoDB [139], OrthoInspector [141], eggNOG [138], or the database Homologene from the NCBI portal.
For plant comparative genomic, we can cite the databases PLAZA [208], GreenPhyl [209], and Phytozome [210]. PLAZA 4.0 contains gene family data, phylogenetic trees, and gene colinearity information. It comprises two instances, one for monocots (Monocots PLAZA 4.5) that includes data from 39 species and one for dicots (Dicots PLAZA 4.0) that includes data from 55 species. The latest PLAZA instance offers one or more REST-full APIs, depending on the Platform software version. GreenPhyl 4 contains gene families and phylogenetic trees from 37 species. It has not been updated since 2015 but contains a section of manually annotated families comprising 2956 clusters. Other interesting sections are transcription factors and families specific to species or phylum (family of homologous genes found only in one species or excluding/including one phylum). Finally, the plant database Phytozome13 (last update in May 2019) contains 184 assembled and annotated genomes. Inparanoid pairwise orthology and paralogy groups have been calculated across all Phytozome proteomes and families of related genes representing the modern descendants of putative ancestral genes have been constructed at key phylogenetic nodes. The dataset can easily be downloaded or mined via a dedicated tool named PhytoMine.
4. Conclusions
To conclude, when considering duplicated genes inside a given species, it appears clear that they represent very different entities when taking into account their mechanism of formation, their fate, and their age. This is particularly important when it comes to their identification and analysis. It is indeed tempting to only detect all genes that are in several copies without taking into account the evolutionary complexity behind them. This is why it is also important to be aware of the different methodological approaches that can be used because this choice will greatly depend on the investigated biological question.
Author Contributions
E.L. and C.R. conceived the review; C.L., E.L., M.L., T.L., and C.R. wrote the different versions of the manuscript; T.L., and M.L. equally contributed to the work. All authors have read and agreed to the published version of the manuscript.
Funding
This work received no external funding.
Acknowledgments
This work was supported by the CNRS, the University Lyon 1, and the Laboratory “Biométrie et Biologie Evolutive”.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ohno, S. Evolution by Gene Duplication; Springer: Berlin/Heidelberg, Germany, 1970; ISBN 978-3-642-86661-6. [Google Scholar]
- Kondrashov, F.A. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc. R. Soc. B Biol. Sci. 2012, 279, 5048–5057. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Maere, S.; Meyer, A. The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 2009, 10, 725–732. [Google Scholar] [CrossRef]
- Vallejo-Marín, M.; Buggs, R.J.A.; Cooley, A.M.; Puzey, J.R. Speciation by genome duplication: Repeated origins and genomic composition of the recently formed allopolyploid species Mimulus peregrinus. Evolution 2015, 69, 1487–1500. [Google Scholar] [CrossRef]
- Ting, C.T.; Tsaur, S.C.; Sun, S.; Browne, W.E.; Chen, Y.C.; Patel, N.H.; Wu, C.I. Gene duplication and speciation in Drosophila: Evidence from the Odysseus locus. Proc. Natl. Acad. Sci. USA 2004, 101, 12232–12235. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Gu, W.; Hurles, M.E.; Lupski, J.R. Copy number variation in human health, disease, and evolution. Annu. Rev. Genomics Hum. Genet. 2009, 10, 451–481. [Google Scholar] [CrossRef] [PubMed]
- Dickerson, J.E.; Robertson, D.L. On the origins of Mendelian disease genes in man: The impact of gene duplication. Mol. Biol. Evol. 2012, 29, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Tollis, M.; Schneider-Utaka, A.K.; Maley, C.C. The Evolution of Human Cancer Gene Duplications across Mammals. Mol. Biol. Evol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Mendivil Ramos, O.; Ferrier, D.E.K. Mechanisms of Gene Duplication and Translocation and Progress towards Understanding Their Relative Contributions to Animal Genome Evolution. Int. J. Evol. Biol. 2012, 2012, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K. Robustness—it’s not where you think it is. Nat. Genet. 2000, 25, 3–4. [Google Scholar] [CrossRef]
- Sharman, A.C. Some new terms for duplicated genes. Semin. Cell Dev. Biol. 1999, 10, 561–563. [Google Scholar] [CrossRef]
- Sonnhammer, E.L.L.; Koonin, E.V. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002, 18, 619–620. [Google Scholar] [CrossRef]
- Koonin, E.V. Orthologs, Paralogs, and Evolutionary Genomics. Annu. Rev. Genet. 2005, 39, 309–338. [Google Scholar] [CrossRef] [PubMed]
- Altenhoff, A.M.; Glover, N.M.; Dessimoz, C. Inferring Orthology and Paralogy. In Evolutionary Genomics; Anisimova, M., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; Volume 1910, pp. 149–175. ISBN 978-1-4939-9073-3. [Google Scholar]
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, J.; Schemske, D.W. Pathways, Mechanisms, and Rates of Polyploid Formation in Flowering Plants. Annu. Rev. Ecol. Syst. 1998, 29, 467–501. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef]
