DNA6mA-MINT: DNA-6mA Modification Identification Neural Tool
Abstract
1. Introduction
2. Benchmark Dataset
3. Methodology
4. Figure of Merits
5. Results and Discussion
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Greer, E.L.; Blanco, M.A.; Gu, L.; Sendinc, E.; Liu, J.; Aristizábal-Corrales, D.; Hsu, C.H.; Aravind, L.; He, C.; Shi, Y. DNA methylation on N6-adenine in C. elegans. Cell 2015, 161, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Huang, H.; Liu, D.; Cheng, Y.; Liu, X.; Zhang, W.; Yin, R.; Zhang, D.; Zhang, P.; Liu, J.; et al. N6-methyladenine DNA modification in Drosophila. Cell 2015, 161, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.Z.; He, C. DNA N6-methyladenine in metazoans: Functional epigenetic mark or bystander? Nat. Struct. Mol. Biol. 2017, 24, 503–506. [Google Scholar] [CrossRef]
- Campbell, J.L.; Kleckner, N. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 1990, 62, 967–979. [Google Scholar] [CrossRef]
- Pukkila, P.J.; Peterson, J.; Herman, G.; Modrich, P.; Meselson, M. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics 1983, 104, 571–582. [Google Scholar] [PubMed]
- Robbins-Manke, J.L.; Zdraveski, Z.Z.; Marinus, M.; Essigmann, J.M. Analysis of global gene expression and double-strand-break formation in DNA adenine methyltransferase-and mismatch repair-deficient Escherichia coli. J. Bacteriol. 2005, 187, 7027–7037. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.L.; Zhu, S.; He, M.; Chen, D.; Zhang, Q.; Chen, Y.; Yu, G.; Liu, J.; Xie, S.Q.; Luo, F.; et al. N6-methyladenine DNA modification in the human genome. Mol. Cell 2018, 71, 306–318. [Google Scholar] [CrossRef]
- Liu, X.; Lai, W.; Li, Y.; Chen, S.; Liu, B.; Zhang, N.; Mo, J.; Lyu, C.; Zheng, J.; Du, Y.R.; et al. N6-methyladenine is incorporated into mammalian genome by DNA polymerase. Cell Res. 2020. [Google Scholar] [CrossRef]
- Dunn, D.; Smith, J. Occurrence of a new base in the deoxyribonucleic acid of a strain of Bacterium coli. Nature 1955, 175, 336–337. [Google Scholar] [CrossRef]
- Bird, A.P. Use of restriction enzymes to study eukaryotic DNA methylation: II. The symmetry of methylated sites supports semi-conservative copying of the methylation pattern. J. Mol. Biol. 1978, 118, 49–60. [Google Scholar] [CrossRef]
- Flusberg, B.A.; Webster, D.R.; Lee, J.H.; Travers, K.J.; Olivares, E.C.; Clark, T.A.; Korlach, J.; Turner, S.W. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 2010, 7, 461. [Google Scholar] [CrossRef] [PubMed]
- Pomraning, K.R.; Smith, K.M.; Freitag, M. Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods 2009, 47, 142–150. [Google Scholar] [CrossRef]
- Liu, B.; Liu, X.; Lai, W.; Wang, H. Metabolically generated stable isotope-labeled deoxynucleoside code for tracing DNA N6-Methyladenine in human cells. Anal. Chem. 2017, 89, 6202–6209. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Luo, G.Z.; Chen, K.; Deng, X.; Yu, M.; Han, D.; Hao, Z.; Liu, J.; Lu, X.; Doré, L.C.; et al. N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell 2015, 161, 879–892. [Google Scholar] [CrossRef]
- Mondo, S.J.; Dannebaum, R.O.; Kuo, R.C.; Louie, K.B.; Bewick, A.J.; LaButti, K.; Haridas, S.; Kuo, A.; Salamov, A.; Ahrendt, S.R.; et al. Widespread adenine N6-methylation of active genes in fungi. Nat. Genet. 2017, 49, 964–968. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Wang, C.; Liu, H.; Zhou, Q.; Liu, Q.; Guo, Y.; Peng, T.; Song, J.; Zhang, J.; Chen, L.; et al. Identification and analysis of adenine N6-methylation sites in the rice genome. Nat. Plants 2018, 4, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Feng, P.; Yang, H.; Ding, H.; Lin, H.; Chen, W.; Chou, K.C. iDNA6mA-PseKNC: Identifying DNA N6-methyladenosine sites by incorporating nucleotide physicochemical properties into PseKNC. Genomics 2019, 111, 96–102. [Google Scholar] [CrossRef]
