Lights and Shadows of TORCH Infection Proteomics
Abstract
1. Background
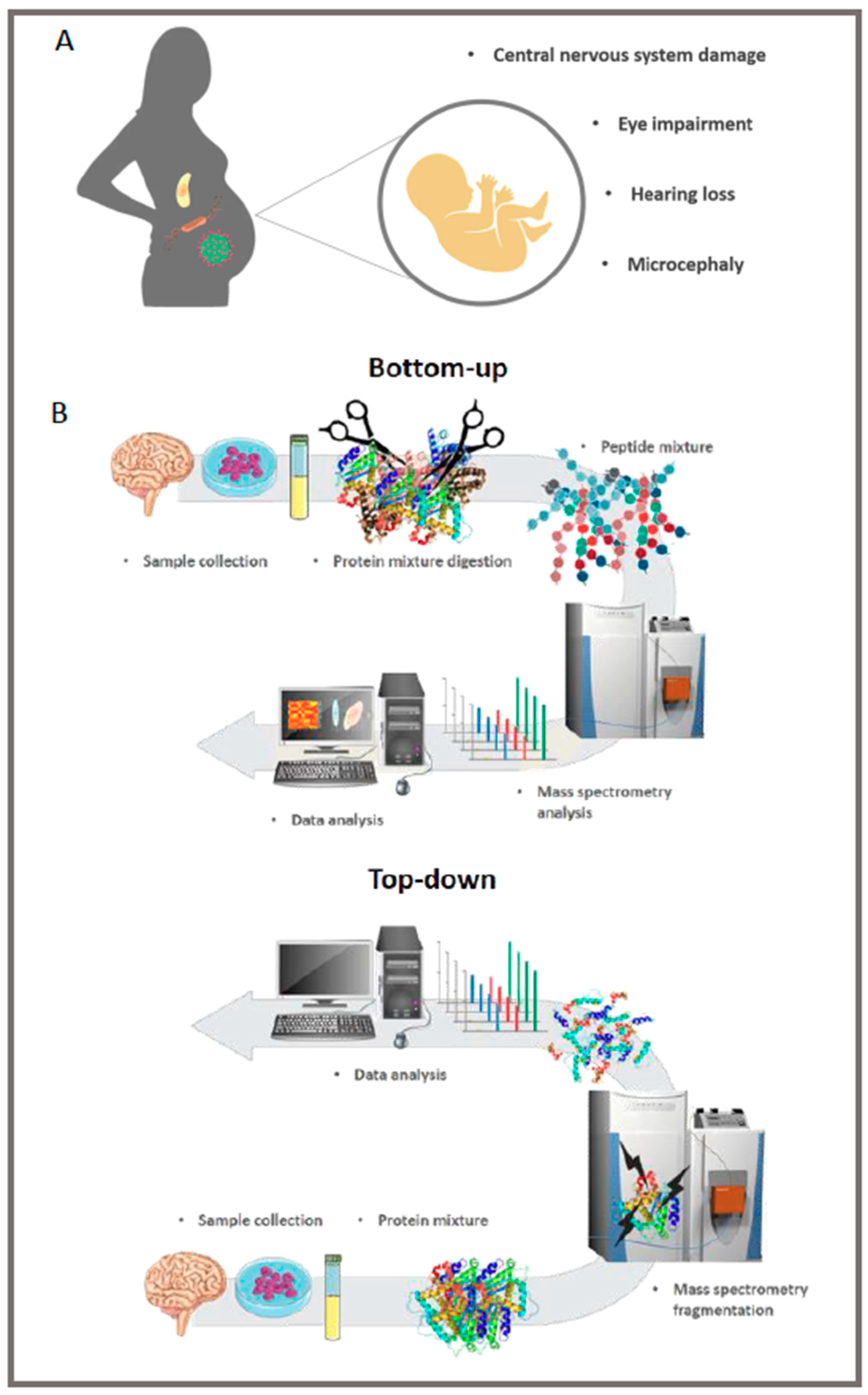
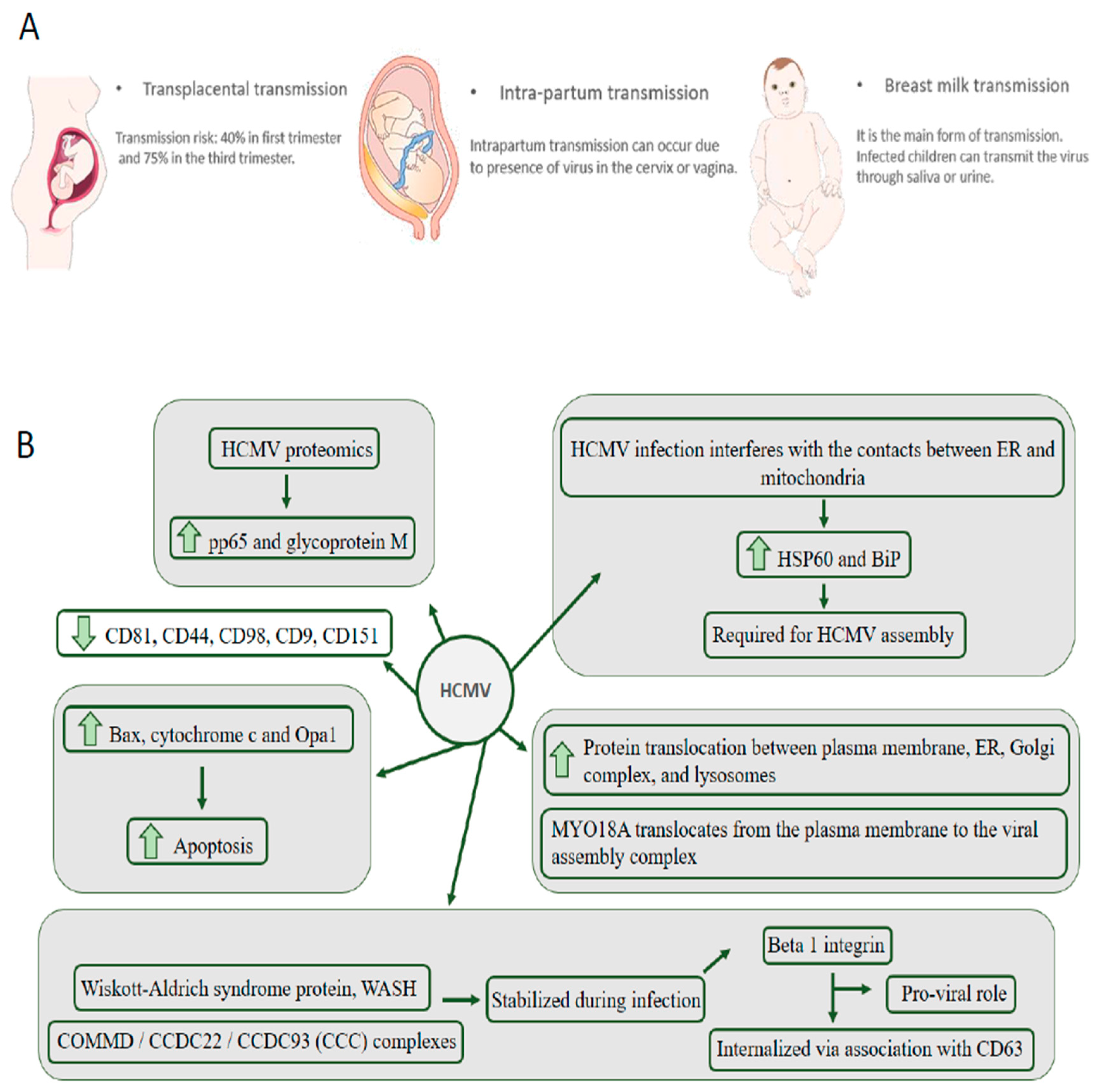
2. HCMV Is the Leading Cause of Congenital Neurological Disease by Transmission through the Placenta from the Mother to the Child
3. ZIKV: In 2015, the World Health Organization Reported Cases of Neurological Disorders in Infants Who Had Their Mothers Exposed to the Virus during Pregnancy
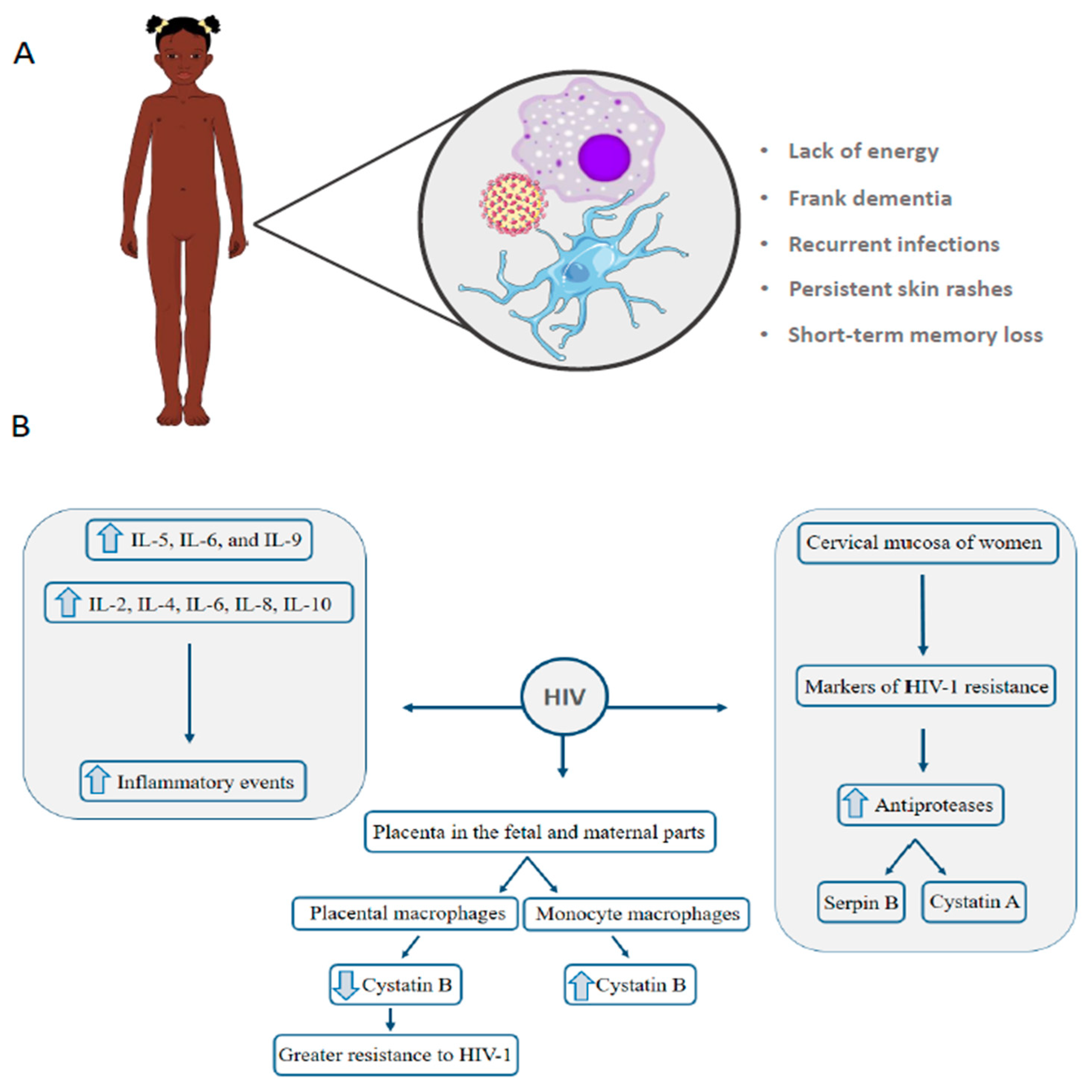
4. HIV: Vertical Transmission Is the Leading Cause of Infection in Children under 13 Years
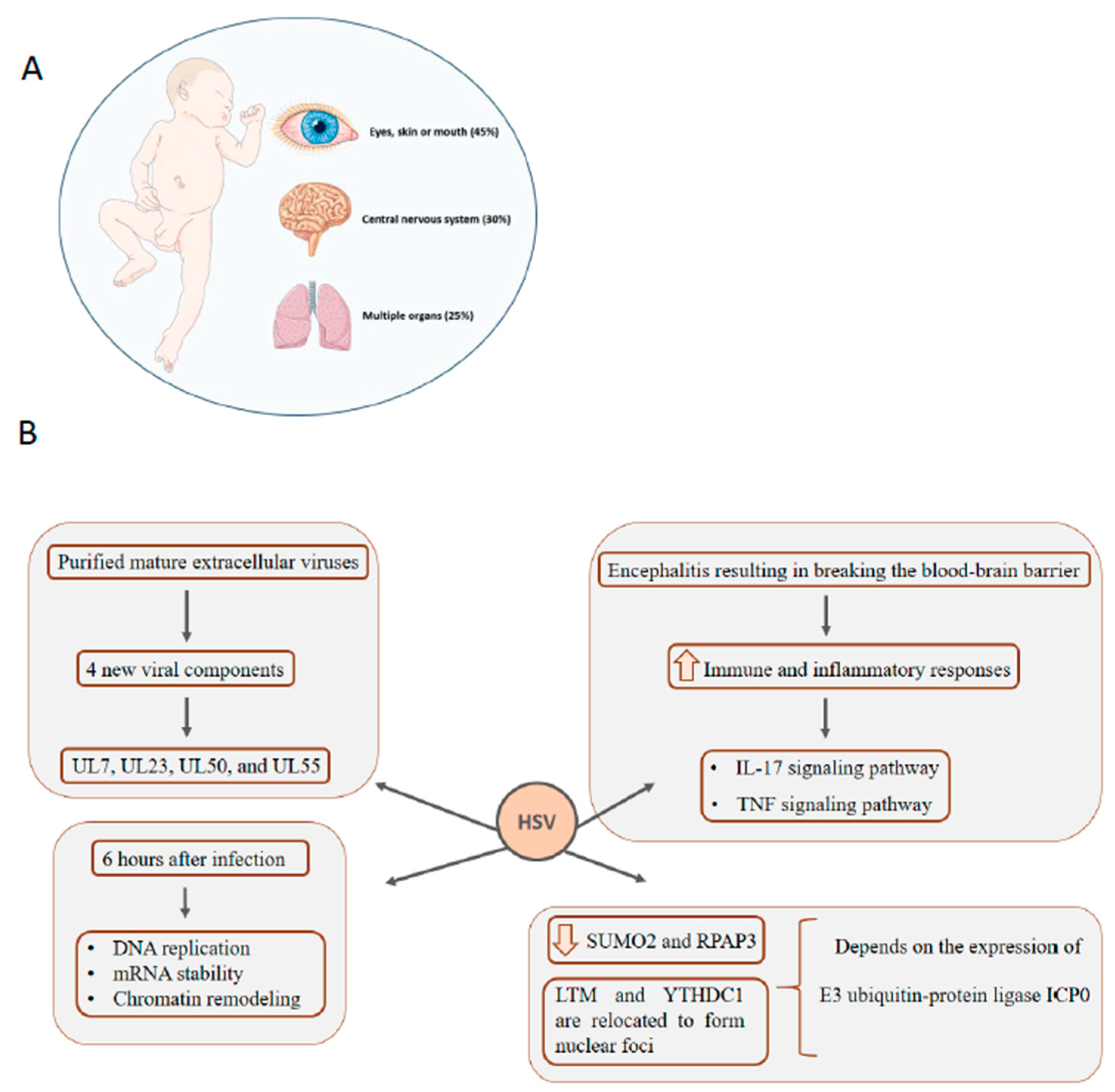
5. HSV: Infection in Newborns Can Affect Multiple Organs, Central Nervous System, Eyes, Skin, and Mouth
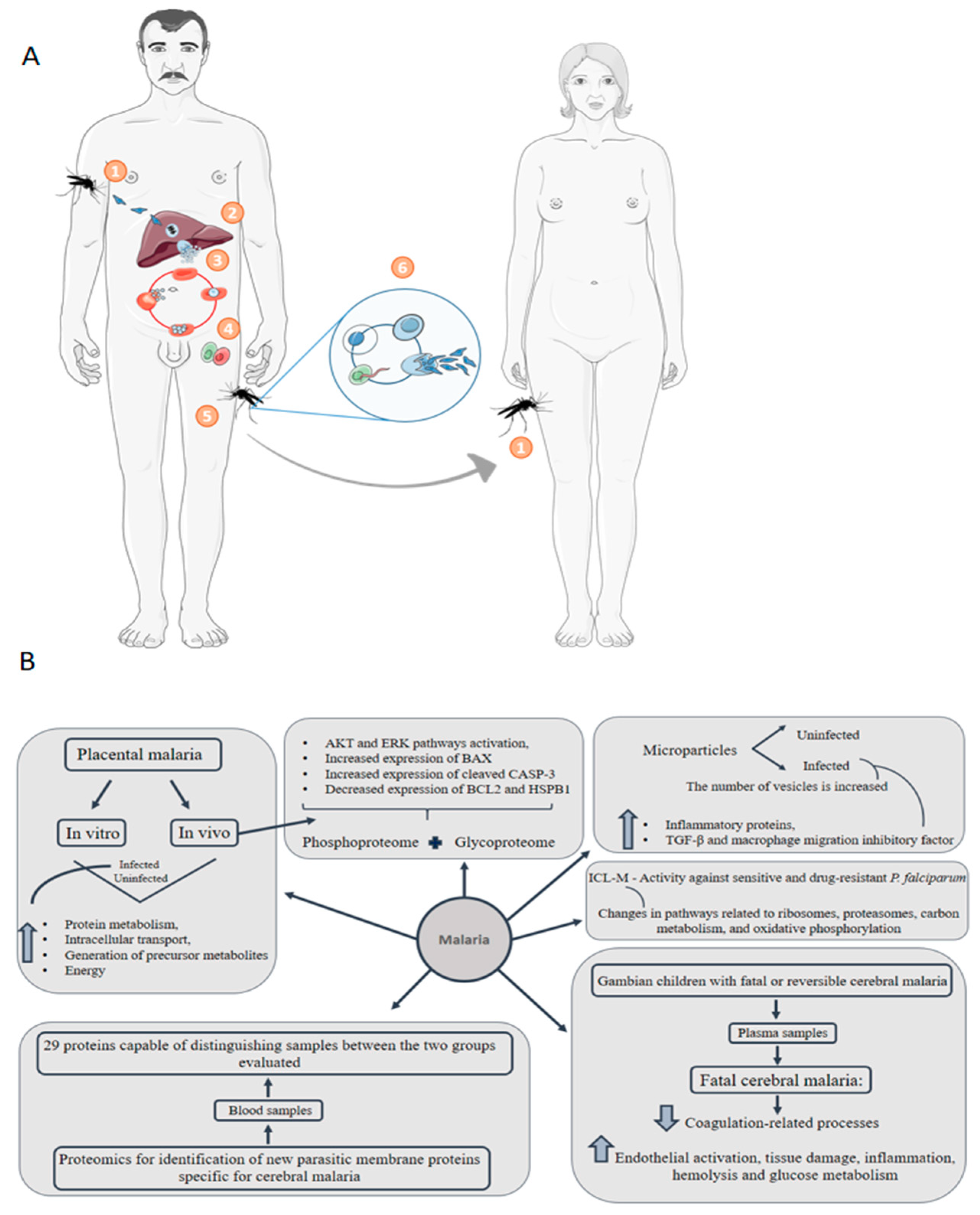
6. Malaria: Congenital Malaria Is Defined When the Parasite Is Identified in the Peripheral Blood of a Neonate in the First Week of Life
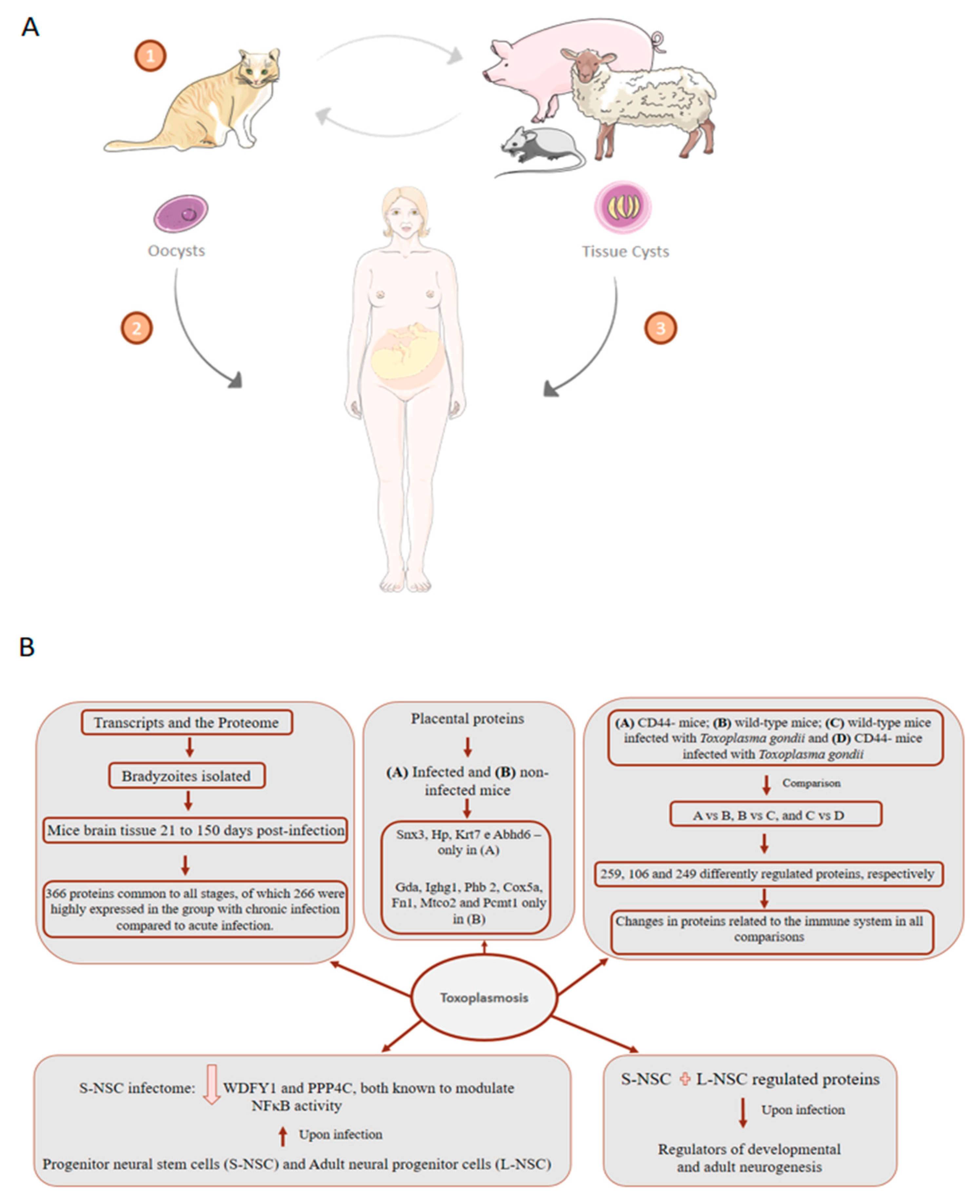
7. Toxoplasmosis: About 75% of Cases of Congenital Toxoplasmosis Have No Clinical Evidence, Making Early Treatment Difficult
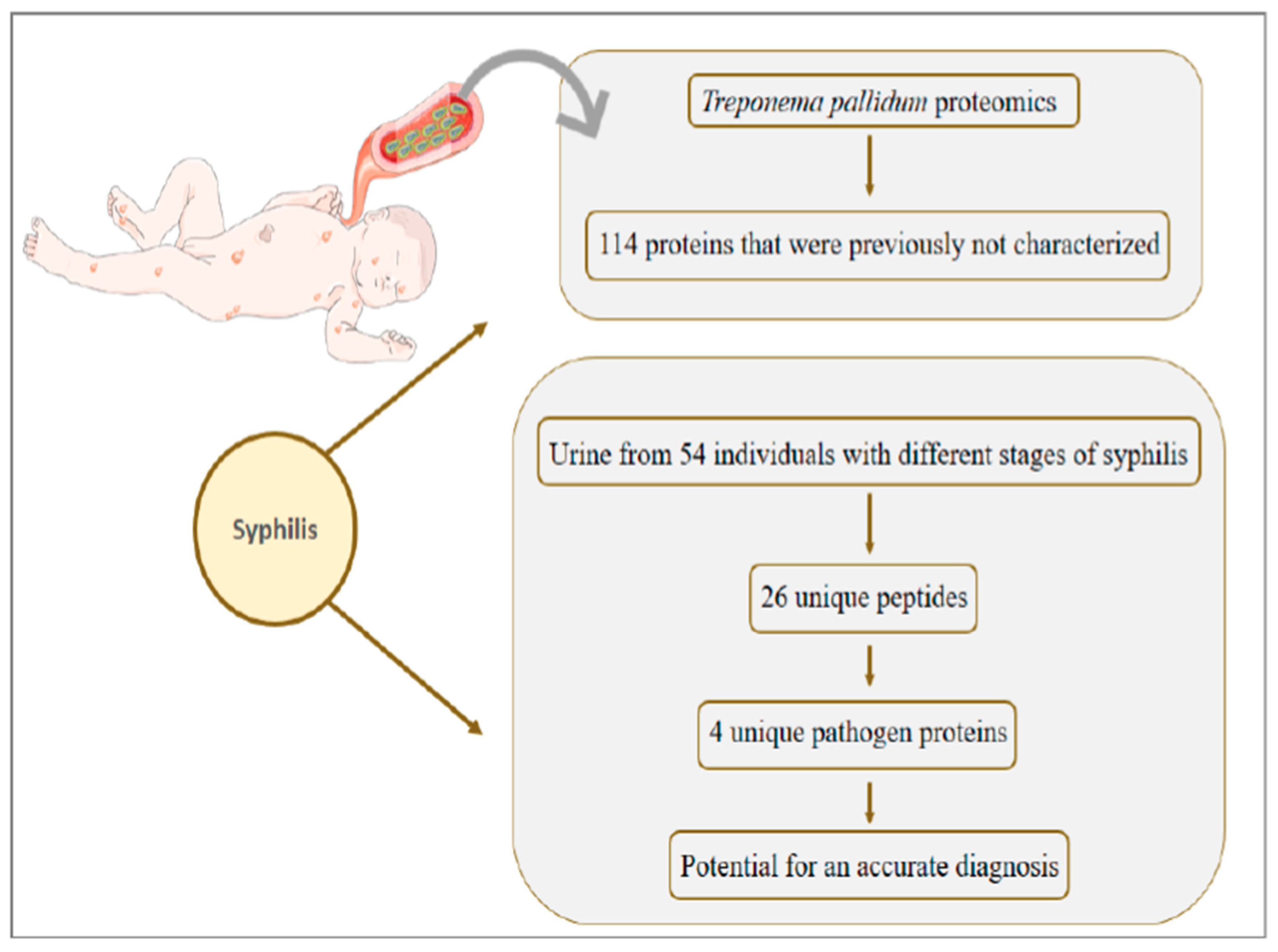
8. Syphilis: Congenital Syphilis Presents about One Million Cases per Year and Is Responsible for more than 300 Thousand Perinatal Deaths
9. Congenital Transmission of Varicella, Rubella, and Parvovirus B19 Has a Gap in Proteomic Studies
10. Critical Points of Proteomics Approaches Applied to Congenital Diseases
11. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pereira, L. Congenital viral infection: Traversing the uterine-placental interface. Annu. Rev. Virol. 2018, 5, 273–299. [Google Scholar] [CrossRef] [PubMed]
- Gude, N.M.; Roberts, C.T.; Kalionis, B.; King, R.G. Growth and function of the normal human placenta. Thromb. Res. 2004, 114, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Racicot, K.; Mor, G. Risks associated with viral infections during pregnancy. J. Clin. Investig. 2017, 127, 1591–1599. [Google Scholar] [CrossRef] [PubMed]
- Campos, G.S.; Bandeira, A.C.; Sardi, S.I. Zika Virus Outbreak, Bahia, Brazil. Emerg. Infect. Dis. 2015, 21, 1885–1886. [Google Scholar] [CrossRef]
- Adams Waldorf, K.M.; Nelson, B.R.; Stencel-Baerenwald, J.E.; Studholme, C.; Kapur, R.P.; Armistead, B.; Walker, C.L.; Merillat, S.; Vornhagen, J.; Tisoncik-Go, J.; et al. Congenital Zika virus infection as a silent pathology with loss of neurogenic output in the fetal brain. Nat. Med. 2018, 24, 368–374. [Google Scholar] [CrossRef]
- Stegmann, B.J.; Carey, J.C. TORCH Infections. Toxoplasmosis, Other (syphilis, varicella-zoster, parvovirus B19), Rubella, Cytomegalovirus (CMV), and Herpes infections. Curr. Womens Health Rep. 2002, 2, 253–258. [Google Scholar]
- Costa, M.L.; de Moraes Nobrega, G.; Antolini-Tavares, A. Key Infections in the Placenta. Obstet. Gynecol. Clin. N. Am. 2020, 47, 133–146. [Google Scholar] [CrossRef]
- Martin, G.P.; Marriott, C.; Kellaway, I.W. The effect of natural surfactants on the pheological properties of mucus. J. Pharm. Pharmacol. 1976, 28, 76. [Google Scholar]
- León-Juárez, M.; Martínez-Castillo, M.; González-García, L.D.; Helguera-Repetto, A.C.; Zaga-Clavellina, V.; García-Cordero, J.; Flores-Pliego, A.; Herrera-Salazar, A.; Vázquez-Martínez, E.R.; Reyes-Muñoz, E. Cellular and molecular mechanisms of viral infection in the human placenta. Pathog. Dis. 2017, 75. [Google Scholar] [CrossRef]
- Koi, H.; Zhang, J.; Makrigiannakis, A.; Getsios, S.; MacCalman, C.D.; Kopf, G.S.; Strauss, J.F.; Parry, S. Differential expression of the coxsackievirus and adenovirus receptor regulates adenovirus infection of the placenta. Biol. Reprod. 2001, 64, 1001–1009. [Google Scholar] [CrossRef]
- Feire, A.L.; Koss, H.; Compton, T. Cellular integrins function as entry receptors for human cytomegalovirus via a highly conserved disintegrin-like domain. Proc. Natl. Acad. Sci. USA 2004, 101, 15470–15475. [Google Scholar] [CrossRef] [PubMed]
- Aagaard, K.M.; Lahon, A.; Suter, M.A.; Arya, R.P.; Seferovic, M.D.; Vogt, M.B.; Hu, M.; Stossi, F.; Mancini, M.A.; Harris, R.A.; et al. Primary Human Placental Trophoblasts are Permissive for Zika Virus (ZIKV) Replication. Sci. Rep. 2017, 7, 41389. [Google Scholar] [CrossRef] [PubMed]
- Parry, S.; Holder, J.; Strauss, J.F. Mechanisms of trophoblast-virus interaction. J. Reprod. Immunol. 1997, 37, 25–34. [Google Scholar] [CrossRef]
- Ranger-Rogez, S.; Alain, S.; Denis, F. Virus des hépatites: Transmission mère-enfant. Pathol. Biol. 2002, 50, 568–575. [Google Scholar] [CrossRef]
- Robinson, D.P.; Klein, S.L. Pregnancy and pregnancy-associated hormones alter immune responses and disease pathogenesis. Horm. Behav. 2012, 62, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Menendez, C. Malaria during pregnancy: A priority area of malaria research and control. Parasitol. Today 1995, 11, 178–183. [Google Scholar] [CrossRef]
- Szekeres-Bartho, J.; Faust, Z.S.; Varga, P.; Szereday, L.; Kelemen, K. The Immunological Pregnancy Protective Effect of Progesterone Is Manifested via Controlling Cytokine Production. Am. J. Reprod. Immunol. 1996, 35, 348–351. [Google Scholar] [CrossRef]
- Taneja, V. Sex Hormones Determine Immune Response. Front. Immunol. 2018, 9, 1931. [Google Scholar] [CrossRef]
- Marzi, M.; Vigano, A.; Trabattoni, D.; Villa, M.L.; Salvaggio, A.; Clerici, E.; Clerici, M. Characterization of type 1 and type 2 cytokine production profile in physiologic and pathologic human pregnancy. Clin. Exp. Immunol. 1996, 106, 127–133. [Google Scholar] [CrossRef]
- Orton, D.; Doucette, A. Proteomic Workflows for Biomarker Identification Using Mass Spectrometry—Technical and Statistical Considerations during Initial Discovery. Proteomes 2013, 1, 109–127. [Google Scholar] [CrossRef]
- Maxwell, K.L.; Frappier, L. Viral Proteomics. Microbiol. Mol. Biol. Rev. 2007, 71, 398–411. [Google Scholar] [CrossRef] [PubMed]