- Feldman, M.; Levy, A.A. Genome Evolution Due to Allopolyploidization in Wheat. Genetics 2012, 192, 763–774. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Yang, J.; Liu, D.; Wang, X.; Ji, C.; Cheng, F.; Liu, B.; Hu, Z.; Chen, S.; Pental, D.; Ju, Y.; et al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 2016, 48, 1225–1232. [Google Scholar] [CrossRef]
- Sun, F.; Fan, G.; Hu, Q.; Zhou, Y.; Guan, M.; Tong, C.; Li, J.; Du, D.; Qi, C.; Jiang, L.; et al. The high-quality genome of Brassica napus cultivar ‘ZS11′ reveals the introgression history in semi-winter morphotype. Plant J. 2017, 92, 452–468. [Google Scholar] [CrossRef]
- Lu, K.; Wei, L.; Li, X.; Wang, Y.; Wu, J.; Liu, M.; Zhang, C.; Chen, Z.; Xiao, Z.; Jian, H.; et al. Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat. Commun. 2019, 10, 1154. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, M. The 2R hypothesis: An update. Curr. Opin. Immunol. 2007, 19, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Wendel, J.F.; Lisch, D.; Hu, G.; Mason, A.S. The long and short of doubling down: Polyploidy, epigenetics, and the temporal dynamics of genome fractionation. Curr. Opin. Genet. Dev. 2018, 49, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Freeling, M.; Scanlon, M.J.; Fowler, J.E. Fractionation and subfunctionalization following genome duplications: Mechanisms that drive gene content and their consequences. Curr. Opin. Genet. Dev. 2015, 35, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Wright, J.E.; Johnson, K.; Hollister, A.; May, B. Meiotic models to explain classical linkage, pseudolinkage, and chromosome pairing in tetraploid derivative salmonid genomes. Isozymes 1983, 10, 239–260. [Google Scholar]
- Sacerdot, C.; Louis, A.; Bon, C.; Berthelot, C.; Roest Crollius, H. Chromosome evolution at the origin of the ancestral vertebrate genome. Genome Biol. 2018, 19, 166. [Google Scholar] [CrossRef]
- Pervaiz, N.; Shakeel, N.; Qasim, A.; Zehra, R.; Anwar, S.; Rana, N.; Xue, Y.; Zhang, Z.; Bao, Y.; Abbasi, A.A. Evolutionary history of the human multigene families reveals widespread gene duplications throughout the history of animals. BMC Evol. Biol. 2019, 19, 128. [Google Scholar] [CrossRef]
- Zhang, J. Evolution by gene duplication: An update. Trends Ecol. Evol. 2003, 18, 292–298. [Google Scholar] [CrossRef]
- Arguello, J.R.; Fan, C.; Wang, W.; Long, M. Origination of chimeric genes through DNA-level recombination. In Gene and Protein Evolution; Karger Publishers: Basel, Switzerland, 2007; Volume 3, pp. 131–146. [Google Scholar] [CrossRef]
- Reams, A.B.; Roth, J.R. Mechanisms of gene duplication and amplification. Cold Spring Harb. Perspect. Biol. 2015, 7, a016592. [Google Scholar] [CrossRef]
- Cook, D.E.; Lee, T.G.; Guo, X.; Melito, S.; Wang, K.; Bayless, A.M.; Wang, J.; Hughes, T.J.; Willis, D.K.; Clemente, T.E.; et al. Copy Number Variation of Multiple Genes at Rhg1 Mediates Nematode Resistance in Soybean. Science 2012, 338, 1206–1209. [Google Scholar] [CrossRef]
- Kono, T.J.Y.; Brohammer, A.B.; McGaugh, S.E.; Hirsch, C.N. Tandem Duplicate Genes in Maize Are Abundant and Date to Two Distinct Periods of Time. G3 Genes Genomes Genet. 2018, 8, 3049–3058. [Google Scholar] [CrossRef]
- Tan, B.C.; Guan, J.C.; Ding, S.; Wu, S.; Saunders, J.W.; Koch, K.E.; McCarty, D.R. Structure and Origin of the White Cap Locus and Its Role in Evolution of Grain Color in Maize. Genetics 2017, 206, 135–150. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Vanguri, S.; Boeke, J.D.; Gabriel, A.; Voytas, D.F. Transposable elements and genome organization: A comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998, 8, 464–478. [Google Scholar] [CrossRef] [PubMed]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Brosius, J. Retroposons—Seeds of evolution. Science 1991, 251, 753. [Google Scholar] [CrossRef] [PubMed]
- Moran, J.V.; DeBerardinis, R.J.; Kazazian, H.H. Exon shuffling by L1 retrotransposition. Science 1999, 283, 1530–1534. [Google Scholar] [CrossRef]
- Elrouby, N.; Bureau, T.E. A novel hybrid open reading frame formed by multiple cellular gene transductions by a plant long terminal repeat retroelement. J. Biol. Chem. 2001, 276, 41963–41968. [Google Scholar] [CrossRef]
- Zhang, Z.; Harrison, P.M.; Liu, Y.; Gerstein, M. Millions of Years of Evolution Preserved: A Comprehensive Catalog of the Processed Pseudogenes in the Human Genome. Genome Res. 2003, 13, 2541–2558. [Google Scholar] [CrossRef]
- Casola, C.; Betrán, E. The Genomic Impact of Gene Retrocopies: What Have We Learned from Comparative Genomics, Population Genomics, and Transcriptomic Analyses? Genome Biol. Evol. 2017, 9, 1351–1373. [Google Scholar] [CrossRef]
- Betrán, E.; Thornton, K.; Long, M. Retroposed new genes out of the X in Drosophila. Genome Res. 2002, 12, 1854–1859. [Google Scholar] [CrossRef]
- Bai, Y.; Casola, C.; Feschotte, C.; Betrán, E. Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in Drosophila. Genome Biol. 2007, 8, R11. [Google Scholar] [CrossRef] [PubMed]