- Liu, Z.; Dong, W.; Jiang, W.; He, Z. csDMA: An improved bioinformatics tool for identifying DNA 6mA modifications via Chou’s 5-step rule. Sci. Rep. 2019, 9, 1–9. [Google Scholar]
- Xu, H.; Hu, R.; Jia, P.; Zhao, Z. 6mA-Finder: A novel online tool for predicting DNA N6-methyladenine sites in genomes. Bioinformatics 2020, 36, 3257–3259. [Google Scholar] [CrossRef]
- Chen, W.; Lv, H.; Nie, F.; Lin, H. i6mA-Pred: Identifying DNA N6-methyladenine sites in the rice genome. Bioinformatics 2019, 35, 2796–2800. [Google Scholar] [CrossRef]
- Rahman, M.K. FastFeatGen: Faster Parallel Feature Extraction from Genome Sequences and Efficient Prediction of DNA N6-Methyladenine Sites. In Proceedings of the International Conference on Computational Advances in Bio and Medical Sciences, Miami, FL, USA, 15–17 November 2019; pp. 52–64. [Google Scholar]
- Kong, L.; Zhang, L. i6mA-DNCP: Computational identification of DNA N6-methyladenine sites in the rice genome using optimized dinucleotide-based features. Genes 2019, 10, 828. [Google Scholar] [CrossRef] [PubMed]
- Rehman, M.U.; Khan, S.H.; Abbas, Z.; Danish Rizvi, S.M. Classification of Diabetic Retinopathy Images Based on Customised CNN Architecture. In Proceedings of the 2019 Amity International Conference on Artificial Intelligence (AICAI), Dubai, UAE, 4–6 February 2019; pp. 244–248. [Google Scholar]
- Rehman, M.U.; Khan, S.H.; Rizvi, S.D.; Abbas, Z.; Zafar, A. Classification of skin lesion by interference of segmentation and convolotion neural network. In Proceedings of the 2018 2nd International Conference on Engineering Innovation (ICEI), Bangkok, Thailand, 5–6 July 2018; pp. 81–85. [Google Scholar]
- Mahmoudi, O.; Wahab, A.; Chong, K.T. iMethyl-Deep: N6 methyladenosine identification of yeast genome with automatic feature extraction technique by using deep learning algorithm. Genes 2020, 11, 529. [Google Scholar] [CrossRef] [PubMed]
- Wahab, A.; Mahmoudi, O.; Kim, J.; Chong, K.T. DNC4mC-Deep: Identification and analysis of DNA N4-methylcytosine sites based on different encoding schemes by using deep learning. Cells 2020, 9, 1756. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Wahab, A.; Nazari, I.; Ryu, J.H.; Chong, K.T. i6mA-DNC: Prediction of DNA N6-Methyladenosine sites in rice genome based on dinucleotide representation using deep learning. Chemom. Intell. Lab. Syst. 2020, 204, 104102. [Google Scholar] [CrossRef]
- Wahab, A.; Ali, S.D.; Tayara, H.; Chong, K.T. iIM-CNN: Intelligent identifier of 6mA sites on different species by using convolution neural network. IEEE Access 2019, 7, 178577–178583. [Google Scholar] [CrossRef]
- Chou, K.C. Some remarks on protein attribute prediction and pseudo amino acid composition. J. Theor. Biol. 2011, 273, 236–247. [Google Scholar] [CrossRef]
- Chou, K.C. Advances in predicting subcellular localization of multi-label proteins and its implication for developing multi-target drugs. Curr. Med. Chem. 2019, 26, 4918–4943. [Google Scholar] [CrossRef]
- Ye, P.; Luan, Y.; Chen, K.; Liu, Y.; Xiao, C.; Xie, Z. MethSMRT: An integrative database for DNA N6-methyladenine and N4-methylcytosine generated by single-molecular real-time sequencing. Nucleic Acids Res. 2016, gkw950. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Chollet, F.; Keras Special Interest Group. Keras: Deep Learning Library for Theano and Tensorflow. 2015, Volume 7. p. T1. Available online: https://keras.io/ (accessed on 1 August 2020).