- Wasinger, V.C.; Cordwell, S.J.; Cerpa-Poljak, A.; Yan, J.X.; Gooley, A.A.; Wilkins, M.R.; Duncan, M.W.; Harris, R.; Williams, K.L.; Humphery-Smith, I. Progress with gene-product mapping of the Mollicutes: Mycoplasma genitalium. Electrophoresis 1995, 16, 1090–1094. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fonslow, B.R.; Shan, B.; Baek, M.-C.; Yates, J.R. Protein analysis by shotgun/bottom-up proteomics. Chem. Rev. 2013, 113, 2343–2394. [Google Scholar] [CrossRef]
- Catherman, A.D.; Skinner, O.S.; Kelleher, N.L. Top Down proteomics: Facts and perspectives. Biochem. Biophys. Res. Commun. 2014, 445, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Toby, T.K.; Fornelli, L.; Kelleher, N.L. Progress in Top-Down Proteomics and the Analysis of Proteoforms. Annu. Rev. Anal. Chem. (Palo Alto Calif.) 2016, 9, 499–519. [Google Scholar] [CrossRef] [PubMed]
- Sidoli, S.; Garcia, B.A. Middle-down proteomics: A still unexploited resource for chromatin biology. Expert Rev. Proteomics 2017, 14, 617–626. [Google Scholar] [CrossRef]
- Aebersold, R.; Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Leney, A.C.; Heck, A.J.R. Native Mass Spectrometry: What is in the Name? J. Am. Soc. Mass Spectrom. 2017, 28, 5–13. [Google Scholar] [CrossRef]
- Braun, P.; Gingras, A.-C. History of protein-protein interactions: From egg-white to complex networks. Proteomics 2012, 12, 1478–1498. [Google Scholar] [CrossRef]
- Stern-Ginossar, N.; Weisburd, B.; Michalski, A.; Le, V.T.K.; Hein, M.Y.; Huang, S.-X.; Ma, M.; Shen, B.; Qian, S.-B.; Hengel, H.; et al. Decoding human cytomegalovirus. Science 2012, 338, 1088–1093. [Google Scholar] [CrossRef]
- Marsico, C.; Kimberlin, D.W. Congenital Cytomegalovirus infection: Advances and challenges in diagnosis, prevention and treatment. Ital. J. Pediatr. 2017, 43, 38. [Google Scholar] [CrossRef]
- Xu, W.-F.; Yuan, T.-M. A review on the prevention and treatment of congenital cytomegalovirus infection in mothers and infants. Zhongguo Dang Dai Er Ke Za Zhi 2018, 20, 870–875. [Google Scholar]
- Cannon, M.J.; Schmid, D.S.; Hyde, T.B. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev. Med. Virol. 2010, 20, 202–213. [Google Scholar] [CrossRef]
- Kenneson, A.; Cannon, M.J. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev. Med. Virol. 2007, 17, 253–276. [Google Scholar] [CrossRef]
- Dahle, A.J.; Fowler, K.B.; Wright, J.D.; Boppana, S.B.; Britt, W.J.; Pass, R.F. Longitudinal investigation of hearing disorders in children with congenital cytomegalovirus. J. Am. Acad. Audiol. 2000, 11, 283–290. [Google Scholar] [PubMed]
- Zuhair, M.; Smit, G.S.A.; Wallis, G.; Jabbar, F.; Smith, C.; Devleesschauwer, B.; Griffiths, P. Estimation of the worldwide seroprevalence of cytomegalovirus: A systematic review and meta-analysis. Rev. Med. Virol. 2019, 29, e2034. [Google Scholar] [CrossRef] [PubMed]
- Reeves, M.; Sinclair, J. Aspects of human cytomegalovirus latency and reactivation. Curr. Top. Microbiol. Immunol. 2008, 325, 297–313. [Google Scholar] [CrossRef] [PubMed]
- Robert, M. Oxygen affinity of haemoglobin (author’s transl). Bull. Physiopathol. Respir. (Nancy) 1975, 11, 79–170. [Google Scholar] [PubMed]
- Brizić, I.; Hiršl, L.; Britt, W.J.; Krmpotić, A.; Jonjić, S. Immune responses to congenital cytomegalovirus infection. Microbes Infect. 2018, 20, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Britt, W.J. Maternal Immunity and the Natural History of Congenital Human Cytomegalovirus Infection. Viruses 2018, 10, 405. [Google Scholar] [CrossRef]
- Alford, C.A.; Hayes, K.; Britt, W. Primary Cytomegalovirus Infection in Pregnancy: Comparison of Antibody Responses to Virus-Encoded Proteins between Women with and without Intrauterine Infection. J. Infect. Dis. 1988, 158, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Lilleri, D.; Kabanova, A.; Revello, M.G.; Percivalle, E.; Sarasini, A.; Genini, E.; Sallusto, F.; Lanzavecchia, A.; Corti, D.; Gerna, G. Fetal Human Cytomegalovirus Transmission Correlates with Delayed Maternal Antibodies to gH/gL/pUL128-130-131 Complex during Primary Infection. PLoS ONE 2013, 8, e59863. [Google Scholar] [CrossRef] [PubMed]
- Pass, R.F.; Anderson, B. Mother-to-Child Transmission of Cytomegalovirus and Prevention of Congenital Infection. J. Pediatr. Infect. Dis. Soc. 2014, 3 (Suppl. 1), S2–S6. [Google Scholar] [CrossRef]
- Sinzger, C.; Digel, M.; Jahn, G. Cytomegalovirus cell tropism. Curr. Top. Microbiol. Immunol. 2008, 325, 63–83. [Google Scholar] [CrossRef] [PubMed]
- Varnum, S.M.; Streblow, D.N.; Monroe, M.E.; Smith, P.; Auberry, K.J.; Pasa-Tolic, L.; Wang, D.; Camp, D.G.; Rodland, K.; Wiley, S.; et al. Identification of proteins in human cytomegalovirus (HCMV) particles: The HCMV proteome. J. Virol. 2004, 78, 10960–10966. [Google Scholar] [CrossRef] [PubMed]
- Jean Beltran, P.M.; Cristea, I.M. The life cycle and pathogenesis of human cytomegalovirus infection: Lessons from proteomics. Expert Rev. Proteomics 2014, 11, 697–711. [Google Scholar] [CrossRef]
- Anderholm, K.M.; Bierle, C.J.; Schleiss, M.R. Cytomegalovirus vaccines: Current status and future prospects. Drugs 2016, 76, 1625–1645. [Google Scholar] [CrossRef]
- Plotkin, S.A.; Boppana, S.B. Vaccination against the human cytomegalovirus. Vaccine 2019, 37, 7437–7442. [Google Scholar] [CrossRef]
- Viswanathan, K.; Verweij, M.C.; John, N.; Malouli, D.; Früh, K. Quantitative membrane proteomics reveals a role for tetraspanin enriched microdomains during entry of human cytomegalovirus. PLoS ONE 2017, 12, e0187899. [Google Scholar] [CrossRef]
- Martinez-Martin, N.; Marcandalli, J.; Huang, C.S.; Arthur, C.P.; Perotti, M.; Foglierini, M.; Ho, H.; Dosey, A.M.; Shriver, S.; Payandeh, J.; et al. An unbiased screen for human cytomegalovirus identifies neuropilin-2 as a central viral receptor. Cell 2018, 174, 1158–1171.e19. [Google Scholar] [CrossRef]