- Toups, M.A.; Hahn, M.W. Retrogenes reveal the direction of sex-chromosome evolution in mosquitoes. Genetics 2010, 186, 763–766. [Google Scholar] [CrossRef] [PubMed]
- Emerson, J.J.; Kaessmann, H.; Betrán, E.; Long, M. Extensive gene traffic on the mammalian X chromosome. Science 2004, 303, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zheng, H.; Fan, C.; Li, J.; Shi, J.; Cai, Z.; Zhang, G.; Liu, D.; Zhang, J.; Vang, S.; et al. High Rate of Chimeric Gene Origination by Retroposition in Plant Genomes. Plant Cell 2006, 18, 1791–1802. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, X.; Tang, H.; Tan, X.; Ficklin, S.P.; Feltus, F.A.; Paterson, A.H. Modes of gene duplication contribute differently to genetic novelty and redundancy, but show parallels across divergent angiosperms. PLoS ONE 2011, 6, e28150. [Google Scholar] [CrossRef]
- Juretic, N.; Hoen, D.R.; Huynh, M.L.; Harrison, P.M.; Bureau, T.E. The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 2005, 15, 1292–1297. [Google Scholar] [CrossRef]
- Le, Q.H.; Wright, S.; Yu, Z.; Bureau, T. Transposon diversity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2000, 97, 7376–7381. [Google Scholar] [CrossRef]
- Yu, Z.; Wright, S.I.; Bureau, T.E. Mutator-like elements in Arabidopsis thaliana. Structure, diversity and evolution. Genetics 2000, 156, 2019–2031. [Google Scholar]
- Kawasaki, S.; Nitasaka, E. Characterization of Tpn1 family in the Japanese morning glory: En/Spm-related transposable elements capturing host genes. Plant Cell Physiol. 2004, 45, 933–944. [Google Scholar] [CrossRef]
- Zabala, G.; Vodkin, L.O. The wp mutation of Glycine max carries a gene-fragment-rich transposon of the CACTA superfamily. Plant Cell 2005, 17, 2619–2632. [Google Scholar] [CrossRef]
- Jiang, N.; Bao, Z.; Zhang, X.; Eddy, S.R.; Wessler, S.R. Pack-MULE transposable elements mediate gene evolution in plants. Nature 2004, 431, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Samonte, R.V.; Eichler, E.E. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 2002, 3, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.H.; Shields, D.C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 1997, 387, 708–713. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.A.; Gu, Z.; Clark, R.A.; Reinert, K.; Samonte, R.V.; Schwartz, S.; Adams, M.D.; Myers, E.W.; Li, P.W.; Eichler, E.E. Recent segmental duplications in the human genome. Science 2002, 297, 1003–1007. [Google Scholar] [CrossRef]
- Koszul, R.; Caburet, S.; Dujon, B.; Fischer, G. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 2004, 23, 234–243. [Google Scholar] [CrossRef]
- Koszul, R.; Dujon, B.; Fischer, G. Stability of large segmental duplications in the yeast genome. Genetics 2006, 172, 2211–2222. [Google Scholar] [CrossRef]
- Fiston-Lavier, A.-S.; Anxolabehere, D.; Quesneville, H. A model of segmental duplication formation in Drosophila melanogaster. Genome Res. 2007, 17, 1458–1470. [Google Scholar] [CrossRef][Green Version]
- Bailey, J.A.; Liu, G.; Eichler, E.E. An Alu transposition model for the origin and expansion of human segmental duplications. Am. J. Hum. Genet. 2003, 73, 823–834. [Google Scholar] [CrossRef]
- She, X.; Cheng, Z.; Zöllner, S.; Church, D.M.; Eichler, E.E. Mouse segmental duplication and copy number variation. Nat. Genet. 2008, 40, 909–914. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Bailey, J.A.; Eichler, E.E. Primate segmental duplications: Crucibles of evolution, diversity and disease. Nat. Rev. Genet. 2006, 7, 552–564. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Ma, D.; Vasseur, L.; You, M. Segmental duplications: Evolution and impact among the current Lepidoptera genomes. BMC Evol. Biol. 2017, 17, 161. [Google Scholar] [CrossRef] [PubMed]
- Hakes, L.; Lovell, S.C.; Oliver, S.G.; Robertson, D.L. Specificity in protein interactions and its relationship with sequence diversity and coevolution. Proc. Natl. Acad. Sci. USA 2007, 104, 7999–8004. [Google Scholar] [CrossRef] [PubMed]
- Wapinski, I.; Pfeffer, A.; Friedman, N.; Regev, A. Natural history and evolutionary principles of gene duplication in fungi. Nature 2007, 449, 54–61. [Google Scholar] [CrossRef]
- Blanc, G.; Wolfe, K.H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 2004, 16, 1679–1691. [Google Scholar] [CrossRef]
- Maere, S.; Bodt, S.D.; Raes, J.; Casneuf, T.; Montagu, M.V.; Kuiper, M.; De Peer, Y.V. Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 5454–5459. [Google Scholar] [CrossRef]
- Hanada, K.; Zou, C.; Lehti-Shiu, M.D.; Shinozaki, K.; Shiu, S.-H. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 2008, 148, 993–1003. [Google Scholar] [CrossRef]
- Rodgers-Melnick, E.; Mane, S.P.; Dharmawardhana, P.; Slavov, G.T.; Crasta, O.R.; Strauss, S.H.; Brunner, A.M.; DiFazio, S.P. Contrasting patterns of evolution following whole genome versus tandem duplication events in Populus. Genome Res. 2012, 22, 95–105. [Google Scholar] [CrossRef]
- Freeling, M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 2009, 60, 433–453. [Google Scholar] [CrossRef]
- Rizzon, C.; Ponger, L.; Gaut, B.S. Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comput. Biol. 2006, 2, e115. [Google Scholar] [CrossRef]