- De Boer, P.T.; Kroese, D.P.; Mannor, S.; Rubinstein, R.Y. A tutorial on the cross-entropy method. Ann. Oper. Res. 2005, 134, 19–67. [Google Scholar] [CrossRef]
- Bottou, L.; Bousquet, O. The tradeoffs of large scale learning. In Advances in Neural Information Processing Systems; Neural Information Processing Systems Foundation (NIPS): Vancouver, BC, Canada, 2008; pp. 161–168. [Google Scholar]
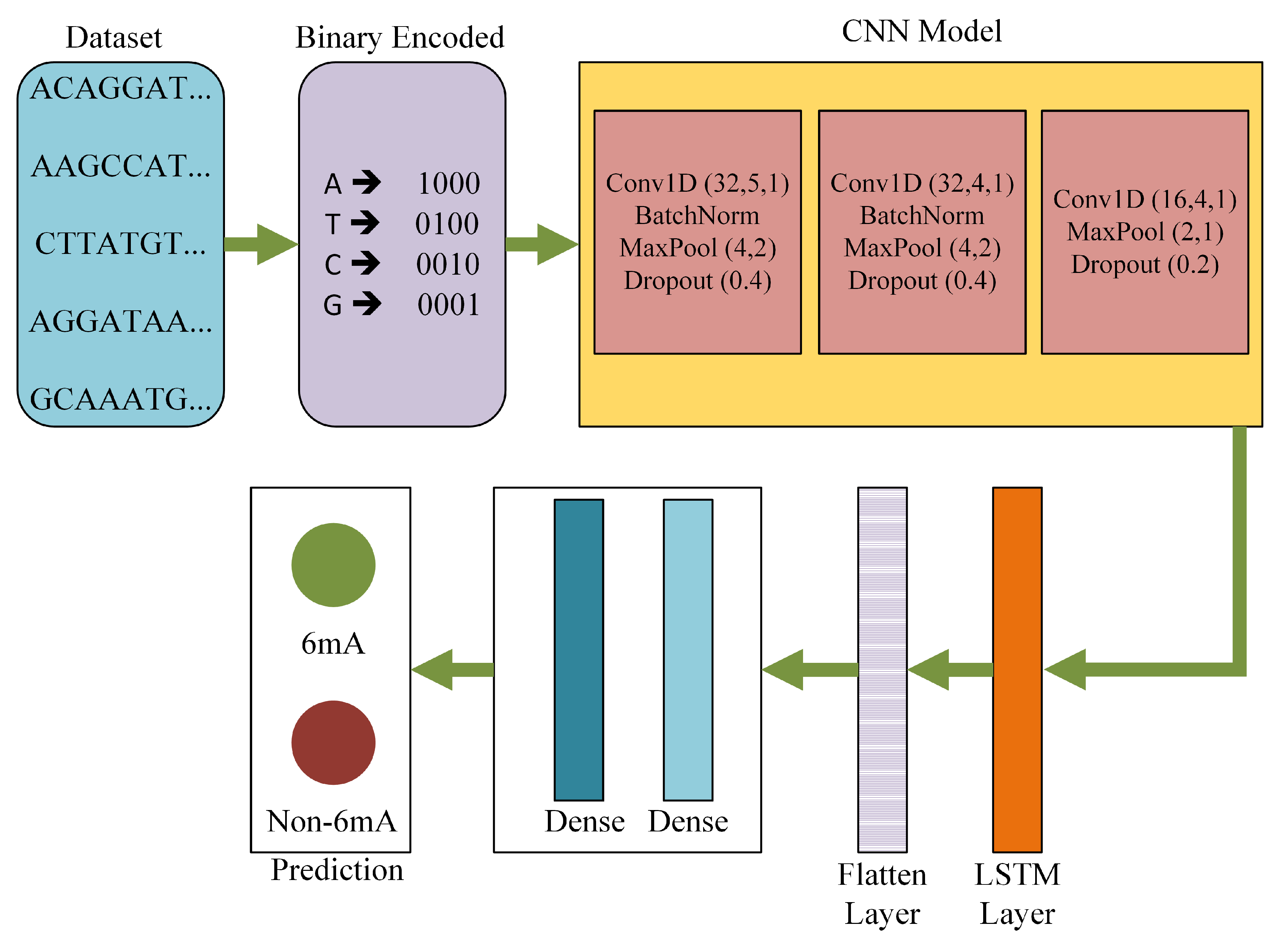
| Layer | Output Shape | Number of Parameters |
|---|---|---|
| Input | (41,4) | - |
| Conv1D (32,5,1) | (37,32) | 672 |
| Batch Normalization | (37,32) | 128 |
| Max Pooling (4,2) | (17,32) | 0 |
| Dropout (0.4) | (17,32) | 0 |
| Conv1D (32,4,1) | (14,32) | 4128 |
| Batch Normalization | (14,32) | 128 |
