Patterns of Sex Chromosome Differentiation in Spiders: Insights from Comparative Genomic Hybridisation
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Chromosome Preparations
2.3. Comparative Genomic Hybridisation (CGH)
2.3.1. Experimental Design and Probe Preparation
2.3.2. CGH
2.4. Fluorescence In Situ Hybridisation (FISH) with Labelled C0t-1 DNA
2.5. Microscopy and Image Analysis
3. Results
3.1. Technical Outcomes and General Patterns of CGH
- Hapten-based labelling as it allowed more reliable and reproducible results than direct FITC-dUTP/Cy3-dUTP labelling.
- 500 ng of each labelled probe per slide, though positive results have been observed also after application of 100 or 300 ng of each genomic probe.
- Application of (at least) ten times more competitive DNA than each genomic probe, accompanied further by a pre-hybridisation step (37 °C, 37 min). Amplification-derived DNA and especially C0t-1 DNA proved to be slightly more efficient than pure female-derived genomic DNA.
- Denaturation of chromosomes in temperatures not higher than 68 °C in mygalomorphs, while araneomorph chromosomes remained mostly unaffected even after denaturation at 72 °C. Denaturation of mygalomorph chromosomes in temperatures higher than 68 °C often led to damage of chromosomes, chromosome loss and artifacts (especially in meiotic plates).
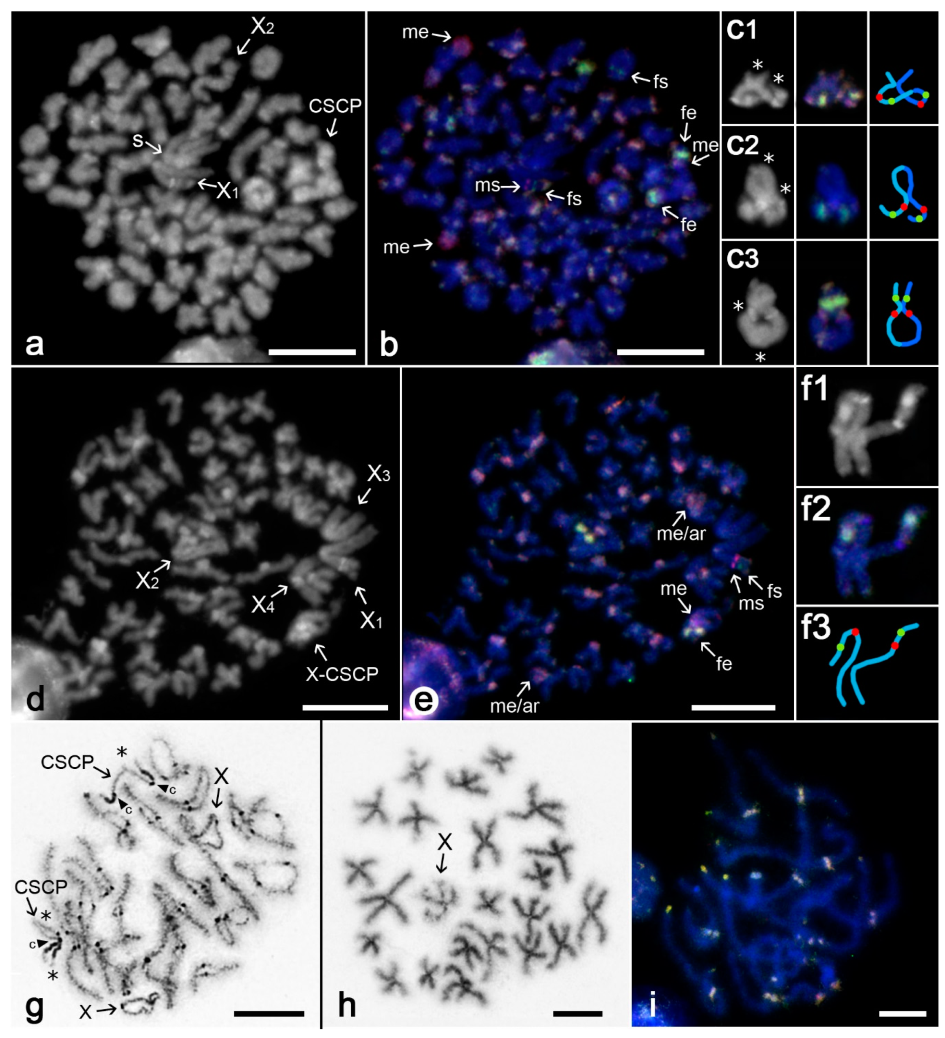
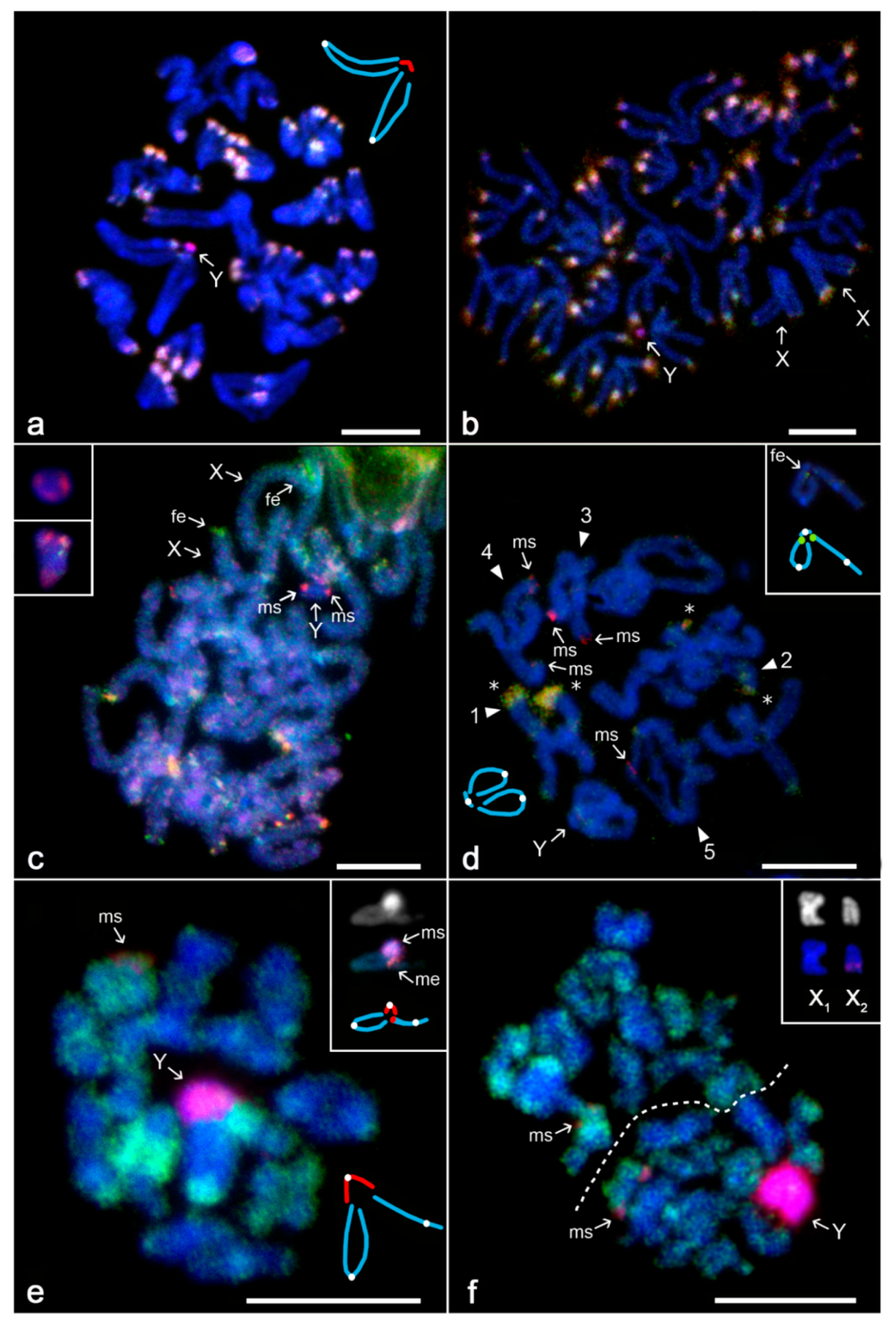
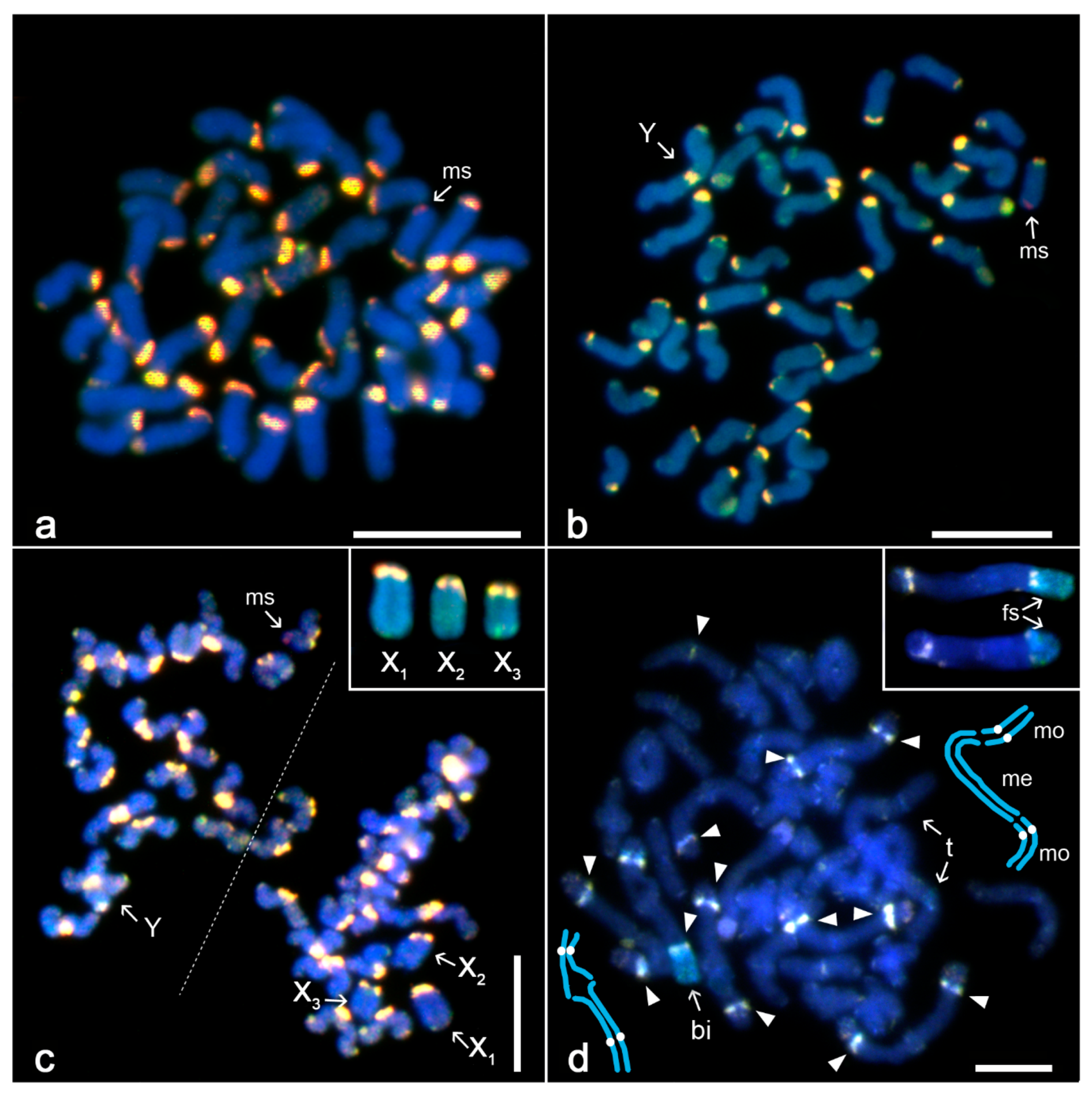
3.2. Differentiation of Mygalomorph Sex Chromosomes
3.3. Differentiation of Sex Chromosomes in Haplogynes with the X1X2Y System
3.4. Differentiation of Neo-Sex Chromosomes
4. Discussion
4.1. Signal Patterns on Autosomes
4.2. Evolution of Sex Chromosomes in Mygalomorphs
4.3. Evolution of the X1X2Y System in Haplogyne Spiders
4.4. Evolution of Neo-Sex Chromosome Systems in Spiders
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ironside, J.E. No amicable divorce? Challenging the notion that sexual antagonism drives sex chromosome evolution. BioEssays 2010, 32, 718–726. [Google Scholar] [CrossRef]