- Acharya, D.; Ghosh, T.C. Global analysis of human duplicated genes reveals the relative importance of whole-genome duplicates originated in the early vertebrate evolution. BMC Genom. 2016, 17, 71. [Google Scholar] [CrossRef] [PubMed]
- Casneuf, T.; De Bodt, S.; Raes, J.; Maere, S.; Van de Peer, Y. Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. Genome Biol. 2006, 7, R13. [Google Scholar] [CrossRef] [PubMed]
- Defoort, J.; Van de Peer, Y.; Carretero-Paulet, L. The Evolution of Gene Duplicates in Angiosperms and the Impact of Protein–Protein Interactions and the Mechanism of Duplication. Genome Biol. Evol. 2019, 11, 2292–2305. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y. Locally duplicated ohnologs evolve faster than nonlocally duplicated ohnologs in Arabidopsis and rice. Genome Biol. Evol. 2013, 5, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Arabidopsis Interactome Mapping Consortium; Dreze, M.; Carvunis, A.R.; Charloteaux, B.; Galli, M.; Pevzner, S.J.; Tasan, M.; Ahn, Y.Y.; Balumuri, P.; Barabási, A.L.; et al. Evidence for network evolution in an Arabidopsis interactome map. Science 2011, 333, 601–607. [Google Scholar] [CrossRef]
- Arsovski, A.A.; Pradinuk, J.; Guo, X.Q.; Wang, S.; Adams, K.L. Evolution of Cis-Regulatory Elements and Regulatory Networks in Duplicated Genes of Arabidopsis. Plant Physiol. 2015, 169, 2982–2991. [Google Scholar] [CrossRef] [PubMed]
- Prince, V.E.; Pickett, F.B. Splitting pairs: The diverging fates of duplicated genes. Nat. Rev. Genet. 2002, 3, 827–837. [Google Scholar] [CrossRef]
- Zou, C.; Lehti-Shiu, M.D.; Thibaud-Nissen, F.; Prakash, T.; Buell, C.R.; Shiu, S.-H. Evolutionary and expression signatures of pseudogenes in Arabidopsis and rice. Plant Physiol. 2009, 151, 3–15. [Google Scholar] [CrossRef]
- Rouquier, S.; Taviaux, S.; Trask, B.J.; Brand-Arpon, V.; Van den Engh, G.; Demaille, J.; Giorgi, D. Distribution of olfactory receptor genes in the human genome. Nat. Genet. 1998, 18, 243–250. [Google Scholar] [CrossRef]
- Quignon, P.; Kirkness, E.; Cadieu, E.; Touleimat, N.; Guyon, R.; Renier, C.; Hitte, C.; André, C.; Fraser, C.; Galibert, F. Comparison of the canine and human olfactory receptor gene repertoires. Genome Biol. 2003, 4, R80. [Google Scholar] [CrossRef]
- Hahn, M.W. Distinguishing among evolutionary models for the maintenance of gene duplicates. J. Hered. 2009, 100, 605–617. [Google Scholar] [CrossRef]
- Innan, H.; Kondrashov, F. The evolution of gene duplications: Classifying and distinguishing between models. Nat. Rev. Genet. 2010, 11, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. The Neutral Theory of Molecular Evolution; Cambridge University Press: Cambridge, UK, 1983; ISBN 978-0-521-31793-1. [Google Scholar]
- Logeman, B.L.; Wood, L.K.; Lee, J.; Thiele, D.J. Gene duplication and neo-functionalization in the evolutionary and functional divergence of metazoan copper transporters Ctr1 and Ctr2. J. Biol. Chem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Escriva, H.; Bertrand, S.; Germain, P.; Robinson-Rechavi, M.; Umbhauer, M.; Cartry, J.; Duffraisse, M.; Holland, L.; Gronemeyer, H.; Laudet, V. Neofunctionalization in vertebrates: The example of retinoic acid receptors. PLoS Genet. 2006, 2, e102. [Google Scholar] [CrossRef] [PubMed]
- Hughes, T.E.; Langdale, J.A.; Kelly, S. The impact of widespread regulatory neofunctionalization on homeolog gene evolution following whole-genome duplication in maize. Genome Res. 2014, 24, 1348–1355. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Chen, Y.; Long, M. Recurrent Tandem Gene Duplication Gave Rise to Functionally Divergent Genes in Drosophila. Mol. Biol. Evol. 2008, 25, 1451–1458. [Google Scholar] [CrossRef]
- Force, A.; Lynch, M.; Pickett, F.B.; Amores, A.; Yan, Y.L.; Postlethwait, J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 1999, 151, 1531–1545. [Google Scholar]
- Piatigorsky, J.; Wistow, G. The recruitment of crystallins: New functions precede gene duplication. Science 1991, 252, 1078–1079. [Google Scholar] [CrossRef]
- Hughes, A.L. The evolution of functionally novel proteins after gene duplication. Proc. R. Soc. Lond. B Biol. Sci. 1994, 256, 119–124. [Google Scholar] [CrossRef]
- Otto, S.P.; Yong, P. The evolution of gene duplicates. Adv. Genet. 2002, 46, 451–483. [Google Scholar] [CrossRef]
- Jackson, P.J.; Douglas, N.R.; Chai, B.; Binkley, J.; Sidow, A.; Barsh, G.S.; Millhauser, G.L. Structural and molecular evolutionary analysis of Agouti and Agouti-related proteins. Chem. Biol. 2006, 13, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Carlson, K.D.; Bhogale, S.; Anderson, D.; Zaragoza-Mendoza, A.; Madlung, A. Subfunctionalization of phytochrome B1/B2 leads to differential auxin and photosynthetic responses. Plant Direct 2020, 4, e00205. [Google Scholar] [CrossRef] [PubMed]
- Vavouri, T.; Semple, J.I.; Lehner, B. Widespread conservation of genetic redundancy during a billion years of eukaryotic evolution. Trends Genet. 2008, 24, 485–488. [Google Scholar] [CrossRef]
- Gout, J.F.; Lynch, M. Maintenance and Loss of Duplicated Genes by Dosage Subfunctionalization. Mol. Biol. Evol. 2015, 32, 2141–2148. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Liao, B.Y.; Chang, A.Y.F.; Zhang, J. Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet. 2010, 26, 425–430. [Google Scholar] [CrossRef]