| Max Pooling (4,2) | (6,32) | 0 |
| Dropout (0.4) | (6,32) | 0 |
| Conv1D (16,4,1) | (3,16) | 2064 |
| Max Pooling (2,1) | (1,16) | 0 |
| Dropout (0.2) | (1,16) | 0 |
| LSTM | (1,4) | 336 |
| Flatten | 4 | 0 |
| Dense | 32 | 160 |
| Dense | 1 | 33 |
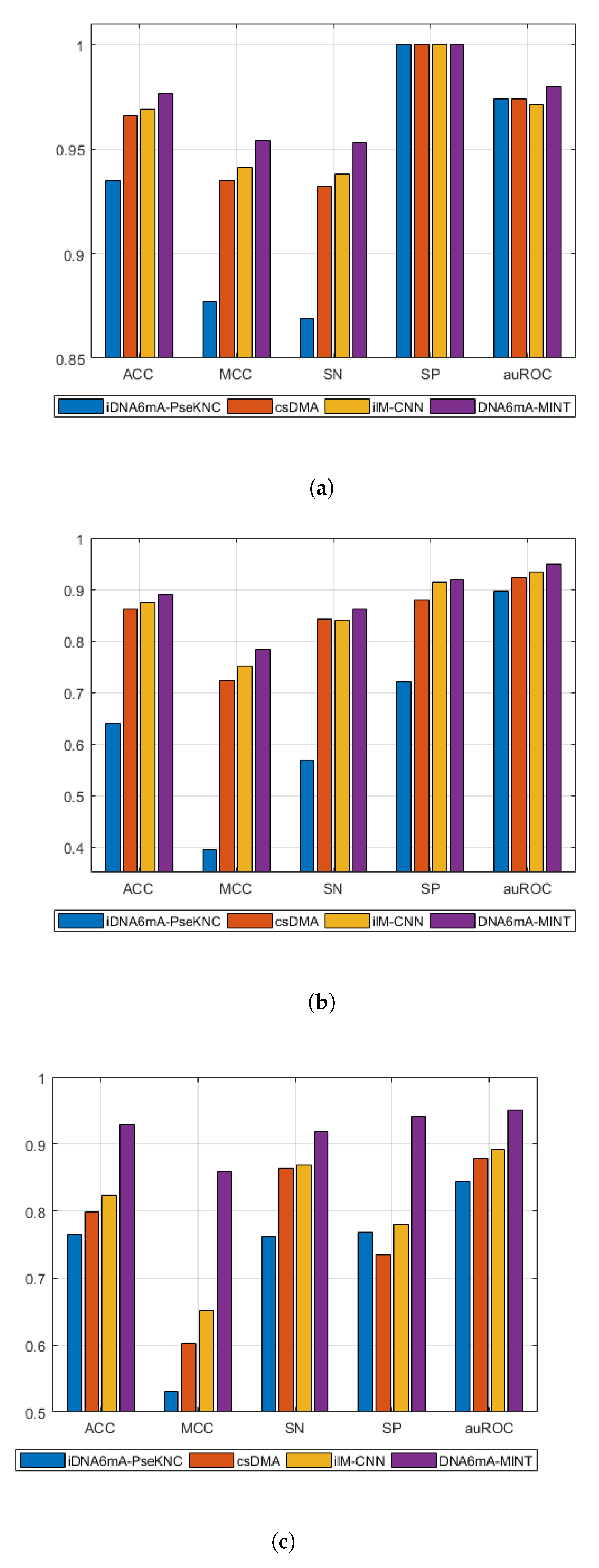
| Model | Species | Folds | SN | SP | ACC | MCC | auROC |
|---|---|---|---|---|---|---|---|
| iDNA6mA-PseKNC | M. musculus | 5 | 0.869 | 1 | 0.935 | 0.877 | 0.974 |
| Rice | 5 | 0.569 | 0.721 | 0.641 | 0.394 | 0.896 | |
| Combined-species | 5 | 0.762 | 0.769 | 0.765 | 0.531 | 0.844 | |
| csDMA | M. musculus | 5 | 0.932 | 1 | 0.966 | 0.935 | 0.974 |
| Rice | 5 | 0.842 | 0.880 | 0.861 | 0.723 | 0.923 | |
| Combined-species | 5 | 0.863 | 0.735 | 0.799 | 0.603 | 0.879 | |