- Ellegren, H. Sex-chromosome evolution: Recent progress and the influence of male and female heterogamety. Nat. Rev. Genet. 2011, 12, 157–166. [Google Scholar] [CrossRef]
- Grossen, C.; Neuenschwander, S.; Perrin, N. The evolution of XY recombination: Sexually antagonistic selection versus deleterious mutation load. Evolution 2012, 66, 3155–3166. [Google Scholar] [CrossRef]
- Mank, J.E. Sex chromosome dosage compensation: Definitely not for everyone. Trends Genet. 2013, 29, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.; Sýkorová, M.; Šíchová, J.; Kůta, V.; Dalíková, M.; Čapková Frydrychová, R.; Neven, L.G.; Sahara, K.; Marec, F. Neo-sex chromosomes and adaptive potential in tortricid pests. Proc. Natl. Acad. Sci. USA 2013, 110, 6931–6936. [Google Scholar] [CrossRef] [PubMed]
- Vicoso, B.; Bachtrog, D. Reversal of an ancient sex chromosome to an autosome in Drosophila. Nature 2013, 499, 332–335. [Google Scholar] [CrossRef] [PubMed]
- Kiuchi, T.; Koga, H.; Kawamoto, M.; Shoji, K.; Sakai, H.; Arai, Y.; Ishihara, G.; Kawaoka, S.; Sugano, S.; Shimada, T.; et al. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 2014, 509, 633–636. [Google Scholar] [CrossRef]
- Schartl, M.; Schmid, M.; Nanda, I. Dynamics of vertebrate sex chromosome evolution: From equal size to giants and dwarfs. Chromosoma 2016, 125, 553–571. [Google Scholar] [CrossRef]
- Dalíková, M.; Zrzavá, M.; Hladová, I.; Nguyen, P.; Šonský, I.; Flegrová, M.; Kubíčková, S.; Voleníková, A.; Kawahara, A.Y.; Peters, R.S.; et al. New insights into the evolution of the W chromosome in Lepidoptera. J. Hered. 2017, 108, 709–719. [Google Scholar] [CrossRef]
- Tomaszkiewicz, M.; Medvedev, P.; Makova, K.D. Y and W chromosome assemblies: Approaches and discoveries. Trends Genet. 2017, 33, 266–282. [Google Scholar] [CrossRef]
- Cavoto, E.; Neuenschwander, S.; Goudet, J.; Perrin, N. Sex-antagonistic genes, XY recombination and feminized Y chromosomes. J. Evol. Biol. 2018, 31, 416–427. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S. Sex Chromosomes and Sex-Linked Genes; Springer: New York, NY, USA, 1967; pp. 1–192. ISBN 978-3-642-88180-0. [Google Scholar]