- Bozidis, P.; Williamson, C.D.; Colberg-Poley, A.M. Mitochondrial and secretory human cytomegalovirus UL37 proteins traffic into mitochondrion-associated membranes of human cells. J. Virol. 2008, 82, 2715–2726. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Williamson, C.D.; Wong, D.S.; Bullough, M.D.; Brown, K.J.; Hathout, Y.; Colberg-Poley, A.M. Quantitative proteomic analyses of human cytomegalovirus-induced restructuring of endoplasmic reticulum-mitochondrial contacts at late times of infection. Mol. Cell. Proteomics 2011, 10, M111.009936. [Google Scholar] [CrossRef] [PubMed]
- Jean Beltran, P.M.; Mathias, R.A.; Cristea, I.M. A portrait of the human organelle proteome in space and time during cytomegalovirus infection. Cell Syst. 2016, 3, 361–373.e6. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Sheng, X.; Murray-Nerger, L.A.; Cristea, I.M. Temporal dynamics of protein complex formation and dissociation during human cytomegalovirus infection. Nat. Commun. 2020, 11, 806. [Google Scholar] [CrossRef] [PubMed]
- Cook, K.C.; Cristea, I.M. Location is everything: Protein translocations as a viral infection strategy. Curr. Opin. Chem. Biol. 2019, 48, 34–43. [Google Scholar] [CrossRef]
- Liu, Z.; Tian, Y.; Wang, B.; Yan, Z.; Qian, D.; Ding, S.; Song, X.; Bai, Z.; Li, L. Serum proteomics with SELDI-TOF-MS in congenital human cytomegalovirus hepatitis. J. Med. Virol. 2007, 79, 1500–1505. [Google Scholar] [CrossRef]
- Kindhauser, M.K.; Allen, T.; Frank, V.; Santhana, R.S.; Dye, C. Zika: The origin and spread of a mosquito-borne virus. Bull. World Health Organ. 2016, 94, 675C–686C. [Google Scholar] [CrossRef]
- Bailey, M.J.; Broecker, F.; Duehr, J.; Arumemi, F.; Krammer, F.; Palese, P.; Tan, G.S. Antibodies elicited by an NS1-based vaccine protect mice against zika virus. mBio 2019, 10. [Google Scholar] [CrossRef]
- Delaney, A.; Mai, C.; Smoots, A.; Cragan, J.; Ellington, S.; Langlois, P.; Breidenbach, R.; Fornoff, J.; Dunn, J.; Yazdy, M.; et al. Population-based surveillance of birth defects potentially related to zika virus infection—15 States and U.S. Territories, 2016. MMWR Morb. Mortal. Wkly. Rep. 2018, 67, 91–96. [Google Scholar] [CrossRef]
- Driggers, R.W.; Ho, C.-Y.; Korhonen, E.M.; Kuivanen, S.; Jääskeläinen, A.J.; Smura, T.; Rosenberg, A.; Hill, D.A.; DeBiasi, R.L.; Vezina, G.; et al. Zika Virus Infection with Prolonged Maternal Viremia and Fetal Brain Abnormalities. N. Engl. J. Med. 2016, 374, 2142–2151. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, F.M.E.; Pietrobon, A.J.; de Oliveira, L.M.; da Oliveira, L.M.S.; Sato, M.N. Maternal-Fetal Interplay in Zika Virus Infection and Adverse Perinatal Outcomes. Front. Immunol. 2020, 11, 175. [Google Scholar] [CrossRef] [PubMed]
- Tonnerre, P.; Melgaço, J.G.; Torres-Cornejo, A.; Pinto, M.A.; Yue, C.; Blümel, J.; de Sousa, P.S.F.; de da Mello, V.M.; Moran, J.; de Filippis, A.M.B.; et al. Evolution of the innate and adaptive immune response in women with acute Zika virus infection. Nat. Microbiol. 2020, 5, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Priyamvada, L.; Suthar, M.S.; Ahmed, R.; Wrammert, J. Humoral immune responses against zika virus infection and the importance of preexisting flavivirus immunity. J. Infect. Dis. 2017, 216, S906–S911. [Google Scholar] [CrossRef] [PubMed]
- Lesteberg, K.E.; Fader, D.S.; Beckham, J.D. Pregnancy alters innate immune responses to Zika virus infection in the genital tract. Immunology 2019. [Google Scholar] [CrossRef]
- Moore, C.A.; Staples, J.E.; Dobyns, W.B.; Pessoa, A.; Ventura, C.V.; da Fonseca, E.B.; Ribeiro, E.M.; Ventura, L.O.; Neto, N.N.; Arena, J.F.; et al. Characterizing the pattern of anomalies in congenital zika syndrome for pediatric clinicians. JAMA Pediatr. 2017, 171, 288–295. [Google Scholar] [CrossRef]
- Mohr, E.L.; Block, L.N.; Newman, C.M.; Stewart, L.M.; Koenig, M.; Semler, M.; Breitbach, M.E.; Teixeira, L.B.C.; Zeng, X.; Weiler, A.M.; et al. Ocular and uteroplacental pathology in a macaque pregnancy with congenital Zika virus infection. PLoS ONE 2018, 13, e0190617. [Google Scholar] [CrossRef]
- Souza, I.N.O.; Barros-Aragão, F.G.Q.; Frost, P.S.; Figueiredo, C.P.; Clarke, J.R. Late neurological consequences of zika virus infection: Risk factors and pharmaceutical approaches. Pharmaceuticals 2019, 12, 60. [Google Scholar] [CrossRef]
- Rice, M.E.; Galang, R.R.; Roth, N.M.; Ellington, S.R.; Moore, C.A.; Valencia-Prado, M.; Ellis, E.M.; Tufa, A.J.; Taulung, L.A.; Alfred, J.M.; et al. Vital Signs: Zika-Associated Birth Defects and Neurodevelopmental Abnormalities Possibly Associated with Congenital Zika Virus Infection—U.S. Territories and Freely Associated States, 2018. MMWR Morb. Mortal. Wkly. Rep. 2018, 67, 858–867. [Google Scholar] [CrossRef]
- Prata-Barbosa, A.; Martins, M.M.; Guastavino, A.B.; da Cunha, A.J.L.A. Effects of Zika infection on growth. J. Pediatr. (Rio J.) 2019, 95 (Suppl. 1), 30–41. [Google Scholar] [CrossRef]
- Van der Linden, V.; Pessoa, A.; Dobyns, W.; Barkovich, A.J.; van der Júnior, H.L.; Filho, E.L.R.; Ribeiro, E.M.; de Leal, M.C.; de Coimbra, P.P.A.; de Aragão, M.F.V.V.; et al. Description of 13 Infants Born During October 2015-January 2016 With Congenital Zika Virus Infection Without Microcephaly at Birth - Brazil. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 1343–1348. [Google Scholar] [CrossRef]
- Smith, D.R.; Hollidge, B.; Daye, S.; Zeng, X.; Blancett, C.; Kuszpit, K.; Bocan, T.; Koehler, J.W.; Coyne, S.; Minogue, T.; et al. Neuropathogenesis of Zika Virus in a Highly Susceptible Immunocompetent Mouse Model after Antibody Blockade of Type I Interferon. PLoS Negl. Trop. Dis. 2017, 11, e0005296. [Google Scholar] [CrossRef] [PubMed]