| ilM-CNN | M. musculus | 5 | 0.938 | 1 | 0.969 | 0.941 | 0.971 |
| Rice | 5 | 0.841 | 0.914 | 0.875 | 0.752 | 0.934 | |
| Combined-species | 5 | 0.869 | 0.780 | 0.824 | 0.651 | 0.892 | |
| 6mA-Finder | M. musculus | 10 | 0.9349 | 1 | 0.9674 | 0.935 | 0.9954 |
| Rice | 10 | - | - | - | - | 0.9394 | |
| Combined-species | 10 | - | - | - | - | 0.9207 | |
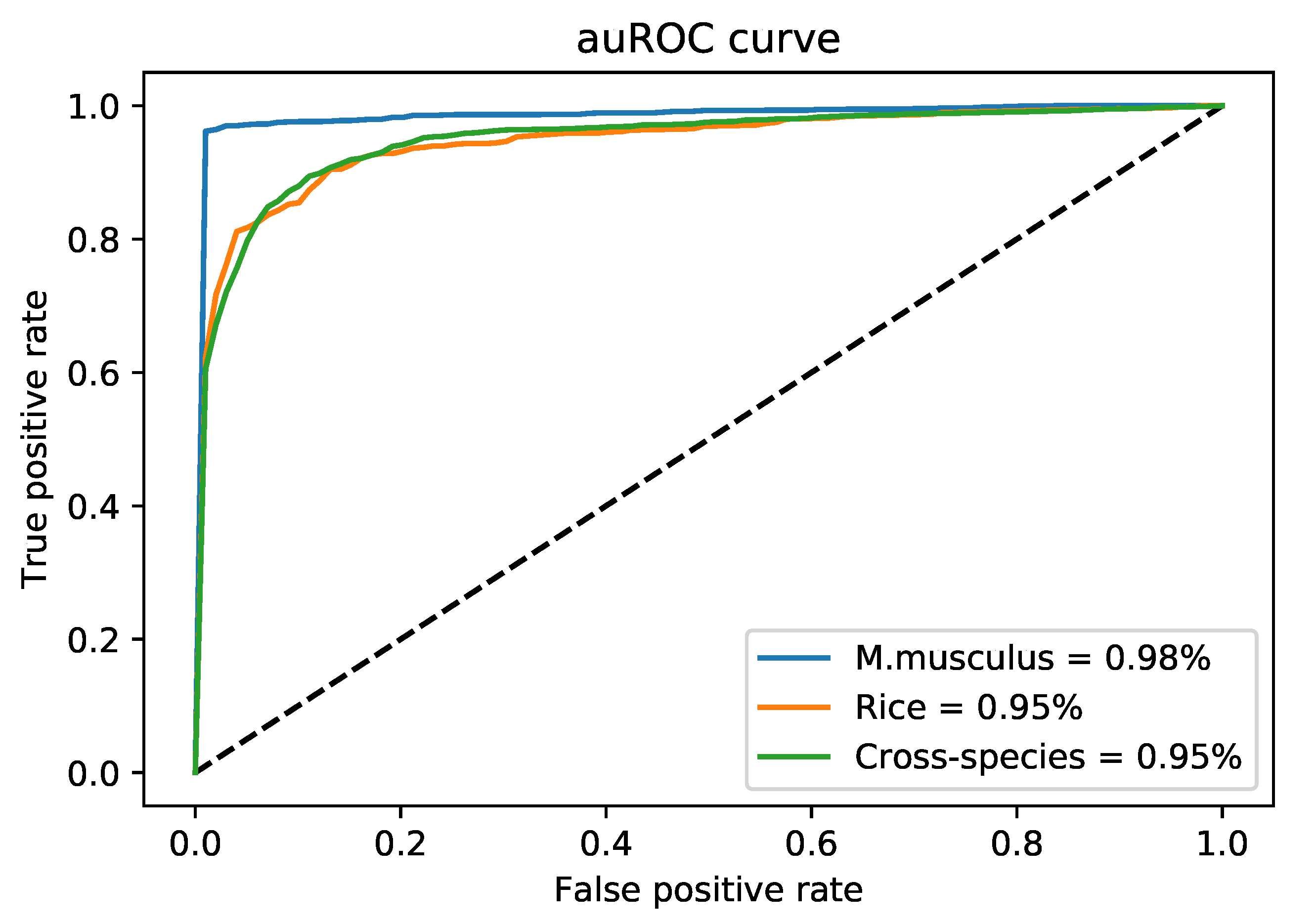
| DNA6mA-MINT | M. musculus | 5 | 0.9531 | 1 | 0.9766 | 0.9543 | 0.980 |
| Rice | 5 | 0.8621 | 0.9195 | 0.8908 | 0.7829 | 0.950 | |
| Combined-species | 5 | 0.9182 | 0.9409 | 0.9295 | 0.8593 | 0.950 | |
| DNA6mA-MINT | M. musculus | 10 | 0.9427 | 1 | 0.9714 | 0.9444 | 0.98 |
| Rice | 10 | 0.9425 | 0.908 | 0.9253 | 0.8511 | 0.950 | |
| Combined-species | 10 | 0.9318 | 0.9321 | 0.932 | 0.8639 | 0.960 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rehman, M.U.; Chong, K.T. DNA6mA-MINT: DNA-6mA Modification Identification Neural Tool. Genes 2020, 11, 898. https://doi.org/10.3390/genes11080898
Rehman MU, Chong KT. DNA6mA-MINT: DNA-6mA Modification Identification Neural Tool. Genes. 2020; 11(8):898. https://doi.org/10.3390/genes11080898
Chicago/Turabian StyleRehman, Mobeen Ur, and Kil To Chong. 2020. "DNA6mA-MINT: DNA-6mA Modification Identification Neural Tool" Genes 11, no. 8: 898. https://doi.org/10.3390/genes11080898
APA StyleRehman, M. U., & Chong, K. T. (2020). DNA6mA-MINT: DNA-6mA Modification Identification Neural Tool. Genes, 11(8), 898. https://doi.org/10.3390/genes11080898