- Charlesworth, D.; Charlesworth, B.; Marais, G. Steps in the evolution of heteromorphic sex chromosomes. Heredity 2005, 95, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.E.; Dean, R.; Zimmer, F.; Mank, J.E. How to make a sex chromosome. Nat. Commun. 2016, 7, 12087. [Google Scholar] [CrossRef] [PubMed]
- Bergero, R.; Charlesworth, D. The evolution of restricted recombination in sex chromosomes. Trends Ecol. Evol. 2009, 24, 94–102. [Google Scholar] [CrossRef]
- Kaiser, V.B.; Bachtrog, D. Evolution of sex chromosomes in insects. Annu. Rev. Genet. 2010, 44, 91–112. [Google Scholar] [CrossRef] [PubMed]
- Blackmon, H.; Ross, L.; Bachtrog, D. Sex determination, sex chromosomes, and karyotype evolution in insects. J. Hered. 2017, 108, 78–93. [Google Scholar] [CrossRef] [PubMed]
- Král, J. Evolution of multiple sex chromosomes in the spider genus Malthonica (Araneae: Agelenidae) indicates unique structure of the spider sex chromosome systems. Chromosome Res. 2007, 15, 863–879. [Google Scholar] [CrossRef]
- Král, J.; Kořínková, T.; Forman, M.; Krkavcová, L. Insights into the meiotic behavior and evolution of multiple sex chromosome systems in spiders. Cytogenet. Genome Res. 2011, 133, 43–66. [Google Scholar] [CrossRef]
- Araujo, D.; Schneider, M.C.; Paula-Neto, E.; Cella, D.M. Sex chromosomes and meiosis in spiders: A review. In Meiosis—Molecular Mechanisms and Cytogenetic Diversity; Swan, A., Ed.; InTechOpen: Rieka, Croatia, 2012; Volume 5, pp. 87–108. ISBN 978-953-51-0118-5. [Google Scholar]
- Král, J.; Kořínková, T.; Krkavcová, L.; Musilová, J.; Forman, M.; Ávila Herrera, I.M.; Haddad, C.R.; Vítková, M.; Henriques, S.; Palacios Vargas, J.G.; et al. Evolution of karyotype, sex chromosomes, and meiosis in mygalomorph spiders (Araneae: Mygalomorphae). Biol. J. Linn. Soc. 2013, 109, 377–408. [Google Scholar] [CrossRef]
- Kořínková, T.; Král, J. Karyotypes, sex chromosomes, and meiotic division in spiders. In Spider Ecophysiology, 1st ed.; Nentwig, W., Ed.; Springer: Berlin, Germany, 2013; pp. 159–169. [Google Scholar] [CrossRef]
- World Spider Catalog. Available online: https://wsc.nmbe.ch/ (accessed on 24 May 2020).
