Genome-Wide Analysis of the Role of NAC Family in Flower Development and Abiotic Stress Responses in Cleistogenes songorica
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of NAC Genes in C. songorica
2.2. Chromosome Localization, Gene Duplication and Syntenic Analysis
2.3. Protein Properties, Conserved Motif, Gene Structure and Phylogenetic Analysis
2.4. Gene Expression Analysis in Various Tissues and under Multiple Stress Treatments
2.5. Coexpression Network Construction and Gene Annotation Analysis
3. Results and Discussion
3.1. Identification of NAC TFs in C. songorica
3.2. Phylogenetic Relationship and Evolutionary Analysis of CsNAC and OsNAC TFs
3.3. CsNAC Gene Structures and Conserved Motifs
3.4. Genomic Locations and Duplication of the NAC Genes in C. songorica
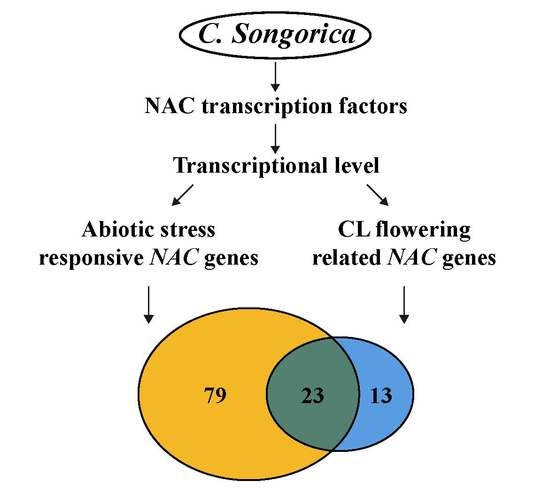
3.5. Expression Profiles and Coexpression Network Analysis of CL Flowering Related CsNAC Genes
3.6. Expression Profiles and Coexpression Network Analysis of Abiotic Stress-Related CsNAC Genes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mittler, R.; Blumwald, E. Genetic engineering for modern agriculture: Challenges and perspectives. Annu. Rev. Plant Biol. 2010, 61, 443–462. [Google Scholar] [CrossRef]
- Wang, H.; Miyazaki, S.; Kawai, K.; Deyholos, M.; Galbraith, D.W.; Bohnert, H.J. Temporal progression of gene expression responses to salt shock in maize roots. Plant Mol. Biol. 2003, 52, 873–891. [Google Scholar] [CrossRef] [PubMed]
- Luscombe, N.M.; Thornton, J.M. Protein–DNA interactions: Amino acid conservation and the effects of mutations on binding specificity. J. Mol. Biol. 2002, 320, 991–1009. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.-Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.; Samaha, R. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Souer, E.; van Houwelingen, A.; Kloos, D.; Mol, J.; Koes, R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef]
- Swati, P.; Pranav Pankaj, S.; Srivastava, P.S.; Manoj, P. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar]
- Seo, P.J.; Kim, S.G.; Park, C.M. Membrane-bound transcription factors in plants. Trends Plant Sci. 2008, 13, 550–556. [Google Scholar] [CrossRef]
- Min, X.; Jin, X.; Zhang, Z.; Wei, X.; Ndayambaza, B.; Wang, Y.; Liu, W. Genome-wideidentification of NAC transcription factor family and functional analysis of the abiotic stress-responsive genes in Medicago sativa L. J. Plant Growth Regul. 2020, 39, 324–337. [Google Scholar] [CrossRef]
- Sablowsk, R.W.M.; Meyerowitz, E.M. A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 1998, 92, 93–103. [Google Scholar] [CrossRef]
- Ren, T.; Wang, J.; Zhao, M.; Gong, X.; Wang, S.; Wang, G.; Zhou, C. Involvement of NAC transcription factor SiNAC1 in a positive feedback loop via ABA biosynthesis and leaf senescence in Foxtail millet. Planta 2018, 247, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Zhai, Q.; Deng, L.; Li, S.; Li, H.; Yan, L.; Huang, Z.; Wang, B.; Jiang, H.; Huang, T. Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell 2014, 26, 3167–3184. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Motomura, T.; Komeda, Y.; Saito, T.; Kato, A. Overexpression of the NAC transcription factor family gene ANAC036 results in a dwarf phenotype in Arabidopsis thaliana. J. Plant Physiol. 2010, 167, 571–577. [Google Scholar] [CrossRef]