- Xin, Q.-L.; Deng, C.-L.; Chen, X.; Wang, J.; Wang, S.-B.; Wang, W.; Deng, F.; Zhang, B.; Xiao, G.; Zhang, L.-K. Quantitative Proteomic Analysis of Mosquito C6/36 Cells Reveals Host Proteins Involved in Zika Virus Infection. J. Virol. 2017, 91, e00554-17. [Google Scholar] [CrossRef] [PubMed]
- Garcez, P.P.; Minardi Nascimento, J.; Mota de Vasconcelos, J.; Madeiro da Costa, R.; Delvecchio, R.; Trindade, P.; Correia Loiola, E.; Higa, L.M.; Cassoli, J.; Vitória, G.; et al. Combined Proteome and Transcriptome Analyses Reveal That Zika Virus Circulating in Brazil Alters Cell Cycle and Neurogenic Programmes in Human Neurospheres. Available online: https://peerj.com/preprints/2033/ (accessed on 9 May 2016).
- Scaturro, P.; Stukalov, A.; Haas, D.A.; Cortese, M.; Draganova, K.; Płaszczyca, A.; Bartenschlager, R.; Götz, M.; Pichlmair, A. An orthogonal proteomic survey uncovers novel Zika virus host factors. Nature 2018, 561, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Rosa-Fernandes, L.; Cugola, F.R.; Russo, F.B.; Kawahara, R.; de Melo Freire, C.C.; Leite, P.E.C.; Bassi Stern, A.C.; Angeli, C.B.; de Oliveira, D.B.L.; Melo, S.R.; et al. Zika Virus Impairs Neurogenesis and Synaptogenesis Pathways in Human Neural Stem Cells and Neurons. Front. Cell. Neurosci. 2019, 13, 64. [Google Scholar] [CrossRef] [PubMed]
- Robbiani, D.F.; Olsen, P.C.; Costa, F.; Wang, Q.; Oliveira, T.Y.; Nery, N.; Aromolaran, A.; do Rosário, M.S.; Sacramento, G.A.; Cruz, J.S.; et al. Risk of Zika microcephaly correlates with features of maternal antibodies. J. Exp. Med. 2019, 216, 2302–2315. [Google Scholar] [CrossRef] [PubMed]
- Allgoewer, K.; Zhao, A.; Maity, S.; Lashua, L.; Ramgopal, M.; Balkaran, B.N.; Liu, L.; Arévalo, M.T.; Ross, T.M.; Choi, H.; et al. High-resolution proteomics identifies potential new markers of Zika and dengue infections. Syst. Biol. 2019. [Google Scholar] [CrossRef]
- Song, G.; Rho, H.-S.; Pan, J.; Ramos, P.; Yoon, K.-J.; Medina, F.A.; Lee, E.M.; Eichinger, D.; Ming, G.; Muñoz-Jordan, J.L.; et al. Multiplexed Biomarker Panels Discriminate Zika and Dengue Virus Infection in Humans. Mol. Cell. Proteomics 2018, 17, 349–356. [Google Scholar] [CrossRef]
- Chen, Z.; Luckay, A.; Sodora, D.L.; Telfer, P.; Reed, P.; Gettie, A.; Kanu, J.M.; Sadek, R.F.; Yee, J.; Ho, D.D.; et al. Human immunodeficiency virus type 2 (HIV-2) seroprevalence and characterization of a distinct HIV-2 genetic subtype from the natural range of simian immunodeficiency virus-infected sooty mangabeys. J. Virol. 1997, 71, 3953–3960. [Google Scholar] [CrossRef]
- Librelotto, C.S.; Gräf, T.; Simon, D.; de Almeida, S.E.M.; Lunge, V.R. HIV-1 epidemiology and circulating subtypes in the countryside of South Brazil. Rev. Soc. Bras. Med. Trop. 2015, 48, 249–257. [Google Scholar] [CrossRef]
- Deeks, S.G.; Overbaugh, J.; Phillips, A.; Buchbinder, S. HIV infection. Nat. Rev. Dis. Primers 2015, 1, 15035. [Google Scholar] [CrossRef]
- Meléndez, L.M.; Colon, K.; Rivera, L.; Rodriguez-Franco, E.; Toro-Nieves, D. Proteomic Analysis of HIV-Infected Macrophages. J. Neuroimmune Pharmacol. 2011, 6, 89–106. [Google Scholar] [CrossRef] [PubMed]
- Bongertz, V. Vertical human immunodeficiency virus type 1—HIV-1–transmission—A review. Mem. Inst. Oswaldo Cruz. 2001, 96, 1–14. [Google Scholar] [CrossRef][Green Version]
- Chu, S.Y.; Buehler, J.W.; Berkelman, R.L. Impact of the human immunodeficiency virus epidemic on mortality in women of reproductive age, United States. JAMA 1990, 264, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.; Santiago, M.R.; Souza, D.A.; Silva, G.E.B.; Chahud, F.; Quintana, S.M.; Mendes-Junior, C.T.; Donadi, E.A.; Fernandes, A.P.M. The role of the placenta in the vertical transmission of HIV-1. Med. (Ribeirao Preto. Online) 2016, 49, 80. [Google Scholar] [CrossRef]
- Cocker, A.T.H.; Shah, N.M.; Raj, I.; Dermont, S.; Khan, W.; Mandalia, S.; Imami, N.; Johnson, M.R. Pregnancy Gestation Impacts on HIV-1-Specific Granzyme B Response and Central Memory CD4 T Cells. Front. Immunol. 2020, 11, 153. [Google Scholar] [CrossRef] [PubMed]
- Wallick, S.C.; Figari, I.S.; Morris, R.E.; Levinson, A.D.; Palladino, M.A. Immunoregulatory role of transforming growth factor beta (TGF-beta) in development of killer cells: Comparison of active and latent TGF-beta 1. J. Exp. Med. 1990, 172, 1777–1784. [Google Scholar] [CrossRef]
- Burgener, A.; Boutilier, J.; Wachihi, C.; Kimani, J.; Carpenter, M.; Westmacott, G.; Cheng, K.; Ball, T.B.; Plummer, F. Identification of Differentially Expressed Proteins in the Cervical Mucosa of HIV-1-Resistant Sex Workers. J. Proteome Res. 2008, 7, 4446–4454. [Google Scholar] [CrossRef]
- Luciano-Montalvo, C.; Ciborowski, P.; Duan, F.; Gendelman, H.E.; Meléndez, L.M. Proteomic Analyses Associate Cystatin B with Restricted HIV-1 Replication in Placental Macrophages. Placenta 2008, 29, 1016–1023. [Google Scholar] [CrossRef]
- García, K.; García, V.; Pérez Laspiur, J.; Duan, F.; Meléndez, L.M. Characterization of the Placental Macrophage Secretome: Implications for Antiviral Activity. Placenta 2009, 30, 149–155. [Google Scholar] [CrossRef][Green Version]
- Soler-García, A.A.; Johnson, D.; Hathout, Y.; Ray, P.E. Iron-related proteins: Candidate urine biomarkers in childhood HIV-associated renal diseases. Clin. J. Am. Soc. Nephrol. 2009, 4, 763–771. [Google Scholar] [CrossRef]
- Whitley, R.J.; Roizman, B. Herpes simplex virus infections. Lancet 2001, 357, 1513–1518. [Google Scholar] [CrossRef]
- Koelle, D.M.; Corey, L. Herpes Simplex: Insights on Pathogenesis and Possible Vaccines. Annu. Rev. Med. 2008, 59, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Looker, K.J. A systematic review of the epidemiology and interaction of herpes simplex virus types 1 and 2. Sex. Trans. Infect. 2005, 81, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Mahnert, N.; Roberts, S.W.; Laibl, V.R.; Sheffield, J.S.; Wendel, G.D. The incidence of neonatal herpes infection. Am. J. Obstet. Gynecol. 2007, 196, e55–e56. [Google Scholar] [CrossRef] [PubMed]