- Coddington, J.A.; Levi, H.W. Systematics and evolution of spiders (Araneae). Annu. Rev. Ecol. Syst. 1991, 22, 565–592. [Google Scholar] [CrossRef]
- Coddington, J.A. Phylogeny and classification of spiders. In Spiders of North America: An Identification Manual; Ubick, D., Paquin, P., Cushing, P.E., Roth, V., Eds.; American Arachnological Society: San Francisco, CA, USA, 2005; pp. 18–24. ISBN 13: 9780977143900. [Google Scholar]
- Suzuki, S. Cytological studies in spiders. III. Studies on the chromosomes of fifty-seven species of spiders belonging to seventeen families with general considerations on chromosomal evolution. J. Sci. Hiroshima Univ. B 1954, 15, 23–136. [Google Scholar]
- White, M.J.D. Animal Cytology and Evolution, 3rd ed.; Cambridge University Press: London, UK, 1973; pp. 1–468. ISBN 9780521292276. [Google Scholar]
- Palacios-Gimenez, O.M.; Cabral-de Mello, D.C. Repetitive DNA chromosomal organization in the cricket Cycloptiloides americanus: A case of the unusual X1X20 sex chromosome system in Orthoptera. Mol. Genet. Genomics 2015, 290, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Postiglioni, A.; Brum-Zorrilla, M. Karyological studies on Uruguayan spiders II. Sex chromosomes in spiders of the genus Lycosa (Araneae-Lycosidae). Genetica 1981, 56, 47–53. [Google Scholar] [CrossRef]
- Maddison, W.P. XXXY sex chromosomes in males of the jumping spider genus Pellenes (Araneae: Salticidae). Chromosoma 1982, 85, 23–37. [Google Scholar] [CrossRef]
- Král, J.; Musilová, J.; Šťáhlavský, F.; Řezáč, M.; Akan, Z.; Edwards, R.L.; Coyle, F.A.; Almerje, C.R. Evolution of the karyotype and sex chromosome systems in basal clades of araneomorph spiders (Araneae: Araneomorphae). Chromosome Res. 2006, 14, 859–880. [Google Scholar] [CrossRef]
- Řezáč, M.; Král, J.; Musilová, J.; Pekár, S. Unusual karyotype diversity in the European spiders of the genus Atypus (Araneae: Atypidae). Hereditas 2006, 143, 123–129. [Google Scholar] [CrossRef]
- Sharp, H.E.; Rowell, D.M. Unprecedented chromosomal diversity and behaviour modify linkage patterns and speciation potential: Structural heterozygosity in an Australian spider. J. Evol. Biol. 2007, 20, 2427–2439. [Google Scholar] [CrossRef]
- Maddison, W.P.; Leduc-Robert, G. Multiple origins of sex chromosome fusions correlated with chiasma localization in Habronattus jumping spiders (Araneae: Salticidae). Evolution 2013, 67, 2258–2272. [Google Scholar] [CrossRef]
- Maddison, W.P.; Maddison, D.R.; Derkarabetian, S.; Hedin, M. Sitticine jumping spiders: Phylogeny, classification, and chromosomes (Araneae, Salticidae, Sitticini). ZooKeys 2020, 925, 1–54. [Google Scholar] [CrossRef]
- Silva, R.W.; Klisiowicz, D.D.R.; Cella, D.M.; Mangili, O.C.; Sbalqueiro, I.J. Differential distribution of constitutive heterochromatin in two species of brown spider: Loxosceles intermedia and L. laeta (Araneae, Sicariidae), from the metropolitan region of Curitiba, PR (Brasil). Acta Biol. Parana. 2002, 31, 123–136. [Google Scholar]
- Král, J.; Forman, M.; Kořínková, T.; Reyes Lerma, A.C.; Haddad, C.R.; Musilová, J.; Řezáč, M.; Ávila Herrera, I.; Thakur, S.; Dippenaar-Schoeman, A.S.; et al. Insights into the karyotype and genome evolution of haplogyne spiders indicate a polyploid origin of lineage with holokinetic chromosomes. Sci. Rep. 2019, 9, 3001. [Google Scholar] [CrossRef] [PubMed]
- Araujo, D.; Schneider, M.C.; Zacaro, A.A.; de Oliveira, E.G.; Martins, R.; Brescovit, A.D.; Knysak, I.; Cella, D.M. Venomous Loxosceles species (Araneae, Haplogynae, Sicariidae) from Brazil: 2n♂ = 23 and X1X2Y sex chromosome system as shared characteristics. Zoolog. Sci. 2020, 37, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Paula-Neto, E.; Cella, D.M.; Araújo, D.; Brescovit, A.D.; Schneider, M.C. Comparative cytogenetic analysis among filistatid spiders (Araneomorphae: Haplogynae). J. Arachnol. 2017, 45, 123–128. [Google Scholar] [CrossRef]
- Cordellier, M.; Schneider, J.M.; Uhl, G.; Posnien, N. Sex differences in spiders: From phenotype to genomics. Dev. Genes Evol. 2020, 230, 155–172. [Google Scholar] [CrossRef]
- Sheffer, M.M.; Hoppe, A.; Krehenwinkel, H.; Uhl, G.; Kuss, A.W.; Jensen, L.; Jensen, C.; Gillespie, R.G.; Hoff, K.J.; Prost, S. Chromosome-level reference genome of the European wasp spider Argiope bruennichi: A resource for studies on range expansion and evolutionary adaptation. bioRxiv 2020. [Google Scholar] [CrossRef]
- Bechsagaard, J.; Schou, M.F.; Vanthournout, B.; Hendrickx, F.; Knudsen, B.; Settepani, V.; Schierup, M.H.; Bilde, T. Evidence for faster X chromosome evolution in spiders. Mol. Biol. Evol. 2019, 36, 1281–1293. [Google Scholar] [CrossRef]
- Traut, W.; Winking, H. Meiotic chromosomes and stages of sex chromosome evolution in fish: Zebrafish, platyfish and guppy. Chromosome Res. 2001, 9, 659–672. [Google Scholar] [CrossRef]
