Mobilization of IMEs Integrated in the oriT of ICEs Involves Their Own Relaxase Belonging to the Rep-Trans Family of Proteins
Abstract
1. Introduction
2. Materials and Methods
2.1. In Silico Search and Analysis of IMEs Encoding a Relaxase or an Integrase Related to the One of IME_SagLMG15084_oriT
2.2. Construction of Phylogenetic Trees
2.3. Bacterial Strains and Growth Conditions
2.4. IME and ICE Tagging with a Resistance Gene and Deletion of Integrase and Relaxase Genes of the Integrative Elements
2.5. Excision Tests and Mating Experiments
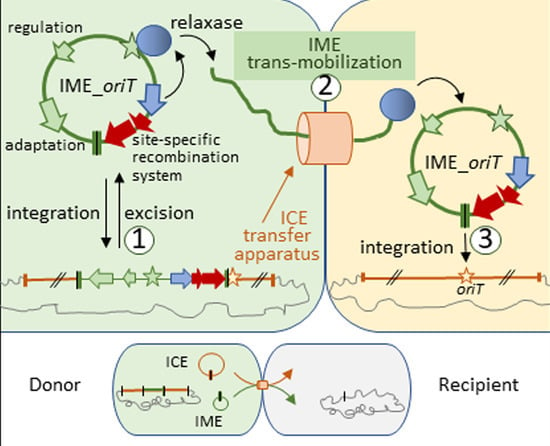
3. Results
3.1. Integrative Mobilizable Elements (IMEs) That Target oriT of Other Integrative Elements (IME_oriTs) Are Widespread in Firmicutes and Carry Different Cargo Genes
3.1.1. Distribution of IME_oriTs in Firmicutes
3.1.2. Chromosomal Integration Sites of IME_oriTs
3.1.3. Cargo Genes of IME_oriTs
3.2. IME_oriTs Identified in S. salivarius Strains L11, L25 and L45 Are Able to Excise from the Chromosome
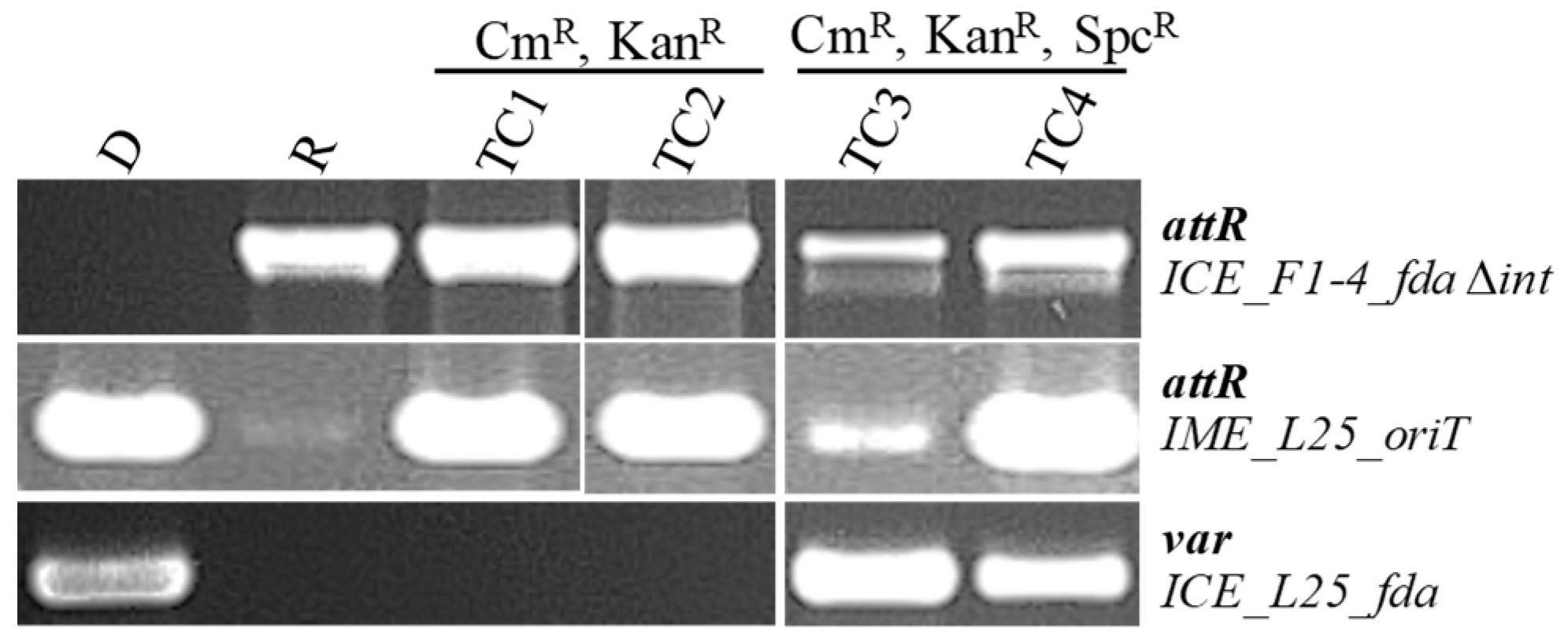
3.3. IME_oriT of S. salivarius Strain L25 (IME_SsalL25_oriT) Can Be Transferred by Conjugation to Another S. salivarius Strain and to S. thermophilus
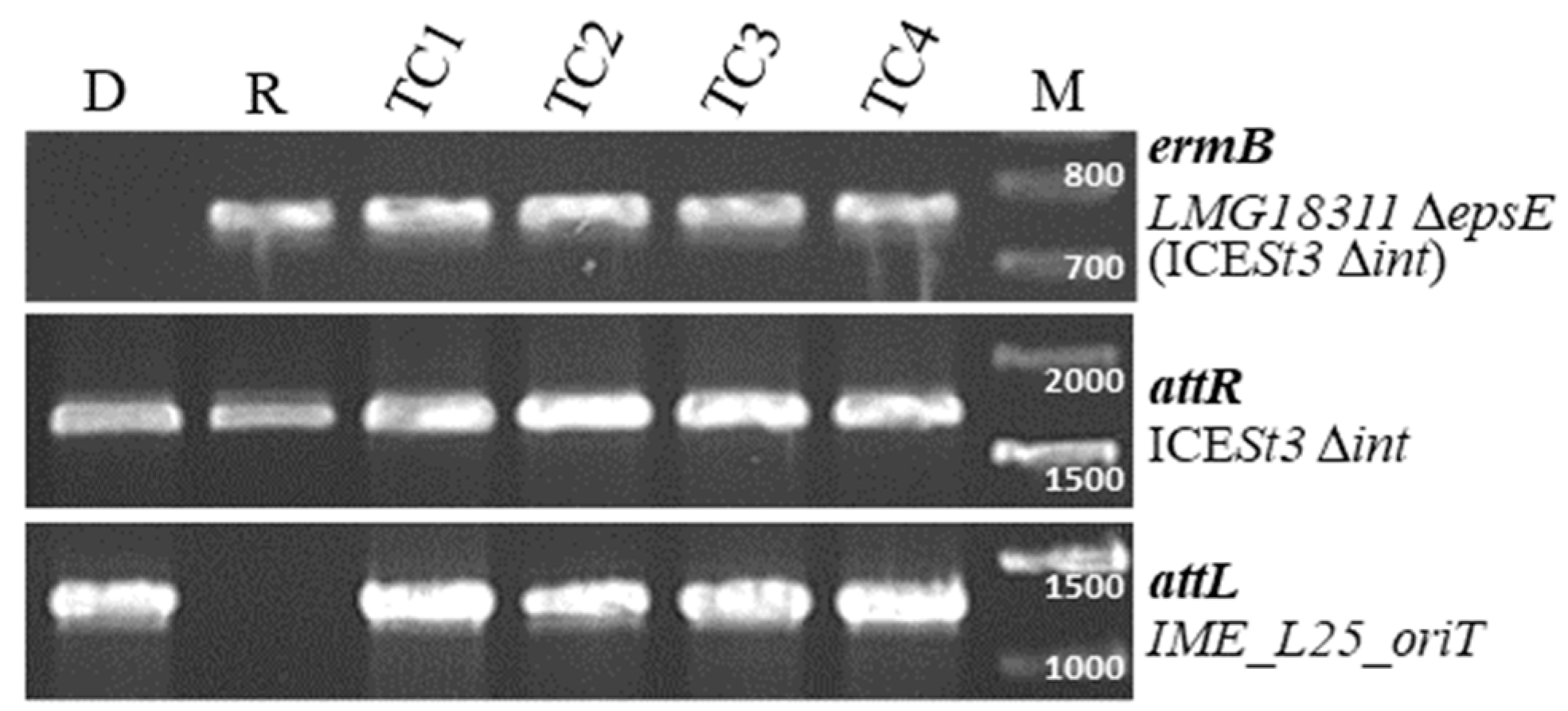
3.4. IME_SsalL25_oriT Can Be Mobilized in Trans
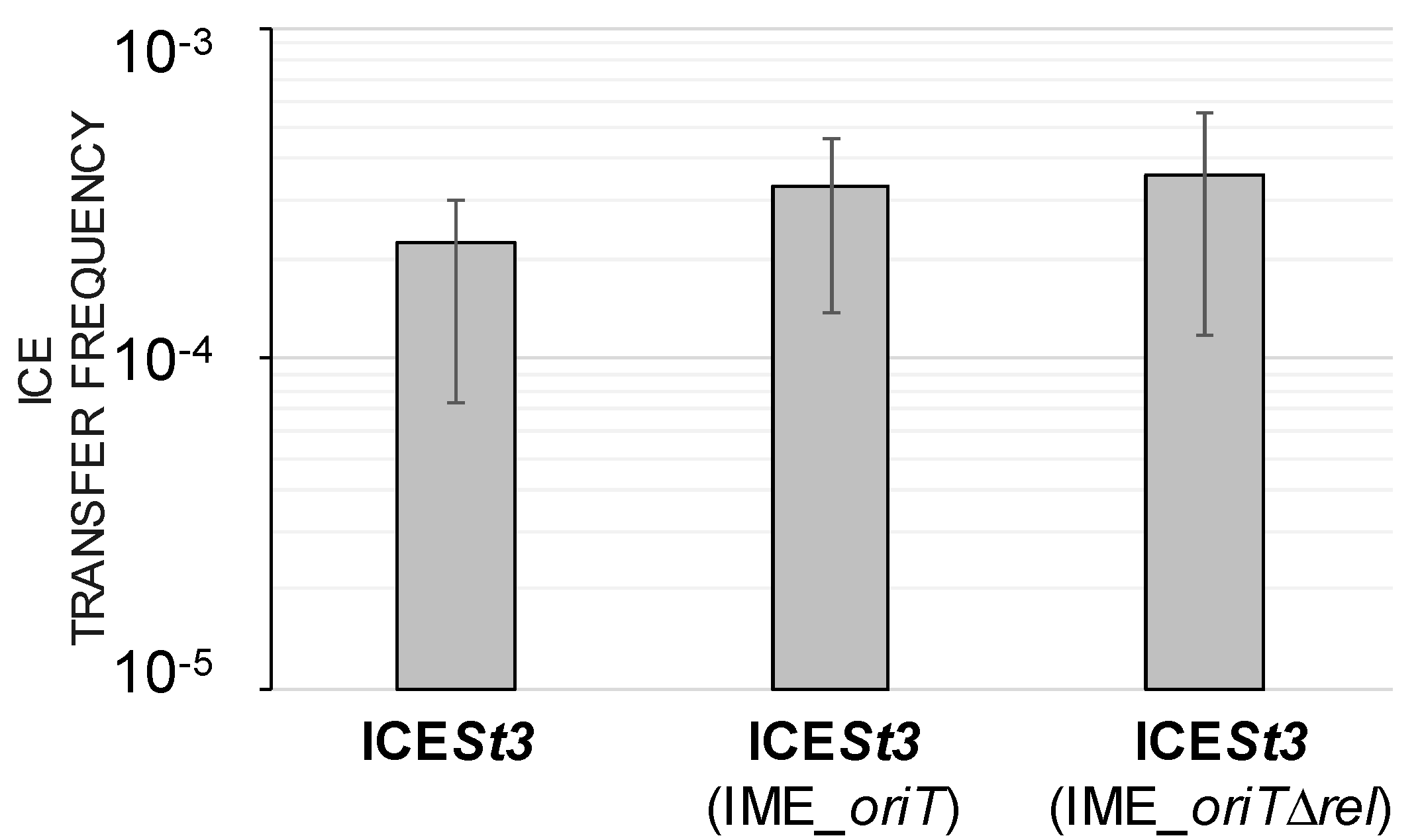
3.5. The Relaxase of IME_SsalL25_oriT Is Needed for IME Mobilization and Does Not Contribute to the Transfer of the Helper ICE
3.6. IME_SsalL25_oriT and Helper ICE Do Not Compete for the Conjugation Machinery
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bellanger, X.; Payot, S.; Leblond-Bourget, N.; Guédon, G. Conjugative and mobilizable genomic islands in bacteria: Evolution and diversity. FEMS Microbiol. Rev. 2014, 38, 720–760. [Google Scholar] [CrossRef] [PubMed]
- Delavat, F.; Miyazaki, R.; Carraro, N.; Pradervand, N.; van der Meer, J.R. The hidden life of integrative and conjugative elements. FEMS Microbiol. Rev. 2017, 41, 512–537. [Google Scholar] [CrossRef] [PubMed]
- Guglielmini, J.; Quintais, L.; Garcillán-Barcia, M.P.; de la Cruz, F.; Rocha, E.P.C. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 2011, 7, e1002222. [Google Scholar] [CrossRef] [PubMed]