- Patel, C.D.; Backes, I.M.; Taylor, S.A.; Jiang, Y.; Marchant, A.; Pesola, J.M.; Coen, D.M.; Knipe, D.M.; Ackerman, M.E.; Leib, D.A. Maternal immunization confers protection against neonatal herpes simplex mortality and behavioral morbidity. Sci. Transl. Med. 2019, 11, eaau6039. [Google Scholar] [CrossRef]
- Purewal, R.; Costello, L.; Garlapati, S.; Mitra, S.; Mitchell, M.; Moffett, K.S. Congenital Herpes Simplex Virus in the Newborn: A Diagnostic Dilemma. J. Ped. Infect. Dis. 2016, 5, e21–e23. [Google Scholar] [CrossRef]
- Fernandes, N.D.; Badri, T. Congenital Herpes Simplex. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Loret, S.; Guay, G.; Lippé, R. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 2008, 82, 8605–8618. [Google Scholar] [CrossRef]
- Takeuchi, K.; Suzumura, E.; Hirata, K.; Majima, Y.; Sakakura, Y. Role of transepithelial ion transport as a determinant of mucus viscoelasticity in chronic inflammation of the maxillary sinus. Acta Otolaryngol. 1991, 111, 1133–1138. [Google Scholar] [CrossRef]
- Liu, H.; Huang, C.-X.; He, Q.; Li, D.; Luo, M.-H.; Zhao, F.; Lu, W. Proteomics analysis of HSV-1-induced alterations in mouse brain microvascular endothelial cells. J. Neurovirol. 2019, 25, 525–539. [Google Scholar] [CrossRef]
- Antrobus, R.; Grant, K.; Gangadharan, B.; Chittenden, D.; Everett, R.D.; Zitzmann, N.; Boutell, C. Proteomic analysis of cells in the early stages of herpes simplex virus type-1 infection reveals widespread changes in the host cell proteome. Proteomics 2009, 9, 3913–3927. [Google Scholar] [CrossRef]
- Berard, A.R.; Coombs, K.M.; Severini, A. Quantification of the host response proteome after herpes simplex virus type 1 infection. J. Proteome Res. 2015, 14, 2121–2142. [Google Scholar] [CrossRef] [PubMed]
- Everett, R.D. HSV-1 biology and life cycle. Methods Mol. Biol. 2014, 1144, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kulej, K.; Avgousti, D.C.; Sidoli, S.; Herrmann, C.; Della Fera, A.N.; Kim, E.T.; Garcia, B.A.; Weitzman, M.D. Time-resolved Global and Chromatin Proteomics during Herpes Simplex Virus Type 1 (HSV-1) Infection. Mol. Cell Proteomics 2017, 16, S92–S107. [Google Scholar] [CrossRef] [PubMed]
- Drayman, N.; Karin, O.; Mayo, A.; Danon, T.; Shapira, L.; Rafael, D.; Zimmer, A.; Bren, A.; Kobiler, O.; Alon, U. Dynamic Proteomics of Herpes Simplex Virus Infection. mBio 2017, 8, e01612-17. [Google Scholar] [CrossRef]
- Sloan, E.; Tatham, M.H.; Groslambert, M.; Glass, M.; Orr, A.; Hay, R.T.; Everett, R.D. Analysis of the SUMO2 Proteome during HSV-1 Infection. PLoS Pathog. 2015, 11, e1005059. [Google Scholar] [CrossRef] [PubMed]
- Ashley, E.A.; Pyae Phyo, A.; Woodrow, C.J. Malaria. Lancet 2018, 391, 1608–1621. [Google Scholar] [CrossRef]
- Bertin, G.I.; Sabbagh, A.; Argy, N.; Salnot, V.; Ezinmegnon, S.; Agbota, G.; Ladipo, Y.; Alao, J.M.; Sagbo, G.; Guillonneau, F.; et al. Proteomic analysis of Plasmodium falciparum parasites from patients with cerebral and uncomplicated malaria. Sci. Rep. 2016, 6, 26773. [Google Scholar] [CrossRef]
- Bloland, P.B.; Williams, H.A.; National Research Council (US) Committee on Population; Program on Forced Migration and Health at the Mailman School of Public Health, C.U. Malaria Control during Mass Population Movements and Natural Disasters; National Academies Press (US): Washington, DC, USA, 2002; ISBN 978-0-309-08615-8. [Google Scholar]
- Phillips, M.A.; Burrows, J.N.; Manyando, C.; van Huijsduijnen, R.H.; Van Voorhis, W.C.; Wells, T.N.C. Malaria. Nat. Rev. Dis. Primers 2017, 3, 17050. [Google Scholar] [CrossRef]
- Aly, A.S.I.; Vaughan, A.M.; Kappe, S.H.I. Malaria parasite development in the mosquito and infection of the mammalian host. Annu. Rev. Microbiol. 2009, 63, 195–221. [Google Scholar] [CrossRef]
- Lacey, R.W. Basic medical microbiology (4th edition). J. Hosp. Infect. 1992, 20, 135–136. [Google Scholar] [CrossRef]
- Bhatia, R.; Rajwaniya, D.; Agrawal, P. Congenital Malaria due to Plasmodium Vivax Infection in a Neonate. Case Rep. Pediatr. 2016, 2016, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Harrington, W.E.; Duffy, P.E. Congenital malaria: Rare but potentially fatal. Pediatr. Health 2008, 2, 235–248. [Google Scholar] [CrossRef]
- Dombrowski, J.G.; de Souza, R.M.; Lima, F.A.; Bandeira, C.L.; Murillo, O.; de Costa, D.S.; Peixoto, E.P.M.; dos Cunha, M.P.; de Zanotto, P.M.A.; Bevilacqua, E.; et al. Association of Malaria Infection During Pregnancy with Head Circumference of Newborns in the Brazilian Amazon. JAMA Netw. Open 2019, 2, e193300. [Google Scholar] [CrossRef] [PubMed]
- Sharma, L.; Shukla, G. Placental Malaria: A New Insight into the Pathophysiology. Front. Med. (Lausanne) 2017, 4, 117. [Google Scholar] [CrossRef]
- Desai, M.; ter Kuile, F.O.; Nosten, F.; McGready, R.; Asamoa, K.; Brabin, B.; Newman, R.D. Epidemiology and burden of malaria in pregnancy. Lancet Infect. Dis. 2007, 7, 93–104. [Google Scholar] [CrossRef]
- Rogerson, S.J.; Hviid, L.; Duffy, P.E.; Leke, R.F.G.; Taylor, D.W. Malaria in pregnancy: Pathogenesis and immunity. Lancet Infect. Dis. 2007, 7, 105–117. [Google Scholar] [CrossRef]
- Darmstadt, G.L.; Zaidi, A.K.M.; Stoll, B.J. Neonatal Infections. In Infectious Diseases of the Fetus and Newborn; Elsevier: Philadelphia, PA, USA, 2011; pp. 24–51. ISBN 978-1-4160-6400-8. [Google Scholar]
- Dobbs, K.R.; Dent, A.E. Plasmodium malaria and antimalarial antibodies in the first year of life. Parasitology 2016, 143, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Odorizzi, P.M.; Feeney, M.E. Impact of In Utero Exposure to Malaria on Fetal T Cell Immunity. Trends Mol. Med. 2016, 22, 877–888. [Google Scholar] [CrossRef]