- Vítková, M.; Fuková, I.; Kubíčková, S.; Marec, F. Molecular divergence of the W chromosomes in pyralid moths (Lepidoptera). Chromosome Res. 2007, 15, 917–930. [Google Scholar] [CrossRef]
- Pokorná, M.; Rens, W.; Rovatsos, M.; Kratochvíl, L. A ZZ/ZW sex chromosome system in the thick-tailed gecko (Underwoodisaurus milii; Squamata: Gekkota: Carphodactylidae), a member of the ancient gecko lineage. Cytogenet. Genome Res. 2014, 142, 190–196. [Google Scholar] [CrossRef]
- Šíchová, J.; Voleníková, A.; Dincă, V.; Nguyen, P.; Vila, R.; Sahara, K.; Marec, F. Dynamic karyotype evolution and unique sex determinatin systems in Leptidea wood white butterflies. BMC Evol. Biol. 2015, 15, 1–16. [Google Scholar] [CrossRef]
- Altmanová, M.; Rovatsos, M.; Kratochvíl, L.; Johnson Pokorná, M. Minute Y chromosomes and karyotype evolution in Madagascan iguanas (Squamata: Iguania: Opluridae). Biol. J. Linn. Soc. 2016, 118, 618–633. [Google Scholar] [CrossRef]
- Montiel, E.E.; Badenhorst, D.; Tamplin, J.; Burke, R.L.; Valenzuela, N. Discovery of the youngest sex chromosomes reveals first case of convergent co-option of ancestral autosomes in turtles. Chromosoma 2017, 126, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Yano, C.F.; Bertollo, L.A.C.; Ezaz, T.; Trifonov, V.; Sember, A.; Liehr, T.; Cioffi, M.B. Highly conserved Z and molecularly diverged W chromosomes in the fish genus Triportheus (Characiformes, Triportheidae). Heredity 2017, 118, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Sember, A.; Bertollo, L.A.C.; Yano, C.F.; Hatanaka, T.; Ráb, P.; de Oliveira, E.A.; Cioffi, M.B. Sex chromosome evolution and genomic divergence in the fish Hoplias malabaricus (Characiformes, Erythrinidae). Front. Genet. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zrzavá, M.; Hladová, I.; Dalíková, M.; Šíchová, J.; Õunap, E.; Kubíčková, S.; Marec, F. Sex chromosomes of the iconic moth Abraxas grossulariata (Lepidoptera, Geometridae) and its congener A. sylvata. Genes 2018, 9, 279. [Google Scholar] [CrossRef] [PubMed]
- Moraes, R.L.; Sember, A.; Bertollo, L.A.C.; de Oliveira, E.A.; Ráb, P.; Hatanaka, T.; Marinho, M.M.F.; Liehr, T.; Al-Rikabi, A.B.H.; Feldberg, E.; et al. Comparative cytogenetics and neo-Y formation in small-sized fish species of the genus Pyrrhulina (Characiformes, Lebiasinidae). Front. Genet. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Dolejš, P.; Kořínková, T.; Musilová, J.; Opatová, V.; Kubcová, L.; Buchar, J.; Král, J. Karyotypes of central European spiders of the genera Arctosa, Tricca, and Xerolycosa (Araneae: Lycosidae). Eur. J. Entomol. 2011, 108, 1–16. [Google Scholar] [CrossRef]
- Winnepenninckx, B.; Backeljau, T.; De Wachter, R. Extraction of high molecular weight DNA from molluscs. Trends Genet. 1993, 9, 407. [Google Scholar] [CrossRef]
- Kubíčková, S.; Černohorská, H.; Musilová, P.; Rubeš, J. The use of laser microdissection for the preparation of chromosome-specific painting probes in farm animals. Vet. Res. 2002, 10, 571–577. [Google Scholar] [CrossRef]
- Britten, R.J.; Graham, D.E.; Neufeld, B.R. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 1974, 29, 363–418. [Google Scholar] [CrossRef]
- Peterson, D.G.; Pearson, W.R.; Stack, S.M. Characterization of the tomato (Lycopsersicon esculentum) genome using in vitro and in situ DNA reassociation. Genome 1998, 41, 346–356. [Google Scholar] [CrossRef]
- Zwick, M.S.; Hanson, R.E.; McKnight, T.D.; Islam-Faridi, M.N.; Stelly, D.M.; Wing, R.A.; Price, H.J. A rapid procedure for the isolation of Cot-1 DNA from plants. Genome 1997, 40, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Traut, W.; Eickhoff, U.; Schorch, J.C. Identification and analysis of sex chromosomes by comparative genomic hybridization (CGH). Methods Cell Sci. 2001, 23, 157–163. [Google Scholar] [CrossRef]
- Symonová, R.; Sember, A.; Majtánová, Z.; Ráb, P. Characterization of fish genomes by GISH and CGH. In Fish Cytogenetic Techniques, 1st ed.; Ozouf-Costaz, C., Pisano, E., Foresti, F., de Almeida, L.F., Eds.; CRC Press: Cleveland, OH, USA, 2015; pp. 118–131. [Google Scholar] [CrossRef]
- Sember, A.; Bohlen, J.; Šlechtová, V.; Altmanová, M.; Symonová, R.; Ráb, P. Karyotype differentiation in 19 species of river loach fishes (Nemacheilidae, Teleostei): Extensive variability associated with rDNA and heterochromatin distribution and its phylogenetic and ecological interpretation. BMC Evol. Biol. 2015, 15, 251. [Google Scholar] [CrossRef] [PubMed]
- Levan, A.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]
- Wheeler, W.C.; Coddington, J.A.; Crowley, L.M.; Dimitrov, D.; Goloboff, P.A.; Griswold, C.E.; Hormiga, G.; Prendini, L.; Ramírez, M.J.; Sierwald, P.; et al. The spider tree of life: Phylogeny of Araneae based on target-gene analyses from an extensive taxon sampling. Cladistics 2017, 33, 574–616. [Google Scholar] [CrossRef]
- Lüddecke, T.; Krehenwinkel, H.; Canning, G.; Glaw, F.; Longhorn, S.J.; Tänzler, R.; Wendt, I.; Vences, M. Discovering the silk road: Nuclear and mitochondrial sequence data resolve the phylogenetic relationships among theraphosid spider subfamilies. Mol. Phylogenet. Evol. 2018, 119, 63–70. [Google Scholar] [CrossRef]