- Burrus, V.; Pavlovic, G.; Decaris, B.; Guédon, G. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid 2002, 48, 77–97. [Google Scholar] [CrossRef]
- Wozniak, R.A.F.; Waldor, M.K. Integrative and conjugative elements: Mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 2010, 8, 552–563. [Google Scholar] [CrossRef]
- Grohmann, E.; Christie, P.J.; Waksman, G.; Backert, S. Type IV secretion in Gram-negative and Gram-positive bacteria: Type IV secretion. Mol. Microbiol. 2018, 107, 455–471. [Google Scholar] [CrossRef]
- Garcillán-Barcia, M.P.; Redondo-Salvo, S.; Vielva, L.; de la Cruz, F. MOBscan: Automated annotation of MOB relaxases. In Horizontal Gene Transfer; de la Cruz, F., Ed.; Springer: New York, NY, USA, 2020; Volume 2075, pp. 295–308. ISBN 978-1-4939-9876-0. [Google Scholar]
- Guglielmini, J.; de la Cruz, F.; Rocha, E.P.C. Evolution of conjugation and type IV secretion systems. Mol. Biol. Evol. 2013, 30, 315–331. [Google Scholar] [CrossRef]
- Grohmann, E.; Muth, G.; Espinosa, M. Conjugative plasmid transfer in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2003, 67, 277–301. [Google Scholar] [CrossRef]
- Guglielmini, J.; Neron, B.; Abby, S.S.; Garcillan-Barcia, M.P.; de la Cruz, F.; Rocha, E.P.C. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res. 2014, 42, 5715–5727. [Google Scholar] [CrossRef]
- Guédon, G.; Libante, V.; Coluzzi, C.; Payot, S.; Leblond-Bourget, N. The obscure world of integrative and mobilizable elements, highly widespread elements that pirate bacterial conjugative systems. Genes 2017, 8, 337. [Google Scholar] [CrossRef]
- Libante, V.; Nombre, Y.; Coluzzi, C.; Staub, J.; Guédon, G.; Gottschalk, M.; Teatero, S.; Fittipaldi, N.; Leblond-Bourget, N.; Payot, S. Chromosomal conjugative and mobilizable elements in Streptococcus suis: Major actors in the spreading of antimicrobial resistance and bacteriocin synthesis genes. Pathogens 2020, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Coluzzi, C.; Guédon, G.; Devignes, M.-D.; Ambroset, C.; Loux, V.; Lacroix, T.; Payot, S.; Leblond-Bourget, N. A glimpse into the world of integrative and mobilizable elements in streptococci reveals an unexpected diversity and novel families of mobilization proteins. Front. Microbiol. 2017, 8, 443. [Google Scholar] [CrossRef] [PubMed]