- Greer, J.M.; Puetz, J.; Thomas, K.R.; Capecchi, M.R. Maintenance of functional equivalence during paralogous HOX gene evolution. Nature 2000, 403, 661–665. [Google Scholar] [CrossRef]
- Dean, E.J.; Davis, J.C.; Davis, R.W.; Petrov, D.A. Pervasive and persistent redundancy among duplicated genes in yeast. PLoS Genet. 2008, 4, e1000113. [Google Scholar] [CrossRef]
- Averof, M.; Dawes, R.; Ferrier, D. Diversification of arthropod HOX genes as a paradigm for the evolution of gene functions. Semin. Cell Dev. Biol. 1996, 7, 539–551. [Google Scholar] [CrossRef]
- Wang, W.; Brunet, F.G.; Nevo, E.; Long, M. Origin of sphinx, a young chimeric RNA gene in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2002, 99, 4448–4453. [Google Scholar] [CrossRef]
- Nisole, S.; Lynch, C.; Stoye, J.P.; Yap, M.W. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. USA 2004, 101, 13324–13328. [Google Scholar] [CrossRef]
- Sayah, D.M.; Sokolskaja, E.; Berthoux, L.; Luban, J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 2004, 430, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. Evolving protein functional diversity in new genes of Drosophila. Proc. Natl. Acad. Sci. USA 2004, 101, 16246–16250. [Google Scholar] [CrossRef] [PubMed]
- Machado, J.P.; Antunes, A. The genomic context of retrocopies increases their chance of functional relevancy in mammals. Genomics 2020, 112, 2410–2417. [Google Scholar] [CrossRef] [PubMed]
- Makino, T.; McLysaght, A. Positionally biased gene loss after whole genome duplication: Evidence from human, yeast, and plant. Genome Res. 2012, 22, 2427–2435. [Google Scholar] [CrossRef]
- Jiang, W.; Liu, Y.; Xia, E.; Gao, L. Prevalent role of gene features in determining evolutionary fates of whole-genome duplication duplicated genes in flowering plants. Plant Physiol. 2013, 161, 1844–1861. [Google Scholar] [CrossRef]
- Pan, D.; Zhang, L. Quantifying the major mechanisms of recent gene duplications in the human and mouse genomes: A novel strategy to estimate gene duplication rates. Genome Biol. 2007, 8, R158. [Google Scholar] [CrossRef]
- Marques-Bonet, T.; Girirajan, S.; Eichler, E.E. The origins and impact of primate segmental duplications. Trends Genet. 2009, 25, 443–454. [Google Scholar] [CrossRef]
- Assis, R.; Bachtrog, D. Neofunctionalization of young duplicate genes in Drosophila. Proc. Natl. Acad. Sci. USA 2013, 110, 17409–17414. [Google Scholar] [CrossRef]
- Pearson, W.R. An introduction to sequence similarity (“homology”) searching. Curr. Protoc. Bioinforma. 2013. [Google Scholar] [CrossRef]
- Shapiro, B.; Hofreiter, M. A paleogenomic perspective on evolution and gene function: New insights from ancient DNA. Science 2014, 343, 1236573. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.S.; Eddy, S.R.; Portugaly, E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinform. 2010, 11, 431. [Google Scholar] [CrossRef]
- Smith, T.F.; Waterman, M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981, 147, 195–197. [Google Scholar] [CrossRef]
- Saebø, P.E.; Andersen, S.M.; Myrseth, J.; Laerdahl, J.K.; Rognes, T. PARALIGN: Rapid and sensitive sequence similarity searches powered by parallel computing technology. Nucleic Acids Res. 2005, 33, W535–W539. [Google Scholar] [CrossRef] [PubMed]
- Rucci, E.; Garcia Sanchez, C.; Botella Juan, G.; Giusti, A.D.; Naiouf, M.; Prieto-Matias, M. SWIMM 2.0: Enhanced Smith-Waterman on Intel’s Multicore and Manycore Architectures Based on AVX-512 Vector Extensions. Int. J. Parallel Program 2019, 47, 296–316. [Google Scholar] [CrossRef]
- Koonin, E.V.; Galperin, M.Y. Sequence—Evolution—Function: Computational Approaches in Comparative Genomics; Kluwer Academic: Boston, MA, USA, 2003; ISBN 978-1-4020-7274-1. [Google Scholar]
- Sander, C.; Schneider, R. Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins 1991, 9, 56–68. [Google Scholar] [CrossRef]
- Rost, B. Twilight zone of protein sequence alignments. Protein Eng. 1999, 12, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Li, W.H.; Gu, Z.; Wang, H.; Nekrutenko, A. Evolutionary analyses of the human genome. Nature 2001, 409, 847–849. [Google Scholar] [CrossRef]
- Blanc, G.; Wolfe, K.H. Widespread Paleopolyploidy in Model Plant Species Inferred from Age Distributions of Duplicate Genes. Plant Cell 2004, 16, 1667–1678. [Google Scholar] [CrossRef]
- Wootton, J.C.; Federhen, S. Statistics of local complexity in amino acid sequences and sequence databases. Comput. Chem. 1993, 17, 149–163. [Google Scholar] [CrossRef]
- Shoja, V.; Zhang, L. A roadmap of tandemly arrayed genes in the genomes of human, mouse, and rat. Mol. Biol. Evol. 2006, 23, 2134–2141. [Google Scholar] [CrossRef] [PubMed]
- Britten, R.J. Almost all human genes resulted from ancient duplication. Proc. Natl. Acad. Sci. USA 2006, 103, 19027–19032. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Zhang, L. Tandemly arrayed genes in vertebrate genomes. Comp. Funct. Genom. 2008, 545269. [Google Scholar] [CrossRef] [PubMed]