- Reis, A.S.; Barboza, R.; Murillo, O.; Barateiro, A.; Peixoto, E.P.M.; Lima, F.A.; Gomes, V.M.; Dombrowski, J.G.; Leal, V.N.C.; Araujo, F.; et al. Inflammasome activation and IL-1 signaling during placental malaria induce poor pregnancy outcomes. Sci. Adv. 2020, 6, eaax6346. [Google Scholar] [CrossRef]
- Kawahara, R.; Rosa-Fernandes, L.; Dos Santos, A.F.; Bandeira, C.L.; Dombrowski, J.G.; Souza, R.M.; Da Fonseca, M.P.; Festuccia, W.T.; Labriola, L.; Larsen, M.R.; et al. Integrated proteomics reveals apoptosis-related mechanisms associated with placental malaria. Mol. Cell. Proteomics 2019, 18, 182–199. [Google Scholar] [CrossRef]
- Antwi-Baffour, S.; Adjei, J.K.; Agyemang-Yeboah, F.; Annani-Akollor, M.; Kyeremeh, R.; Asare, G.A.; Gyan, B. Proteomic analysis of microparticles isolated from malaria positive blood samples. Proteome Sci. 2016, 15, 5. [Google Scholar] [CrossRef] [PubMed]
- Moussa, E.M.; Huang, H.; Thézénas, M.L.; Fischer, R.; Ramaprasad, A.; Sisay-Joof, F.; Jallow, M.; Pain, A.; Kwiatkowski, D.; Kessler, B.M.; et al. Proteomic profiling of the plasma of Gambian children with cerebral malaria. Malar. J. 2018, 17, 337. [Google Scholar] [CrossRef] [PubMed]
- Fried, M.; Wendler, J.P.; Mutabingwa, T.K.; Duffy, P.E. Mass spectrometric analysis ofPlasmodium falciparum erythrocyte membrane protein-1 variants expressed by placental malaria parasites. Proteomics 2004, 4, 1086–1093. [Google Scholar] [CrossRef] [PubMed]
- Gonzales Hurtado, P.A.; Morrison, R.; Ribeiro, J.M.C.; Magale, H.; Attaher, O.; Diarra, B.S.; Mahamar, A.; Barry, A.; Dicko, A.; Duffy, P.E.; et al. Proteomics Pipeline for Identifying Variant Proteins in Plasmodium falciparum Parasites Isolated from Children Presenting with Malaria. J. Proteome Res. 2019, 18, 3831–3839. [Google Scholar] [CrossRef] [PubMed]
- Tse, E.G.; Korsik, M.; Todd, M.H. The past, present and future of anti-malarial medicines. Malar. J. 2019, 18, 93. [Google Scholar] [CrossRef] [PubMed]
- Rujimongkon, K.; Mungthin, M.; Tummatorn, J.; Ampawong, S.; Adisakwattana, P.; Boonyuen, U.; Reamtong, O. Proteomic analysis of Plasmodium falciparum response to isocryptolepine derivative. PLoS ONE 2019, 14, e0220871. [Google Scholar] [CrossRef]
- Dunay, I.R.; Gajurel, K.; Dhakal, R.; Liesenfeld, O.; Montoya, J.G. Treatment of toxoplasmosis: Historical perspective, animal models, and current clinical practice. Clin. Microbiol. Rev. 2018, 31, e00057-17. [Google Scholar] [CrossRef] [PubMed]
- Saadatnia, G.; Golkar, M. A review on human toxoplasmosis. Scand. J. Infect. Dis. 2012, 44, 805–814. [Google Scholar] [CrossRef]
- Montoya, J.G.; Liesenfeld, O. Toxoplasmosis. Lancet 2004, 363, 1965–1976. [Google Scholar] [CrossRef]
- Alvarado-Esquivel, C.; Pacheco-Vega, S.J.; Hernández-Tinoco, J.; Centeno-Tinoco, M.M.; Beristain-García, I.; Sánchez-Anguiano, L.F.; Liesenfeld, O.; Rábago-Sánchez, E.; Berumen-Segovia, L.O. Miscarriage history and Toxoplasma gondii infection: A cross-sectional study in women in Durango City, Mexico. Eur. J. Microbiol. Immunol. (Bp.) 2014, 4, 117–122. [Google Scholar] [CrossRef]
- Freeman, K.; Oakley, L.; Pollak, A.; Buffolano, W.; Petersen, E.; Semprini, A.E.; Salt, A.; Gilbert, R. European Multicentre Study on Congenital Toxoplasmosis Association between congenital toxoplasmosis and preterm birth, low birthweight and small for gestational age birth. BJOG 2005, 112, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Chávez, F.; Cañedo-Solares, I.; Ortiz-Alegría, L.B.; Flores-García, Y.; Luna-Pastén, H.; Figueroa-Damián, R.; Mora-González, J.C.; Correa, D. Maternal Immune Response During Pregnancy and Vertical Transmission in Human Toxoplasmosis. Front. Immunol. 2019, 10, 285. [Google Scholar] [CrossRef] [PubMed]
- McAuley, J.B. Congenital Toxoplasmosis. J. Pediatr. Infect. Dis. Soc. 2014, 3 (Suppl. 1), S30–S35. [Google Scholar] [CrossRef] [PubMed]
- Robert-Gangneux, F.; Darde, M.-L. Epidemiology of and Diagnostic Strategies for Toxoplasmosis. Clin. Microbiol. Rev. 2012, 25, 264–296. [Google Scholar] [CrossRef]
- Chowdhury, M.N. Toxoplasmosis: A review. J. Med. 1986, 17, 373–396. [Google Scholar]
- Ngô, H.M.; Zhou, Y.; Lorenzi, H.; Wang, K.; Kim, T.-K.; Zhou, Y.; El Bissati, K.; Mui, E.; Fraczek, L.; Rajagopala, S.V.; et al. Toxoplasma Modulates Signature Pathways of Human Epilepsy, Neurodegeneration & Cancer. Sci. Rep. 2017, 7, 11496. [Google Scholar] [CrossRef]
- Hu, Y.-H.; Zhang, Y.; Jiang, L.-Q.; Wang, S.; Lei, C.-Q.; Sun, M.-S.; Shu, H.-B.; Liu, Y. WDFY1 mediates TLR3/4 signaling by recruiting TRIF. EMBO Rep. 2015, 16, 447–455. [Google Scholar] [CrossRef]
- Le Belle, J.E.; Orozco, N.M.; Paucar, A.A.; Saxe, J.P.; Mottahedeh, J.; Pyle, A.D.; Wu, H.; Kornblum, H.I. Proliferative neural stem cells have high endogenous ROS levels that regulate self-renewal and neurogenesis in a PI3K/Akt-dependant manner. Cell Stem Cell 2011, 8, 59–71. [Google Scholar] [CrossRef]
- Schlüter, D.; Barragan, A. Advances and Challenges in Understanding Cerebral Toxoplasmosis. Front. Immunol. 2019, 10, 242. [Google Scholar] [CrossRef]
- Garfoot, A.L.; Wilson, G.M.; Coon, J.J.; Knoll, L.J. Proteomic and transcriptomic analyses of early and late-chronic Toxoplasma gondii infection shows novel and stage specific transcripts. BMC Genom. 2019, 20, 859. [Google Scholar] [CrossRef]
- Yang, J.; Du, F.; Zhou, X.; Wang, L.; Li, S.; Fang, R.; Zhao, J. Brain proteomic differences between wild-type and CD44- mice induced by chronic Toxoplasma gondii infection. Parasitol. Res. 2018, 117, 2623–2633. [Google Scholar] [CrossRef] [PubMed]
- Jiao, F.; Zhang, D.; Jiang, M.; Mi, J.; Liu, X.; Zhang, H.; Hu, Z.; Xu, X.; Hu, X. Label-free proteomic analysis of placental proteins during Toxoplasma gondii infection. J. Proteom. 2017, 150, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Doggett, J.S.; Nilsen, A.; Forquer, I.; Wegmann, K.W.; Jones-Brando, L.; Yolken, R.H.; Bordon, C.; Charman, S.A.; Katneni, K.; Schultz, T.; et al. Endochin-like quinolones are highly efficacious against acute and latent experimental toxoplasmosis. Proc. Natl. Acad. Sci. USA 2012, 109, 15936–15941. [Google Scholar] [CrossRef] [PubMed]