- Lao, J.; Guédon, G.; Lacroix, T.; Charron-Bourgoin, F.; Libante, V.; Loux, V.; Chiapello, H.; Payot, S.; Leblond-Bourget, N. Abundance, diversity and role of ICEs and IMEs in adaptation of Streptococcus salivarius to the environment. Genes 2020, in press. [Google Scholar]
- Puymège, A.; Bertin, S.; Guédon, G.; Payot, S. Analysis of Streptococcus agalactiae pan-genome for prevalence, diversity and functionality of integrative and conjugative or mobilizable elements integrated in the tRNALys CTT gene. Mol. Genet. Genom. 2015, 1727–1740. [Google Scholar] [CrossRef]
- Malbruny, B.; Werno, A.M.; Murdoch, D.R.; Leclercq, R.; Cattoir, V. Cross-resistance to lincosamides, streptogramins A, and pleuromutilins due to the lsa(C) gene in Streptococcus agalactiae UCN70. Antimicrob. Agents Chemother. 2011, 55, 1470–1474. [Google Scholar] [CrossRef]
- Douarre, P.-E.; Sauvage, E.; Poyart, C.; Glaser, P. Host specificity in the diversity and transfer of lsa resistance genes in group B Streptococcus. J. Antimicrob. Chemother. 2015, 70, 3205–3213. [Google Scholar] [CrossRef]
- Carver, T.; Berriman, M.; Tivey, A.; Patel, C.; Böhme, U.; Barrell, B.G.; Parkhill, J.; Rajandream, M.-A. Artemis and ACT: Viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 2008, 24, 2672–2676. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993. [Google Scholar] [CrossRef]
- Dahmane, N.; Libante, V.; Charron-Bourgoin, F.; Guédon, E.; Guédon, G.; Leblond-Bourget, N.; Payot, S. Diversity of integrative and conjugative elements of Streptococcus salivarius and their intra-and interspecies transfer. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Bellanger, X.; Roberts, A.P.; Morel, C.; Choulet, F.; Pavlovic, G.; Mullany, P.; Decaris, B.; Guédon, G. Conjugative transfer of the integrative conjugative elements ICESt1 and ICESt3 from Streptococcus thermophilus. J. Bacteriol. 2009, 191, 2764–2775. [Google Scholar] [CrossRef] [PubMed]