- Kim, Y.S.; Kim, S.G.; Park, J.E.; Park, H.Y.; Lim, M.H.; Chua, N.H.; Park, C.M. A membrane-mound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 2006, 18, 3132–3144. [Google Scholar] [CrossRef]
- Kim, J.H.; Woo, H.R.; Kim, J.; Lim, P.O.; Lee, I.C.; Choi, S.H.; Hwang, D.; Nam, H.G. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 2009, 323, 1053–1057. [Google Scholar] [CrossRef]
- Zhong, R.; Lee, C.; Ye, Z. Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci. 2010, 15, 625–631. [Google Scholar] [CrossRef]
- Zhang, L.; Yao, L.; Zhang, N.; Yang, J.; Zhu, X.; Tang, X.; Calderón-Urrea, A.; Si, H. Lateral root development in potato is mediated by stu-mi164 regulation of NAC transcription factor. Front. Plant Sci. 2018, 9, 383. [Google Scholar] [CrossRef]
- Kim, S.G.; Lee, A.K.; Yoon, H.K.; Park, C.M. A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. Plant J. 2008, 55, 77–88. [Google Scholar] [CrossRef]
- Zhang, Z.; Dong, J.; Ji, C.; Wu, Y.; Messing, J. NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. Proc. Natl. Acad. Sci. USA 2019, 116, 11223–11228. [Google Scholar] [CrossRef]
- Shan, W.; Kuang, J.; Chen, L.; Xie, H.; Peng, H.; Xiao, Y.; Li, X.; Chen, W.; He, Q.; Chen, J. Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. J. Exp. Bot. 2012, 63, 5171–5187. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Derkx, A.; Liu, D.; Buchner, P.; Hawkesford, M. Overexpression of a NAC transcription factor delays leaf senescence and increases grain nitrogen concentration in wheat. Plant Biol. 2015, 17, 904–913. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.D.; Seo, P.J.; Yoon, H.K.; Park, C.M. The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 2011, 23, 2155–2168. [Google Scholar] [CrossRef] [PubMed]
- Block, A.; Toruño, T.Y.; Elowsky, C.G.; Zhang, C.; Steinbrenner, J.; Beynon, J.; Alfano, J.R. The Pseudomonas syringae type III effector HopD1 suppresses effector-triggered immunity, localizes to the endoplasmic reticulum, and targets the Arabidopsis transcription factor NTL9. New Phytol. 2014, 201, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Zhang, H.; Huang, L.; Li, D.; Song, F. Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front. Plant Sci. 2016, 7, 4. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, H.; Cai, J.; Bi, Y.; Li, D.; Song, F. Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol. 2019, 19, 278. [Google Scholar] [CrossRef]
- Qu, Y.; Duan, M.; Zhang, Z.; Dong, J.; Wang, T. Overexpression of the Medicago falcata NAC transcription factor MfNAC3 enhances cold tolerance in Medicago truncatula. Environ. Exp. Bot. 2016, 129, 67–76. [Google Scholar] [CrossRef]
- Fang, Y.; Liao, K.; Du, H.; Xu, Y.; Song, H.; Li, X.; Xiong, L. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 2015, 66, 6803–6817. [Google Scholar] [CrossRef]
- Hao, Y.; Wei, W.; Song, Q.; Chen, H.; Zhang, Y.; Wang, F.; Zou, H.; Lei, G.; Tian, A.; Zhang, W. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011, 68, 302–313. [Google Scholar] [CrossRef]
- Wang, B.; Wei, J.; Song, N.; Wang, N.; Zhao, J.; Kang, Z. A novel wheat NAC transcription factor, TaNAC30, negatively regulates resistance of wheat to stripe rust. J. Integr. Plant Biol. 2018, 60, 432–443. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Wei, X.; Tai, J.; Jia, C.; Hu, X.; Trethewey, J.A.K. Planting density and irrigation timing affects seed yield sustainability. Agron. J. 2014, 106, 1690–1696. [Google Scholar] [CrossRef]