- Makino, T.; McLysaght, A. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc. Natl. Acad. Sci. USA 2010, 107, 9270–9274. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.P.; Arora, J.; Isambert, H. Identification of Ohnolog Genes Originating from Whole Genome Duplication in Early Vertebrates, Based on Synteny Comparison across Multiple Genomes. PLoS Comput. Biol. 2015, 11, e1004394. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.Y.; El-Gebali, S.; Fraser, M.I.; et al. InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019, 47, D351–D360. [Google Scholar] [CrossRef]
- Kuzniar, A.; Van Ham, R.C.H.J.; Pongor, S.; Leunissen, J.A.M. The quest for orthologs: Finding the corresponding gene across genomes. Trends Genet. 2008, 24, 539–551. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Koonin, E.V.; Lipman, D.J. A genomic perspective on protein families. Science 1997, 278, 631–637. [Google Scholar] [CrossRef]
- Remm, M.; Storm, C.E.; Sonnhammer, E.L. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001, 314, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, F.; Sonnhammer, E.L.L. Hieranoid: Hierarchical orthology inference. J. Mol. Biol. 2013, 425, 2072–2081. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.J.; Julien, P.; Kuhn, M.; Von Mering, C.; Muller, J.; Doerks, T.; Bork, P. eggNOG: Automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008, 36, D250–D254. [Google Scholar] [CrossRef] [PubMed]
- Kriventseva, E.V.; Tegenfeldt, F.; Petty, T.J.; Waterhouse, R.M.; Simão, F.A.; Pozdnyakov, I.A.; Ioannidis, P.; Zdobnov, E.M. OrthoDB v8: Update of the hierarchical catalog of orthologs and the underlying free software. Nucleic Acids Res. 2015, 43, D250–D256. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Stoeckert, C.J.; Roos, D.S. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef]
- Linard, B.; Thompson, J.D.; Poch, O.; Lecompte, O. OrthoInspector: Comprehensive orthology analysis and visual exploration. BMC Bioinform. 2011, 12, 11. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef]
- Train, C.-M.; Glover, N.M.; Gonnet, G.H.; Altenhoff, A.M.; Dessimoz, C. Orthologous Matrix (OMA) algorithm 2.0: More robust to asymmetric evolutionary rates and more scalable hierarchical orthologous group inference. Bioinformatics 2017, 33, i75–i82. [Google Scholar] [CrossRef]
- Dalquen, D.A.; Dessimoz, C. Bidirectional best hits miss many orthologs in duplication-rich clades such as plants and animals. Genome Biol. Evol. 2013, 5, 1800–1806. [Google Scholar] [CrossRef]
- Li, H.; Coghlan, A.; Ruan, J.; Coin, L.J.; Hériché, J.K.; Osmotherly, L.; Li, R.; Liu, T.; Zhang, Z.; Bolund, L.; et al. TreeFam: A curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006, 34, D572–D580. [Google Scholar] [CrossRef]
- Poptsova, M.S.; Gogarten, J.P. BranchClust: A phylogenetic algorithm for selecting gene families. BMC Bioinform. 2007, 8, 120. [Google Scholar] [CrossRef] [PubMed]
- Penel, S.; Arigon, A.M.; Dufayard, J.F.; Sertier, A.S.; Daubin, V.; Duret, L.; Gouy, M.; Perrière, G. Databases of homologous gene families for comparative genomics. BMC Bioinform. 2009, 10 (Suppl. 6), S3. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Capella-Gutierrez, S.; Pryszcz, L.P.; Denisov, I.; Kormes, D.; Marcet-Houben, M.; Gabaldón, T. PhylomeDB v3.0: An expanding repository of genome-wide collections of trees, alignments and phylogeny-based orthology and paralogy predictions. Nucleic Acids Res. 2011, 39, D556–D560. [Google Scholar] [CrossRef] [PubMed]
- Storm, C.E.V.; Sonnhammer, E.L.L. Automated ortholog inference from phylogenetic trees and calculation of orthology reliability. Bioinformatics 2002, 18, 92–99. [Google Scholar] [CrossRef]
- Berglund-Sonnhammer, A.C.; Steffansson, P.; Betts, M.J.; Liberles, D.A. Optimal Gene Trees from Sequences and Species Trees Using a Soft Interpretation of Parsimony. J. Mol. Evol. 2006, 63, 240–250. [Google Scholar] [CrossRef]
- Van der Heijden, R.T.J.M.; Snel, B.; Van Noort, V.; Huynen, M.A. Orthology prediction at scalable resolution by phylogenetic tree analysis. BMC Bioinform. 2007, 8, 83. [Google Scholar] [CrossRef]
- Goodman, M.; Czelusniak, J.; Moore, G.W.; Romero-Herrera, A.E.; Matsuda, G. Fitting the Gene Lineage into its Species Lineage, a Parsimony Strategy Illustrated by Cladograms Constructed from Globin Sequences. Syst. Biol. 1979, 28, 132–163. [Google Scholar] [CrossRef]
- Åkerborg, Ö.; Sennblad, B.; Arvestad, L.; Lagergren, J. Simultaneous Bayesian gene tree reconstruction and reconciliation analysis. Proc. Natl. Acad. Sci. USA 2009, 106, 5714–5719. [Google Scholar] [CrossRef]
- Liu, D.; Hunt, M.; Tsai, I.J. Inferring synteny between genome assemblies: A systematic evaluation. BMC Bioinform. 2018, 19, 26. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Haug-Baltzell, A.; Stephens, S.A.; Davey, S.; Scheidegger, C.E.; Lyons, E. SynMap2 and SynMap3D: Web-based whole-genome synteny browsers. Bioinformatics 2017, 33, 2197–2198. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Proost, S.; Fostier, J.; De Witte, D.; Dhoedt, B.; Demeester, P.; Van de Peer, Y.; Vandepoele, K. i-ADHoRe 3.0—Fast and sensitive detection of genomic homology in extremely large data sets. Nucleic Acids Res. 2012, 40, e11. [Google Scholar] [CrossRef] [PubMed]