- Soler, N.; Robert, E.; Chauvot de Beauchêne, I.; Monteiro, P.; Libante, V.; Maigret, B.; Staub, J.; Ritchie, D.W.; Guédon, G.; Payot, S.; et al. Characterization of a relaxase belonging to the MOBT family, a widespread family in Firmicutes mediating the transfer of ICEs. Mob. DNA 2019, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Dahmane, N.; Robert, E.; Deschamps, J.; Meylheuc, T.; Delorme, C.; Briandet, R.; Leblond-Bourget, N.; Guédon, E.; Payot, S. Impact of cell surface molecules on conjugative transfer of the integrative and conjugative element ICESt3 of Streptococcus thermophilus. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef]
- Gardan, R.; Besset, C.; Guillot, A.; Gitton, C.; Monnet, V. The oligopeptide transport system is essential for the development of natural competence in Streptococcus thermophilus strain LMD-9. J. Bacteriol. 2009, 191, 4647–4655. [Google Scholar] [CrossRef]
- Carr, S.B.; Phillips, S.E.V.; Thomas, C.D. Structures of replication initiation proteins from staphylococcal antibiotic resistance plasmids reveal protein asymmetry and flexibility are necessary for replication. Nucleic Acids Res. 2016, 44, 2417–2428. [Google Scholar] [CrossRef]
- Smyth, D.S.; Robinson, D.A. Integrative and sequence characteristics of a novel genetic element, ICE6013, in Staphylococcus aureus. J. Bacteriol. 2009, 191, 5964–5975. [Google Scholar] [CrossRef]
- Wright, L.D.; Grossman, A.D. Autonomous replication of the conjugative transposon Tn916. J. Bacteriol. 2016, 198, 3355–3366. [Google Scholar] [CrossRef]
- Lee, C.A.; Grossman, A.D. Identification of the origin of transfer (oriT) and DNA relaxase required for conjugation of the integrative and conjugative element ICEBs1 of Bacillus subtilis. J. Bacteriol. 2007, 189, 7254–7261. [Google Scholar] [CrossRef]
- Carraro, N.; Libante, V.; Morel, C.; Decaris, B.; Charron-Bourgoin, F.; Leblond, P.; Guédon, G. Differential regulation of two closely related integrative and conjugative elements from Streptococcus thermophilus. BMC Microbiol. 2011, 11, 238. [Google Scholar] [CrossRef]