- Yan, Q.; Wu, F.; Yan, Z.; Li, J.; Ma, T.; Zhang, Y.; Zhao, Y.; Wang, Y.; Zhang, J. Differential co-expression networks of long non-coding RNAs and mRNAs in Cleistogenes songorica under water stress and during recovery. BMC Plant Biol. 2019, 19, 23. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Wu, F.; Ma, T.; Yan, Q.; Zong, X.; Li, J.; Zhao, Y.; Kanzana, G.; Zhang, J. Analysis of six transcription factor families explores transcript divergence of cleistogamous and chasmogamous flowers in Cleistogenes songorica. DNA Cell Biol. 2020, 39, 273–288. [Google Scholar] [CrossRef]
- Zhang, J.; Duan, Z.; Jahufer, Z.; An, S.; Wang, Y. Stress-inducible expression of a Cleistogenes songorica ALDH gene enhanced drought tolerance in transgenic Arabislopsis thaliana. Plant Omics 2014, 7, 438–444. [Google Scholar]
- Duan, Z.; Zhang, D.; Zhang, J.; Di, H.; Wu, F.; Hu, X.; Meng, X.; Luo, K.; Zhang, J.; Wang, Y. Co-transforming bar and CsALDH genes enhanced resistance to herbicide and drought and salt stress in transgenic alfalfa (Medicago sativa L.). Front. Plant Sci. 2015, 6, 1115. [Google Scholar] [CrossRef]
- Wu, F.; Zhang, D.; Muvunyi, B.P.; Yan, Q.; Zhang, Y.; Yan, Z.; Cao, M.; Wang, Y.; Zhang, J. Analysis of microRNA reveals cleistogamous and chasmogamous floret divergence in dimorphic plant. Sci. Rep. 2018, 8, 6287. [Google Scholar] [CrossRef]
- Yan, Q.; Wu, F.; Ma, T.; Zong, X.; Ma, Q.; Li, J.; Zhao, Y.; Wang, Y.; Zhang, J. Comprehensive analysis of bZIP transcription factors uncovers their roles during dimorphic floret differentiation and stress response in Cleistogenes songorica. BMC Genom. 2019, 20, 760. [Google Scholar] [CrossRef]
- Muvunyi, B.P.; Yan, Q.; Wu, F.; Min, X.; Yan, Z.; Kanzana, G.; Wang, Y.; Zhang, J. Mining late embryogenesis abundant (LEA) family genes in Cleistogenes songorica, a xerophyte perennial desert plant. Int. J. Mol. Sci. 2018, 19, 3430. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef]
- Nishiyama, R.; Watanabe, Y.; Tran, L.-S.P.; Le, D.T.; Shinozaki, K.; Mochida, K.; Yamaguchi-Shinozaki, K. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 2011, 18, 263–276. [Google Scholar]
- Wang, N.; Zheng, Y.; Xin, H.; Fang, L.; Li, S. Comprehensive analysis of NAC domain transcription factor gene family in Vitis vinifera. Plant Cell Rep. 2013, 32, 61–75. [Google Scholar] [CrossRef]
- Zhu, G.; Chen, G.; Zhu, J.; Zhu, Y.; Lu, X.; Li, X.; Hu, Y.; Yan, Y. Molecular characterization and expression profiling of NAC transcription factors in Brachypodium distachyon L. PLoS ONE 2015, 10, e0139794. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; You, J.; Xie, K.; Xie, W.; Xiong, L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genom. 2008, 280, 547–563. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Zhu, W.; Silva, J.C.; Gu, X.; Buell, C.R. Intron gain and loss in segmentally duplicated genes in rice. Genome Biol. 2006, 7, R41. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Qi, G.; Kong, Y.; Kong, D.; Gao, Q.; Zhou, G. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 2010, 10, 145. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Song, Q.; Chen, H.; Zou, H.; Wei, W.; Kang, X.; Ma, B.; Zhang, W.; Zhang, J.; Chen, S. Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta 2010, 232, 1033–1043. [Google Scholar] [CrossRef]