- Rödelsperger, C.; Dieterich, C. CYNTENATOR: Progressive Gene Order Alignment of 17 Vertebrate Genomes. PLoS ONE 2010, 5, e8861. [Google Scholar] [CrossRef] [PubMed]
- Drillon, G.; Carbone, A.; Fischer, G. SynChro: A Fast and Easy Tool to Reconstruct and Visualize Synteny Blocks along Eukaryotic Chromosomes. PLoS ONE 2014, 9, e92621. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Kozik, A.; Chan, B.; Michelmore, R.; Young, N.D. DiagHunter and GenoPix2D: Programs for genomic comparisons, large-scale homology discovery and visualization. Genome Biol. 2003, 4, R68. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, P.P.; Chakravarty, S.; Vision, T.J. Fast identification and statistical evaluation of segmental homologies in comparative maps. Bioinformatics 2003, 19, i74–i80. [Google Scholar] [CrossRef]
- Soderlund, C.; Nelson, W.; Shoemaker, A.; Paterson, A. SyMAP: A system for discovering and viewing syntenic regions of FPC maps. Genome Res. 2006, 16, 1159–1168. [Google Scholar] [CrossRef]
- Sinha, A.U.; Meller, J. Cinteny: Flexible analysis and visualization of synteny and genome rearrangements in multiple organisms. BMC Bioinform. 2007, 8, 82. [Google Scholar] [CrossRef]
- Haas, B.J.; Delcher, A.L.; Wortman, J.R.; Salzberg, S.L. DAGchainer: A tool for mining segmental genome duplications and synteny. Bioinformatics 2004, 20, 3643–3646. [Google Scholar] [CrossRef]
- Hampson, S.; McLysaght, A.; Gaut, B.; Baldi, P. LineUp: Statistical detection of chromosomal homology with application to plant comparative genomics. Genome Res. 2003, 13, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shi, X.; Li, Z.; Zhu, Q.; Kong, L.; Tang, W.; Ge, S.; Luo, J. Statistical inference of chromosomal homology based on gene colinearity and applications to Arabidopsis and rice. BMC Bioinform. 2006, 7, 447. [Google Scholar] [CrossRef]
- Pham, S.K.; Pevzner, P.A. DRIMM-Synteny: Decomposing genomes into evolutionary conserved segments. Bioinformatics 2010, 26, 2509–2516. [Google Scholar] [CrossRef] [PubMed]
- Paten, B.; Herrero, J.; Beal, K.; Fitzgerald, S.; Birney, E. Enredo and Pecan: Genome-wide mammalian consistency-based multiple alignment with paralogs. Genome Res. 2008, 18, 1814–1828. [Google Scholar] [CrossRef]
- Xu, A.W.; Moret, B.M.E. GASTS: Parsimony Scoring under Rearrangements. In Algorithms in Bioinformatics, Proceedings of the 11th International Workshop, WABI 2011, Saarbrücken, Germany, 5–7 September 2011; Przytycka, T.M., Sagot, M.F., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 351–363. [Google Scholar]
- Zhou, L.; Feng, B.; Yang, N.; Tang, J. Ancestral reconstruction with duplications using binary encoding and probabilistic model. In Proceedings of the 7th International conference on Bioinformatics and Computational Biology, Honolulu, HI, USA, 9–11 March 2015; pp. 97–104. [Google Scholar]
- Yang, N.; Hu, F.; Zhou, L.; Tang, J. Reconstruction of Ancestral Gene Orders Using Probabilistic and Gene Encoding Approaches. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Feng, B.; Zhou, L.; Tang, J. Ancestral Genome Reconstruction on Whole Genome Level. Curr. Genom. 2017, 18, 306–315. [Google Scholar] [CrossRef]
- Lucas, J.M.; Muffato, M.; Crollius, H.R. PhylDiag: Identifying complex synteny blocks that include tandem duplications using phylogenetic gene trees. BMC Bioinform. 2014, 15. [Google Scholar] [CrossRef]
- Hachiya, T.; Osana, Y.; Popendorf, K.; Sakakibara, Y. Accurate identification of orthologous segments among multiple genomes. Bioinformatics 2009, 25, 853–860. [Google Scholar] [CrossRef]
- Baek, J.H.; Kim, J.; Kim, C.K.; Sohn, S.H.; Choi, D.; Ratnaparkhe, M.B.; Kim, D.W.; Lee, T.H. MultiSyn: A Webtool for Multiple Synteny Detection and Visualization of User’s Sequence of Interest Compared to Public Plant Species. Evol. Bioinform. 2016. [Google Scholar] [CrossRef]
- Zeng, X.; Nesbitt, M.J.; Pei, J.; Wang, K.; Vergara, I.A.; Chen, N. OrthoCluster: A new tool for mining synteny blocks and applications in comparative genomics. In Advances in database technology, Proceedings of the 11th international conference on Extending database technology, Nantes, France, 25–29 March 2008; Association for Computing Machinery: New York, NY, USA, 2008; pp. 656–667. [Google Scholar]
- Fostier, J.; Proost, S.; Dhoedt, B.; Saeys, Y.; Demeester, P.; Van de Peer, Y.; Vandepoele, K. A greedy, graph-based algorithm for the alignment of multiple homologous gene lists. Bioinformatics 2011, 27, 749–756. [Google Scholar] [CrossRef][Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Simillion, C.; Vandepoele, K.; Saeys, Y.; Van de Peer, Y. Building Genomic Profiles for Uncovering Segmental Homology in the Twilight Zone. Genome Res. 2004, 14, 1095–1106. [Google Scholar] [CrossRef] [PubMed]
- Lipman, D.J.; Pearson, W.R. Rapid and sensitive protein similarity searches. Science 1985, 227, 1435–1441. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Pevzner, P.; Tesler, G. Genome rearrangements in mammalian evolution: Lessons from human and mouse genomes. Genome Res. 2003, 13, 37–45. [Google Scholar] [CrossRef]