- Wawrzyniak, P.; Płucienniczak, G.; Bartosik, D. The different faces of rolling-circle replication and its multifunctional initiator proteins. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Lee, C.A.; Thomas, J.; Grossman, A.D. The Bacillus subtilis conjugative transposon ICEBs1 mobilizes plasmids lacking dedicated mobilization functions. J. Bacteriol. 2012, 194, 3165–3172. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.A.; Babic, A.; Grossman, A.D. Autonomous plasmid-like replication of a conjugative transposon. Mol. Microbiol. 2010, 75, 268–279. [Google Scholar] [CrossRef] [PubMed]
- Stapleton, P.; Pike, R.; Mullany, P.; Lucas, V.; Roberts, G.; Rowbury, R.; Wilson, M.; Richards, H. Mercuric resistance genes in gram-positive oral bacteria. FEMS Microbiol. Lett. 2004, 236, 213–220. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Islam, M.R.; Nagao, J.; Zendo, T.; Sonomoto, K. Antimicrobial mechanism of lantibiotics. Biochem. Soc. Trans. 2012, 40, 1528–1533. [Google Scholar] [CrossRef]
- Joakim Saris, P.E.; Immonen, T.; Reis, M.; Sahl, H.-G. Immunity to lantibiotics. Antonie Van Leeuwenhoek 1996, 69, 151–159. [Google Scholar] [CrossRef]
- Mielecki, D.; Wrzesiński, M.; Grzesiuk, E. Inducible repair of alkylated DNA in microorganisms. Mutat. Res. Rev. Mutat. Res. 2015, 763, 294–305. [Google Scholar] [CrossRef]
- Douard, G.; Praud, K.; Cloeckaert, A.; Doublet, B. The Salmonella genomic island 1 is specifically mobilized in trans by the IncA/C multidrug resistance plasmid family. PLoS ONE 2010, 5, e15302. [Google Scholar] [CrossRef]
- Kiss, J.; Papp, P.P.; Szabó, M.; Farkas, T.; Murányi, G.; Szakállas, E.; Olasz, F. The master regulator of IncA/C plasmids is recognized by the Salmonella genomic island SGI1 as a signal for excision and conjugal transfer. Nucleic Acids Res. 2015, 43, 8735–8745. [Google Scholar] [CrossRef]
- Carraro, N.; Matteau, D.; Luo, P.; Rodrigue, S.; Burrus, V. The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet. 2014, 10. [Google Scholar] [CrossRef]
- Harmer, C.J.; Hamadan, M.; Ambrose, S.J.; Hall, R.M. Destabilization of IncA and IncC plasmids by SGI1 and SGI2 type Salmonella genomic islands. Plasmid 2016, 87–88, 51–57. [Google Scholar] [CrossRef]