- Liu, M.; Ma, Z.; Sun, W.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide analysis of the NAC transcription factor family in tartary buckwheat (Fagopyrum tataricum). BMC Genom. 2019, 20, 113. [Google Scholar] [CrossRef]
- Thiel, T.; Graner, A.; Waugh, R.; Grosse, I.; Close, T.J.; Stein, N. Evidence and evolutionary analysis of ancient whole-genome duplication in barley predating the divergence from rice. BMC Evol. Biol. 2009, 9, 209. [Google Scholar] [CrossRef]
- Fan, K.; Li, F.; Chen, J.; Li, Z.; Lin, W.; Cai, S.; Liu, J.; Lin, W. Asymmetric evolution and expansion of the NAC transcription factor in polyploidized cotton. Front. Plant Sci. 2018, 9, 47. [Google Scholar] [CrossRef]
- Gong, X.; Zhao, L.; Song, X.; Lin, Z.; Gu, B.; Yan, J.; Zhang, S.; Tao, S.; Huang, X. Genome-wide analyses and expression patterns under abiotic stress of NAC transcription factors in white pear (Pyrus bretschneideri). BMC Plant Biol. 2019, 19, 161. [Google Scholar] [CrossRef]
- Culley, T.M.; Klooster, M.R. The cleistogamous breeding system: A review of its frequency, evolution, and ecology in angiosperms. Bot. Rev. 2007, 73, 1–30. [Google Scholar] [CrossRef]
- Shibuya, K.; Shimizu, K.; Niki, T.; Ichimura, K. Identification of a NAC transcription factor, EPHEMERAL 1, that controls petal senescence in Japanese morning glory. Plant J. 2014, 79, 1044–1051. [Google Scholar] [CrossRef] [PubMed]
- Hendelman, A.; Stav, R.; Zemach, H.; Arazi, T. The tomato NAC transcription factor SlNAM2 is involved in flower-boundary morphogenesis. J. Exp. Bot. 2013, 64, 5497–5507. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Kim, S.Y.; Park, C.M. A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Planta 2007, 226, 647–654. [Google Scholar] [CrossRef]
- Luo, Y.; Hu, J.; Li, L.; Luo, Y.; Wang, P.; Song, B. Genome-wide analysis of gene expression reveals gene regulatory networks that regulate chasmogamous and cleistogamous flowering in Pseudostellaria heterophylla (Caryophyllaceae). BMC Genom. 2016, 17, 382. [Google Scholar] [CrossRef]









© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zong, X.; Yan, Q.; Wu, F.; Ma, Q.; Zhang, J. Genome-Wide Analysis of the Role of NAC Family in Flower Development and Abiotic Stress Responses in Cleistogenes songorica. Genes 2020, 11, 927. https://doi.org/10.3390/genes11080927
Zong X, Yan Q, Wu F, Ma Q, Zhang J. Genome-Wide Analysis of the Role of NAC Family in Flower Development and Abiotic Stress Responses in Cleistogenes songorica. Genes. 2020; 11(8):927. https://doi.org/10.3390/genes11080927
Chicago/Turabian StyleZong, Xifang, Qi Yan, Fan Wu, Qian Ma, and Jiyu Zhang. 2020. "Genome-Wide Analysis of the Role of NAC Family in Flower Development and Abiotic Stress Responses in Cleistogenes songorica" Genes 11, no. 8: 927. https://doi.org/10.3390/genes11080927
APA StyleZong, X., Yan, Q., Wu, F., Ma, Q., & Zhang, J. (2020). Genome-Wide Analysis of the Role of NAC Family in Flower Development and Abiotic Stress Responses in Cleistogenes songorica. Genes, 11(8), 927. https://doi.org/10.3390/genes11080927