- Fernandez, R.; Kallal, R.J.; Dimitrov, D.; Ballesteros, J.A.; Arnedo, M.A.; Giribet, G.; Hormiga, G. Phylogenomics, diversification dynamics, and comparative transcriptomics across the spider tree of life. Curr. Biol. 2018, 28, 1489–1497. [Google Scholar] [CrossRef]
- Chirino, M.G.; Fourastie, M.F.; Centeno, N.D.; Bressa, M.J. Unusual chromosome polymorphism and heterochromatin variation in the Argentinean population of the necrophagous fly Lucilia sericata (Diptera: Calliphoridae), comparison with other populations and evolutionary aspects. Eur. J. Entomol. 2020, 117, 295–301. [Google Scholar] [CrossRef]
- Garrido-Ramos, M.A. Satellite DNA: An evolving topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef]
- Fotsig, S.F.; Margoliash, J.; Wang, C.; Saini, S.; Yanicky, R.; Shleizer-Burko, S.; Goren, A.; Gymrek, M.T. The impact of short tandem repeat variation on gene expression. Nat. Genet. 2019, 51, 1652–1659. [Google Scholar] [CrossRef] [PubMed]
- Ananiev, E.V.; Chamberlin, M.A.; Klaiber, J.; Svitashev, S. Microsatellite megatracts in the maize (Zea mays L.) genome. Genome 2005, 48, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.F.; Skaletsky, H.; Koutseva, N.; Pyntikova, T.; Page, D.C. Sex chromosome-to-autosome transposition events counter Y-chromosome gene loss in mammals. Genome Biol. 2015, 16, 104. [Google Scholar] [CrossRef] [PubMed]
- Murata, C.; Kuroki, Y.; Imoto, I.; Kuroiwa, A. Ancestral Y-linked genes were maintained by translocation to the X and Y chromosomes fused to an autosomal pair in the Okinawa spiny rat Tokudaia muenninki. Chromosome Res. 2016, 24, 407–419. [Google Scholar] [CrossRef]
- Tobler, R.; Nolte, V.; Schlötterer, C. High rate of translocation-based gene birth on the Drosophila Y chromosome. Proc. Natl. Acad. Sci. USA 2017, 114, 201706502. [Google Scholar] [CrossRef]
- Willems, T.; Gymrek, M.; Poznik, G.D.; Tyler-Smith, C. The 1000 Genomes Project Chromosome Y Group, Erlich, Y. 2016. Population-scale sequencing data enable precise estimates of Y-STR mutation rates. Am. J. Hum. Genet. 2016, 98, 919–933. [Google Scholar] [CrossRef]
- Jablonka, E.; Lamb, M.J. Meiotic pairing constraints and the activity of sex chromosomes. J. Theor. Biol. 1988, 133, 23–36. [Google Scholar] [CrossRef]
- McKee, B.D.; Handel, M.A. Sex chromosomes, recombination, and chromatin conformation. Chromosoma 1993, 102, 71–80. [Google Scholar] [CrossRef]
- Noronha, R.C.R.; Nagamachi, C.Y.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Pieczarka, J.C. Neo-XY body: An analysis of XY1Y2 meiotic behavior in Carollia (Chiroptera, Phyllostomidae) by chromosome painting. Cytogenet. Genome Res. 2009, 124, 37–43. [Google Scholar] [CrossRef]
- Garrison, N.L.; Rodriguez, J.; Agnarsson, I.; Coddington, J.A.; Griswold, C.E.; Hamilton, C.A.; Hedin, M.; Kocot, K.M.; Ledford, J.M.; Bond, J.E. Spider phylogenomics: Untangling the Spider Tree of Life. PeerJ 2016, 4, e1719. [Google Scholar] [CrossRef]
- Silva, D. Estudio cariotípico de Loxosceles laeta (Araneae: Loxoscelidae). Rev. Perúana Entomol. 1988, 31, 9–12. [Google Scholar]
- Sumner, A.T. Chromosome Banding; Unwin Hyman: London, UK, 1990. [Google Scholar]
- Matsunaga, S. Junk DNA promotes sex chromosome evolution. Heredity 2009, 102, 525–526. [Google Scholar] [CrossRef] [PubMed]
- Moreira-Filho, O.; Bertollo, L.A.C.; Galetti, P.M., Jr. Distribution of sex chromosome mechanisms in neotropical fish and description of a ZZ/ZW system in Parodon hilarii (Parodontidae). Caryologia 1993, 46, 115–125. [Google Scholar] [CrossRef][Green Version]
- Shibata, F.; Hizume, M.; Kuroki, Y. Chromosome painting of Y chromosomes and isolation of a Y chromosome-specific repetitive sequence in the dioecious plant Rumex acetosa. Chromosoma 1999, 108, 266–270. [Google Scholar] [CrossRef]
- Schmid, M.; Feichtinger, W.; Steinlein, C.; Rupprecht, T.; Haaf, T.; Kaiser, H. Chromosome banding in Amphibia. XXIII. Giant W sex chromosomes and extremely small genomes in Eleutherodactylus euphronides and Eleutherodactylus shrevei (Anura, Leptodactylidae). Cytogenet. Genome Res. 2002, 97, 81–94. [Google Scholar] [CrossRef]
- De Oliveira, R.R.; Feldberg, E.; Dos Anjos, M.B.; Zuanon, J. Karyotype characterization and ZZ/ZW sex chromosome heteromorphism in two species of the catfish genus Ancistrus Kner, 1854 (Siluriformes: Loricariidae) from the Amazon basin. Neotrop. Ichthyol. 2007, 5, 301–306. [Google Scholar] [CrossRef]
- Kejnovský, E.; Hobza, R.; Čermák, T.; Kubát, Z.; Vyskot, B. The role of repetitive DNA in structure and evolution of sex chromosomes in plants. Heredity 2009, 102, 533–541. [Google Scholar] [CrossRef]
- Sousa, A.; Fuchs, J.; Renner, S.S. Molecular cytogenetics (FISH, GISH) of Coccinia grandis: A ca. 3 myr-old species of Cucurbitaceae with the largest Y/autosome divergence in flowering plants. Cytogenet. Genome Res. 2013, 139, 107–118. [Google Scholar] [CrossRef]