- Huguet, K.T.; Gonnet, M.; Doublet, B.; Cloeckaert, A. A toxin antitoxin system promotes the maintenance of the IncA/C-mobilizable Salmonella genomic island 1. Sci. Rep. 2016, 6, 32285. [Google Scholar] [CrossRef] [PubMed]
- Chaffanel, F.; Charron-Bourgoin, F.; Libante, V.; Leblond-Bourget, N.; Payot, S. Resistance genes and genetic elements associated with antibiotic resistance in clinical and commensal isolates of Streptococcus salivarius. Appl. Environ. Microbiol. 2015, 81, 4155–4163. [Google Scholar] [CrossRef] [PubMed]


| Bacterial Species | Strain Name (Name of the ICESt3-Related ICE 1, Which Hosts or Not an IME_oriT 1 Carried by the Strain) | Genotype 1 | Reference |
|---|---|---|---|
| Streptococcus salivarius | L11(ICE_SsalL11_fda ::IME_SsalL11_oriT) | Wild type strain carrying ICE_SsalL11_fda hosting IME_ SsalL11_oriT | [14] |
| S. salivarius | L25(ICE_SsalL25_fda ::IME_SsalL25_oriT) | Strain carrying ICE_SsalL25_fda labeled with a SpcR gene and hosting IME_ SsalL25_oriT labeled with a KanR gene | [14] and this work |
| S. salivarius | L45(ICE_SsalL45_rpsI ::IME_SsalL45_oriT) | Wild type strain carrying ICE_SsalL45_rpsI hosting IME_SsalL45_oriT | [14] |
| S. salivarius | F1-4(ICE_SsaF1-4_fda Δint) | Strain carrying ICE_SsaF1-4_fda with its integrase gene replaced by a CmR gene | [21] and this work |
| Streptococcus thermophilus | LMG18311(ICESt3) | Strain carrying ICESt3 integrated in fda and labeled with a CmR or SpcR gene | [22] |
| S. thermophilus | LMG18311(ICESt3Δint) | Strain carrying ICESt3 integrated in fda and labeled with a SpcR gene and with its integrase gene replaced by a CmR gene | This work |
| S. thermophilus | LMG18311(ICESt3Δrel) | Strain carrying ICESt3 integrated in fda and labeled with a CmR gene and deleted for its relaxase gene | [23] |
| S. thermophilus | LMG18311(ICESt3 ::IME_SsalL25_oriT) | Strain carrying ICESt3 integrated in fda and labeled with a CmR gene and hosting IME_SsalL25_oriT KanR | This work |