- Swidan, F.; Rocha, E.P.C.; Shmoish, M.; Pinter, R.Y. An Integrative Method for Accurate Comparative Genome Mapping. PLoS Comput. Biol. 2006, 2. [Google Scholar] [CrossRef]
- Paten, B.; Earl, D.; Nguyen, N.; Diekhans, M.; Zerbino, D.; Haussler, D. Cactus: Algorithms for genome multiple sequence alignment. Genome Res. 2011, 21, 1512–1528. [Google Scholar] [CrossRef]
- Paten, B.; Diekhans, M.; Earl, D.; St. John, J.; Ma, J.; Suh, B.; Haussler, D. Cactus Graphs for Genome Comparisons. In Research in Computational Molecular Biology, Proceedings of the 14th Annual International Conference, RECOMB 2010, Lisbon, Portugal, 25–28 April 2010; Berger, B., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 410–425. [Google Scholar]
- Zhang, L.; Gaut, B.S. Does Recombination Shape the Distribution and Evolution of Tandemly Arrayed Genes (TAGs) in the Arabidopsis thaliana Genome? Genome Res. 2003, 13, 2533–2540. [Google Scholar] [CrossRef]
- Morgulis, A.; Gertz, E.M.; Schäffer, A.A.; Agarwala, R. A fast and symmetric DUST implementation to mask low-complexity DNA sequences. J. Comput. Biol. 2006, 13, 1028–1040. [Google Scholar] [CrossRef]
- Wootton, J.C.; Federhen, S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996, 266, 554–571. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Frith, M.C. A new repeat-masking method enables specific detection of homologous sequences. Nucleic Acids Res. 2011, 39, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Olson, D.; Wheeler, T. ULTRA: A Model Based Tool to Detect Tandem Repeats. In Proceedings of the 9th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, Washington, DC, USA, 29 August–1 September 2018; Association for Computing Machinery: Washington, DC, USA, 2018; pp. 37–46. [Google Scholar]
- Soylev, A.; Le, T.M.; Amini, H.; Alkan, C.; Hormozdiari, F. Discovery of tandem and interspersed segmental duplications using high-throughput sequencing. Bioinformatics 2019, 35, 3923–3930. [Google Scholar] [CrossRef] [PubMed]
- Genovese, L.M.; Mosca, M.M.; Pellegrini, M.; Geraci, F. Dot2dot: Accurate whole-genome tandem repeats discovery. Bioinformatics 2019, 35, 914–922. [Google Scholar] [CrossRef] [PubMed]
- Audemard, E.; Schiex, T.; Faraut, T. Detecting long tandem duplications in genomic sequences. BMC Bioinform. 2012, 13, 83. [Google Scholar] [CrossRef]
- Zheng, D.; Gerstein, M.B. A computational approach for identifying pseudogenes in the ENCODE regions. Genome Biol. 2006, 7 (Suppl. 1), S13. [Google Scholar] [CrossRef]
- Despons, L.; Baret, P.V.; Frangeul, L.; Louis, V.L.; Durrens, P.; Souciet, J.-L. Genome-wide computational prediction of tandem gene arrays: Application in yeasts. BMC Genom. 2010, 11, 56. [Google Scholar] [CrossRef]
- Herrero, J.; Muffato, M.; Beal, K.; Fitzgerald, S.; Gordon, L.; Pignatelli, M.; Vilella, A.J.; Searle, S.M.J.; Amode, R.; Brent, S.; et al. Ensembl comparative genomics resources. Database 2016, 2016. [Google Scholar] [CrossRef]
- Lee, J.; Hong, W.; Cho, M.; Sim, M.; Lee, D.; Ko, Y.; Kim, J. Synteny Portal: A web-based application portal for synteny block analysis. Nucleic Acids Res. 2016, 44, W35–W40. [Google Scholar] [CrossRef]
- Muffato, M.; Louis, A.; Poisnel, C.E.; Crollius, H.R. Genomicus: A database and a browser to study gene synteny in modern and ancestral genomes. Bioinformatics 2010, 26, 1119–1121. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, L.; Suh, B.B.; Raney, B.J.; Burhans, R.C.; Kent, W.J.; Blanchette, M.; Haussler, D.; Miller, W. Reconstructing contiguous regions of an ancestral genome. Genome Res. 2006, 16, 1557–1565. [Google Scholar] [CrossRef]
- Loots, G.; Ovcharenko, I. ECRbase: Database of evolutionary conserved regions, promoters, and transcription factor binding sites in vertebrate genomes. Bioinformatics 2007, 23, 122–124. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.P.; Vergara, I.A.; Frech, C.; Chen, Q.; Zeng, X.; Pei, J.; Chen, N. OrthoClusterDB: An online platform for synteny blocks. BMC Bioinform. 2009, 10, 192. [Google Scholar] [CrossRef] [PubMed]
- Sonnhammer, E.L.L.; Östlund, G. InParanoid 8: Orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 2015, 43, D234–D239. [Google Scholar] [CrossRef] [PubMed]
- Miele, V.; Penel, S.; Duret, L. Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinform. 2011, 12, 116. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Altenhoff, A.M.; Glover, N.M.; Train, C.-M.; Kaleb, K.; Warwick Vesztrocy, A.; Dylus, D.; De Farias, T.M.; Zile, K.; Stevenson, C.; Long, J.; et al. The OMA orthology database in 2018: Retrieving evolutionary relationships among all domains of life through richer web and programmatic interfaces. Nucleic Acids Res. 2018, 46, D477–D485. [Google Scholar] [CrossRef]
- Van Bel, M.; Diels, T.; Vancaester, E.; Kreft, L.; Botzki, A.; Van de Peer, Y.; Coppens, F.; Vandepoele, K. PLAZA 4.0: An integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 2018, 46, D1190–D1196. [Google Scholar] [CrossRef]
- Conte, M.G.; Gaillard, S.; Lanau, N.; Rouard, M.; Périn, C. GreenPhylDB: A database for plant comparative genomics. Nucleic Acids Res. 2008, 36, D991–D998. [Google Scholar] [CrossRef][Green Version]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).