- Poltronieri, J.; Marquioni, V.; Bertollo, L.A.C.; Kejnovský, E.; Molina, W.F.; Liehr, T.; Cioffi, M.B. Comparative chromosomal mapping of microsatellites in Leporinus species (Characiformes, Anostomidae): Unequal accumulation on the W chromosomes. Cytogenet. Genome Res. 2013, 142, 40–45. [Google Scholar] [CrossRef]
- Viana, P.F.; Ezaz, T.; Marajó, L.; Ferreira, M.; Zuanon, J.; Cioffi, M.B.; Bertollo, L.A.C.; Gross, M.C.; Feldberg, E. Genomic organization of repetitive DNAs and differentiation of an XX/XY sex chromosome system in the Amazonian puffer fish, Colomesus asellus (Tetraodontiformes). Cytogenet. Genome Res. 2017, 153, 96–104. [Google Scholar] [CrossRef]
- Wolf, K.W. How meiotic cells deal with non-exchange chromosomes. BioEssays 1994, 16, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Fuková, I.; Nguyen, P.; Marec, F. Codling moth cytogenetics: Karyotype, chromosomal location of rDNA, and molecular differentiation of sex chromosomes. Genome 2005, 48, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Šíchová, J.; Nguyen, P.; Dalíková, M.; Marec, F. Chromosomal evolution in tortricid moths: Conserved karyotypes with diverged features. PLoS ONE 2013, 8, e64520. [Google Scholar] [CrossRef]
- Uno, Y.; Nishida, C.; Yoshimoto, S.; Ito, M.; Oshima, Y.; Yokoyama, S.; Nakamura, M.; Matsuda, Y. Diversity in the origins of sex chromosomes in anurans inferred from comparative mapping of sexual differentiation genes for three species of the Raninae and Xenopodinae. Chromosome Res. 2008, 16, 999–1011. [Google Scholar] [CrossRef] [PubMed]
- Green, J.E.; Dalíková, M.; Sahara, K.; Marec, F.; Akam, M. XX/XY system of sex determination in the geophilomorph centipede Strigamia maritima. PLoS ONE 2016, 11, e0150292. [Google Scholar] [CrossRef]
- Augstenová, B.; Johnson Pokorná, M.; Altmanová, M.; Frynta, D.; Rovatsos, M.; Kratochvíl, L. ZW, XY, and yet ZW: Sex chromosome evolution in snakes even more complicated. Evolution 2018, 72, 1701–1707. [Google Scholar] [CrossRef] [PubMed]
- Kato, A.; Vega, J.M.; Han, F.; Lamb, J.C.; Bircher, J.A. Advances in plant chromosome identification and cytogenetic techniques. Curr. Opin. Plant. Biol. 2005, 8, 148–154. [Google Scholar] [CrossRef]
- Markova, M.; Vyskot, B. New horizons of genomic in situ hybridization. Cytogenet. Genome Res. 2010, 126, 368–375. [Google Scholar] [CrossRef]
- Lahn, B.T.; Page, D.C. Four evolutionary strata on the human X chromosome. Science 1999, 286, 964–967. [Google Scholar] [CrossRef]
- Toder, R.; Wienberg, J.; Voullaire, L.; O’Brien, P.C.M.; Maccarone, P.; Marshall Graves, J.A. Shared DNA sequences between the X and Y chromosomes in the tammar wallaby–Evidence for independent additions to eutherian and marsupial sex chromosomes. Chromosoma 1997, 106, 94–98. [Google Scholar] [CrossRef]
- Lisachov, A.P.; Makunin, A.I.; Giovannotti, M.; Pereira, J.C.; Druzhkova, A.S.; Caputo Barucchi, V.; Ferguson-Smith, M.A.; Trifonov, V.A. Genetic content of the neo-sex chromosomes in Ctenonotus and Norops (Squamata, Actyloidae) and degeneration of the Y chromosome as revealed by high-throughput sequencing of individual chromosomes. Cytogenet. Genome Res. 2019, 157, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Blackmon, H.; Demuth, J.P. The fragile Y hypothesis: Y chromosome aneuploidy as a selective pressure in sex chromosome and meiotic mechanism evolution. BioEssays 2015, 37, 942–950. [Google Scholar] [CrossRef] [PubMed]
- Rovatsos, M.; Farkačová, K.; Altmanová, M.; Johnson Pokorná, M.; Kratochvíl, L. The rise and fall of differentiated sex chromosomes in geckos. Mol. Ecol. 2019, 28, 3042–3052. [Google Scholar] [CrossRef] [PubMed]
- Fuková, I.; Traut, W.; Vítková, M.; Nguyen, P.; Kubíčková, S.; Marec, F. Probing the W chromosome of the codling moth, Cydia pomonella, with sequences from microdissected sex chromatin. Chromosoma 2007, 116, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Mongue, A.J.; Nguyen, P.; Voleníková, A.; Walters, J.R. Neo-sex chromosomes in the monarch butterfly, Danaus plexippus. G3 (Bethesda) 2017, 7, g3.300187.2017. [Google Scholar] [CrossRef]
- Charlesworth, B.; Sniegowski, P.; Stephan, W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 1994, 371, 215–220. [Google Scholar] [CrossRef]
- Ávila Herrera, I.M.; Král, J.; Pastuchová, M.; Forman, M.; Musilová, J.; Kořínková, T.; Šťáhlavský, F.; Zrzavá, M.; Nguyen, P.; Koubová, M.; et al. Evolutionary pattern of karyotypes, nucleolus organizer regions, sex chromosomes, and meiosis in pholcid spiders (Araneae: Pholcidae): Implications for reconstructing karyotype evolution of araneomorph spiders. BMC Evol. Biol. submitted.
- Araujo, D.; Schneider, M.C.; Paula-Neto, E.; Cella, D.M. The Spider Cytogenetic Database. Available online: www.arthropodacytogenetics.bio.br/spiderdatabase (accessed on 17 June 2020).