| S. thermophilus | LMG18311(ICESt3 ::IME_SsalL25_oriTΔrel) | Strain carrying ICESt3 integrated in fda and labeled with a CmR gene and hosting IME_SsalL25_oriT KanR deleted for its relaxase gene | This work |
| S. thermophilus | LMG18311(ICESt3 Δint::IME_SsalL25_oriT) | Strain carrying ICESt3 integrated in fda and SpcR with its integrase gene replaced by a CmR gene and hosting IME_SsalL25_oriT KanR | This work |
| S. thermophilus | LMG18311(ICESt3 Δrel::IME_SsalL25_oriT) | Strain carrying ICE_St3 integrated in fda and CmR deleted for its relaxase gene and hosting IME_SsalL25_oriT KanR | This work |
| S. thermophilus | LMG18311 ΔepsE | Strain deficient in exopolysaccharide production, EryR | [24] |
| S. thermophilus | LMG18311 ΔepsE(ICESt3Δint) | Strain deficient in exopolysaccharide production, EryR and carrying ICESt3 SpcR with its integrase gene replaced by a CmR gene | This work |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Libante, V.; Sarica, N.; Mohamad Ali, A.; Gapp, C.; Oussalah, A.; Guédon, G.; Leblond-Bourget, N.; Payot, S. Mobilization of IMEs Integrated in the oriT of ICEs Involves Their Own Relaxase Belonging to the Rep-Trans Family of Proteins. Genes 2020, 11, 1004. https://doi.org/10.3390/genes11091004
Libante V, Sarica N, Mohamad Ali A, Gapp C, Oussalah A, Guédon G, Leblond-Bourget N, Payot S. Mobilization of IMEs Integrated in the oriT of ICEs Involves Their Own Relaxase Belonging to the Rep-Trans Family of Proteins. Genes. 2020; 11(9):1004. https://doi.org/10.3390/genes11091004
Chicago/Turabian StyleLibante, Virginie, Nazim Sarica, Abbas Mohamad Ali, Chloé Gapp, Anissa Oussalah, Gérard Guédon, Nathalie Leblond-Bourget, and Sophie Payot. 2020. "Mobilization of IMEs Integrated in the oriT of ICEs Involves Their Own Relaxase Belonging to the Rep-Trans Family of Proteins" Genes 11, no. 9: 1004. https://doi.org/10.3390/genes11091004
APA StyleLibante, V., Sarica, N., Mohamad Ali, A., Gapp, C., Oussalah, A., Guédon, G., Leblond-Bourget, N., & Payot, S. (2020). Mobilization of IMEs Integrated in the oriT of ICEs Involves Their Own Relaxase Belonging to the Rep-Trans Family of Proteins. Genes, 11(9), 1004. https://doi.org/10.3390/genes11091004