- Van Dooren, G.G.; Stimmler, L.M.; McFadden, G.I. Metabolic maps and functions of the Plasmodium mitochondrion. FEMS Microbiol. Rev. 2006, 30, 596–630. [Google Scholar] [CrossRef]
- Seidi, A.; Muellner-Wong, L.S.; Rajendran, E.; Tjhin, E.T.; Dagley, L.F.; Aw, V.Y.; Faou, P.; Webb, A.I.; Tonkin, C.J.; van Dooren, G.G. Elucidating the mitochondrial proteome of Toxoplasma gondii reveals the presence of a divergent cytochrome c oxidase. eLife 2018, 7, e38131. [Google Scholar] [CrossRef]
- Xiao, J.; Yolken, R.H. Strain hypothesis of Toxoplasma gondii infection on the outcome of human diseases. Acta Physiol. (Oxf.) 2015, 213, 828–845. [Google Scholar] [CrossRef]
- Zhou, D.-H.; Wang, Z.-X.; Zhou, C.-X.; He, S.; Elsheikha, H.M.; Zhu, X.-Q. Comparative proteomic analysis of virulent and avirulent strains of Toxoplasma gondii reveals strain-specific patterns. Oncotarget 2017, 8, 80481–80491. [Google Scholar] [CrossRef]
- Kojima, N.; Klausner, J.D. An Update on the Global Epidemiology of Syphilis. Curr. Epidemiol. Rep. 2018, 5, 24–38. [Google Scholar] [CrossRef]
- Hook, E.W. Syphilis. Lancet 2017, 389, 1550–1557. [Google Scholar] [CrossRef]
- Peeling, R.W.; Mabey, D.; Kamb, M.L.; Chen, X.-S.; Radolf, J.D.; Benzaken, A.S. Syphilis. Nat. Rev. Dis. Primers 2017, 3, 17073. [Google Scholar] [CrossRef]
- De Cerqueira, L.R.P.; Monteiro, D.L.M.; Taquette, S.R.; Rodrigues, N.C.P.; Trajano, A.J.B.; de Souza, F.M.; Araújo, B.D.M. The magnitude of syphilis: From prevalence to vertical transmission. Rev. Inst. Med. Trop. Sao Paulo 2017, 59, e78. [Google Scholar] [CrossRef] [PubMed]
- The Lancet, null Congenital syphilis in the USA. Lancet 2018, 392, 1168. [CrossRef]
- Bowen, V.; Su, J.; Torrone, E.; Kidd, S.; Weinstock, H. Increase in incidence of congenital syphilis—United States, 2012–2014. MMWR Morb. Mortal. Wkly. Rep. 2015, 64, 1241–1245. [Google Scholar] [CrossRef] [PubMed]
- Cooper, J.M.; Sánchez, P.J. Congenital syphilis. Semin. Perinatol. 2018, 42, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Samson, G.R.; Beatty, D.W.; Malan, A.F. Immune studies in infants with congenital syphilis. Clin. Exp. Immunol. 1990, 81, 315–318. [Google Scholar] [CrossRef] [PubMed]
- Arnold, S.R.; Ford-Jones, E.L. Congenital syphilis: A guide to diagnosis and management. Paediatr. Child Health 2000, 5, 463–469. [Google Scholar] [CrossRef]
- Osbak, K.K.; Houston, S.; Lithgow, K.V.; Meehan, C.J.; Strouhal, M.; Šmajs, D.; Cameron, C.E.; Van Ostade, X.; Kenyon, C.R.; Van Raemdonck, G.A. Characterizing the Syphilis-Causing Treponema pallidum ssp. pallidum Proteome Using Complementary Mass Spectrometry. PLoS Negl. Trop. Dis. 2016, 10, e0004988. [Google Scholar] [CrossRef]
- Ratnam, S. The laboratory diagnosis of syphilis. Can. J. Infect. Dis. Med. Microbiol. 2005, 16, 45–51. [Google Scholar] [CrossRef]
- Osbak, K.K.; Van Raemdonck, G.A.; Dom, M.; Cameron, C.E.; Meehan, C.J.; Deforce, D.; Ostade, X.V.; Kenyon, C.R.; Dhaenens, M. Candidate Treponema pallidum biomarkers uncovered in urine from individuals with syphilis using mass spectrometry. Future Microbiol. 2018, 13, 1497–1510. [Google Scholar] [CrossRef]
- Abdel-Razeq, S.S.; Cross, S.N.; Lipkind, H.S.; Copel, J.A. Cytomegalovirus, Rubella, Toxoplasmosis, Herpes Simplex Virus, and Varicella. In Obstetric Imaging: Fetal Diagnosis and Care; Elsevier: Minneapolis, MN, USA, 2018; pp. 666–681.e3. ISBN 978-0-323-44548-1. [Google Scholar]
- David, S.; Khandhar, P.B. Double-Blind Study. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Baron, S. (Ed.) Medical Microbiology, 4th ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996; ISBN 978-0-9631172-1-2. [Google Scholar]
- Enders, G.; Bolley, I.; Miller, E.; Cradock-Watson, J.; Ridehalgh, M. Consequences of varicella and herpes zoster in pregnancy: Prospective study of 1739 cases. Lancet 1994, 343, 1548–1551. [Google Scholar] [CrossRef]
- Koren, G. Risk of varicella infection during late pregnancy. Can. Fam. Phys. 2003, 49, 1445–1446. [Google Scholar]
- Harger, J.H.; Ernest, J.M.; Thurnau, G.R.; Moawad, A.; Thom, E.; Landon, M.B.; Paul, R.; Miodovnik, M.; Dombrowski, M.; Sibai, B.; et al. Frequency of congenital varicella syndrome in a prospective cohort of 347 pregnant women. Obstet. Gynecol. 2002, 100, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Savarese, I.; De Carolis, M.P.; Costa, S.; De Rosa, G.; De Carolis, S.; Lacerenza, S.; Romagnoli, C. Atypical manifestations of congenital parvovirus B19 infection. Eur. J. Pediatr. 2008, 167, 1463–1466. [Google Scholar] [CrossRef] [PubMed]
- Heegaard, E.D.; Brown, K.E. Human parvovirus B19. Clin. Microbiol. Rev. 2002, 15, 485–505. [Google Scholar] [CrossRef] [PubMed]
- Lassen, J.; Bager, P.; Wohlfahrt, J.; Bottiger, B.; Melbye, M. Parvovirus B19 infection in pregnancy and subsequent morbidity and mortality in offspring. Int. J. Epidemiol. 2013, 42, 1070–1076. [Google Scholar] [CrossRef]
- Vílchez, J.A.; Albaladejo-Otón, M.D. New Trends in Biomarkers and Diseases: An Overview. Bentham Sci. Publ. 2017, 1, 63–64. [Google Scholar] [CrossRef]
- Li, D.; Chan, D.W. Proteomic cancer biomarkers from discovery to approval: It’s worth the effort. Expert Rev. Proteom. 2014, 11, 135–136. [Google Scholar] [CrossRef]
- Finehout, E.J.; Franck, Z.; Choe, L.H.; Relkin, N.; Lee, K.H. Cerebrospinal fluid proteomic biomarkers for Alzheimer’s disease. Ann. Neurol. 2007, 61, 120–129. [Google Scholar] [CrossRef]
- Santamaria, C.; Chatelain, E.; Jackson, Y.; Miao, Q.; Ward, B.J.; Chappuis, F.; Ndao, M. Serum biomarkers predictive of cure in Chagas disease patients after nifurtimox treatment. BMC Infect. Dis. 2014, 14, 302. [Google Scholar] [CrossRef]