- Cuñado, N.; Navajas-Pérez, R.; de la Herrán, R.; Rejón, C.R.; Rejón, M.R.; Santos, J.L.; Garrido-Ramos, M.A. The evolution of sex chromosomes in the genus Rumex (Polygonaceae): Identification of a new species with heteromorphic sex chromosomes. Chromosome Res. 2007, 15, 825–832. [Google Scholar] [CrossRef]
- Mariotti, B.; Manzano, S.; Kejnovský, E.; Vyskot, B.; Jamilena, M. Accumulation of Y-specific satellite DNAs during the evolution of Rumex acetosa sex chromosomes. Mol. Genet. Genom. 2009, 281, 249–259. [Google Scholar] [CrossRef]
- Bachtrog, D. Y-chromosome evolution: Emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 2013, 14, 113–124. [Google Scholar] [CrossRef]
- Jetybayev, I.Y.; Bugrov, A.G.; Ünal, M.; Buleu, O.G.; Rubtsov, N.B. Molecular cytogenetic analysis reveals the existence of two independent neo-XY sex chromosome systems in Anatolian Pamphagidae grasshoppers. BMC Evol. Biol. 2017, 17, 20. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Gimenez, O.M.; Dias, G.B.; de Lima, L.G.; Kuhn, G.C.E.S.; Ramos, É.; Martins, C.; Cabral-de-Mello, D.C. High-throughput analysis of the satellitome revealed enormous diversity of satellite DNAs in the neo-Y chromosome of the cricket Eneoptera surinamensis. Sci. Rep. 2017, 7, 6422. [Google Scholar] [CrossRef] [PubMed]
- Gazoni, T.; Haddad, C.F.B.; Narimatsu, H.; Cabral-de-Mello, D.C.; Lyra, M.L.; Parise-Maltempi, P.P. More sex chromosomes than autosomes in the Amazonian frog Leptodactylus pentadactylus. Chromosoma 2018, 127, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Sember, A.; Zhu, Q.; Oliveira, E.A.; Liehr, T.; Al-Rikabi, A.B.H.; Xiao, Z.; Song, H.; Cioffi, M.B. Deciphering the origin and evolution of the X1X2Y system in two closely-related Oplegnathus species (Oplegnathidae and Centrarchiformes). Int. J. Mol. Sci. 2019, 20, 3571. [Google Scholar] [CrossRef] [PubMed]


| Species | Primary Clade: Family | Number and Stage of Male Specimens a | Locality or Source | 2n ♂ | Male Sex Chromosome System | Reference |
|---|---|---|---|---|---|---|
| Atrophothele socotrana | Mygalomorphae: Barychelidae | 1 AD (1 AD) | Yemen, Socotra Isl., Firmihin plateau | 68 | X1X2X3Y + CSCP? | this study |
| Linothele megatheloides | Mygalomorphae: Dipluridae | 1 SAD, 1 AD | breeding | 86 | X1X2X3X4X5X6 + CSCP | [21] |
| Grammostola aff. porteri | Mygalomorphae: Theraphosidae | 1 AD, 1 SAD (5 AD) | Chile, Limarí province, Coquimbo area, Punitaqui | 72 | X1X2 + 2 CSCP | this study |
| Poecilotheria formosa | Mygalomorphae: Theraphosidae | 1 AD | breeding | 110 | X1X2X3X4 + CSCP | [19] |
| Pterinochilus lugardi | Mygalomorphae: Theraphosidae | 1 AD, 1 SAD (2 AD) | breeding | 23 | X + CSCP | this study |
| Kukulcania aff. hibernalis | Araneomorphae, Haplogynae: Filistatidae | 2 AD | breeding | 25 | X1X2Y | this study |
| Loxosceles simillima | Araneomorphae, Haplogynae: Sicariidae | 2 AD, 1 SAD | Republic of South Africa, Free State, Ndumo Game Reserve | 19 | X1X2Y | [31] b |
| Loxosceles laeta | Araneomorphae, Haplogyne: Sicariidae | 1 AD, 1 SAD | breeding | 23 | X1X2Y | this study |
| Pholcus phalangioides | Araneomorphae, Haplogynae: Pholcidae | 2 SAD, 2 AD | Czech Republic, Prague | 25 | X1X2Y | [31] |
| Tegenaria ferruginea | Araneomorphae, Entelegynae: Agelenidae | 3 AD | Czech Republic, Prague | 40 | neo-X1X2X3X4X5Y | [18] c |
| 1 AD | Greece, Macedonia, 1 km west of Pentalofos | 40 | neo-X1X2X3X4X5Y | this study |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sember, A.; Pappová, M.; Forman, M.; Nguyen, P.; Marec, F.; Dalíková, M.; Divišová, K.; Doležálková-Kaštánková, M.; Zrzavá, M.; Sadílek, D.; et al. Patterns of Sex Chromosome Differentiation in Spiders: Insights from Comparative Genomic Hybridisation. Genes 2020, 11, 849. https://doi.org/10.3390/genes11080849
Sember A, Pappová M, Forman M, Nguyen P, Marec F, Dalíková M, Divišová K, Doležálková-Kaštánková M, Zrzavá M, Sadílek D, et al. Patterns of Sex Chromosome Differentiation in Spiders: Insights from Comparative Genomic Hybridisation. Genes. 2020; 11(8):849. https://doi.org/10.3390/genes11080849
Chicago/Turabian StyleSember, Alexandr, Michaela Pappová, Martin Forman, Petr Nguyen, František Marec, Martina Dalíková, Klára Divišová, Marie Doležálková-Kaštánková, Magda Zrzavá, David Sadílek, and et al. 2020. "Patterns of Sex Chromosome Differentiation in Spiders: Insights from Comparative Genomic Hybridisation" Genes 11, no. 8: 849. https://doi.org/10.3390/genes11080849
APA StyleSember, A., Pappová, M., Forman, M., Nguyen, P., Marec, F., Dalíková, M., Divišová, K., Doležálková-Kaštánková, M., Zrzavá, M., Sadílek, D., Hrubá, B., & Král, J. (2020). Patterns of Sex Chromosome Differentiation in Spiders: Insights from Comparative Genomic Hybridisation. Genes, 11(8), 849. https://doi.org/10.3390/genes11080849