- Weekes, M.P.; Tomasec, P.; Huttlin, E.L.; Fielding, C.A.; Nusinow, D.; Stanton, R.J.; Wang, E.C.Y.; Aicheler, R.; Murrell, I.; Wilkinson, G.W.G.; et al. Quantitative temporal viromics: An approach to investigate host-pathogen interaction. Cell 2014, 157, 1460–1472. [Google Scholar] [CrossRef]
| Disease | Intracranial Calcifications | Hearing Loss | Eye Impairment | Microcephaly | Bone Lesions | CNS Damage |
|---|---|---|---|---|---|---|
| Toxoplasmosis | + | - | + | + | - | + |
| Syphilis | - | - | - | + | + | + |
| ZIKV | + | + | + | + | + | + |
| HIV | - | - | - | - | + | + |
| Varicella | - | - | + | - | - | + |
| CVM | + | + | + | + | - | + |
| HSV | - | + | + | + | + | + |
| Rubella | + | + | + | + | - | + |
| Disease | Matrix | MS Approach | Total Identifications | Reference |
|---|---|---|---|---|
| HCMV | Human serum | Label-free quantification with SELDI-TOF-MS | Not available | [56] |
| primary human fetal foreskin fibroblasts | TMT quantification and LC-MS/MS on Orbitrap Elite and Fusion | >8000 cellular proteins and 139 canonical and 14 ORFs viral proteins | [177] | |
| ARPE-19 and Expi293F cells | Easy nLC 1000 HPLC system coupled to an Orbitrap Elite mass spectrometer | 1297 | [50] | |
| Purified HCMV AD169 virions | Label-free quantification on a Finnigan LCQ ion trap MS | 59 | [45] | |
| MRC5 human lung fibroblasts | Label-free quantification and TMT labeling on a LTQ-Orbitrap XL | 4000 host and 100 viral proteins | [53] | |
| HFFs cells | SILAC labeling with 2D–LC-MS/MS (MudPIT) on a LCQ Deca XP Plus mass | 504 | [49] | |
| HFFs cells | SILAC labeling with LC-MS/MS on a LTQ Orbitrap | 1719 | [52] | |
| MRC5 cells | TMT labeling with nLC-MS/MS on a Q-Exactive HF | 5300 | [54] | |
| ZIKV | HeLa and HFFs cells | iTRAQ labeling with LC–MS/MS on a TripleTOF 5600 | 3544 | [72] |
| NPCs and iPSCs | TMT labeling with nLC-MS/MS on a Q-Exactive HF-Hybrid Quadrupole-Orbitrap | 6080 | [75] | |
| Neurospheres | Label-free quantification on a 2D-RP/RP Synapt G2-Si mass spectrometer | Not available | [73] | |
| NPCs and SK-N-BEB2 cell line | Label-free quantification with AP–LC–MS/MS on a LTQ-Orbitrap XL and Orbitrap Q Exactive HF | 386 ZIKV-interacting proteins and 1216 phosphorylation sites | [74] | |
| Human serum | Label-free quantification with EASY-nLC 1000 on a Q Exactive High | 300 | [77] | |
| HIV | Vaginal discharge | Label-free quantification with 2D-DIGE Nanoflow LC/MSMS on a QStar XL Qq-TOF | 72 protein spots with change in volume | [88] |
| Monocytes and placental macrophages | Label-free quantification with SELDI-TOF and (LC MS/MS) | Not available | [89] | |
| Placenta | Label-free quantification with LC–MS/MS on a LTQ XL | Not available | [90] | |
| HSV | Purified virions | Label-free quantification with ESI-MS/MS on a QTRAP 4000 linear ion trap mass spectrometer | 37 | [99] |
| HEp-2 cells line | Label-free quantification with 2-DE and LC-MS/MS on a Q-TOF 1 Mass Spectrometer | 103 protein spot changes | [102] | |
| HEK293 cells | SILAC labeling with LC-MS/MS on a Q-Star Elite mass | At 4 hpi, 2178; At 24 hpi, 1947; At 10 hpi, 2099 | [103] | |
| HFF cells | Label-free quantification with LC-MS/MS on a Orbitrap Fusion Tribrid mass spectrometer | 4000 | [105] | |
| bEnd.3 cells | TMT labeling with nanoLC-MS/MS on a Q-Exactive Orbitrap | 6761 | [101] | |
| Malaria | Human blood | Label-free quantification with LC-MS/MS on a Linear Trap Quadrupole-Orbitrap Velos | 1527 | [109] |
| Human plasma | Label-free quantification with 2D LC-MS on a LTQ ion trap | 1806 | [125] | |
| Human plasma | Label-free quantification with Nano-LC–MS/MS on a LTQ-Orbitrap Velos | 504 | [126] | |
| Human blood | Label-free quantification on a LTQ Orbitrap Velos | Not available | [128] | |
| Infected placentas | TMT labeling with nano-LC-MS/MS on a Orbitrap Fusion | 2946 | [124] | |
| Human erythrocytes cell culture | Label-free quantification on a micrOTOF-Q | 668 | [130] | |
| Toxoplasmosis | Cysts from brain and muscle tissues of pigs | iTRAQ labeling with LC–MS/MS on a Q Exactive Orbitrap | 2551 | [151] |
| Primary, neuronal and monocytic stem cells | iTRAQ labeling with LC/MS/MS on a LTQ Orbitrap Velos | 4367 | [140] | |
| Brain mice | iTRAQ labeling with 2D-LC-MS/MS on a Orbitrap LC-MS | 2612 | [145] | |
| Brain mice | Label-free quantification with LC-MS/MS on a Q-IT-OT Fusion Lumos | 1683 | [144] | |
| T. gondii-infected and -uninfected placentas of pregnant mice | Label-free quantification on a Q-Exactive Plus Orbitrap mass | 792 | [146] | |
| Mitochondria from parasites | Label-free quantification on a Q-Exactive Orbitrap | 400 | [149] | |
| Syphilis | Urine | Label-free quantification on a 2D-LC-MALDI TOF/TOF and LC/ESI-IM-Q-TOF/HDMS | Not available | [163] |
| DAL-1 strain bacteria isolated from rabbits | Label-free quantification on a MALDI-TOF/TOF and ESI-LTQ-Orbitrap | 557 | [161] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Macedo-da-Silva, J.; Marinho, C.R.F.; Palmisano, G.; Rosa-Fernandes, L. Lights and Shadows of TORCH Infection Proteomics. Genes 2020, 11, 894. https://doi.org/10.3390/genes11080894
Macedo-da-Silva J, Marinho CRF, Palmisano G, Rosa-Fernandes L. Lights and Shadows of TORCH Infection Proteomics. Genes. 2020; 11(8):894. https://doi.org/10.3390/genes11080894
Chicago/Turabian StyleMacedo-da-Silva, Janaina, Claudio Romero Farias Marinho, Giuseppe Palmisano, and Livia Rosa-Fernandes. 2020. "Lights and Shadows of TORCH Infection Proteomics" Genes 11, no. 8: 894. https://doi.org/10.3390/genes11080894
APA StyleMacedo-da-Silva, J., Marinho, C. R. F., Palmisano, G., & Rosa-Fernandes, L. (2020). Lights and Shadows of TORCH Infection Proteomics. Genes, 11(8), 894. https://doi.org/10.3390/genes11080894







