Network Analysis Identifies Gene Regulatory Network Indicating the Role of RUNX1 in Human Intervertebral Disc Degeneration
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of IDD-Related Genes
2.2. Gene Ontology (GO) and Pathway Enrichment Analyses
2.3. Integrated Protein Network Analysis
2.4. Network Module Analysis
2.5. Prediction of Regulatory Networks of Transcription Factors (TFs)
2.6. Validation of Target Genes Using Publicly Available Microarray Datasets
3. Results
3.1. Identification of IDD-Related Genes
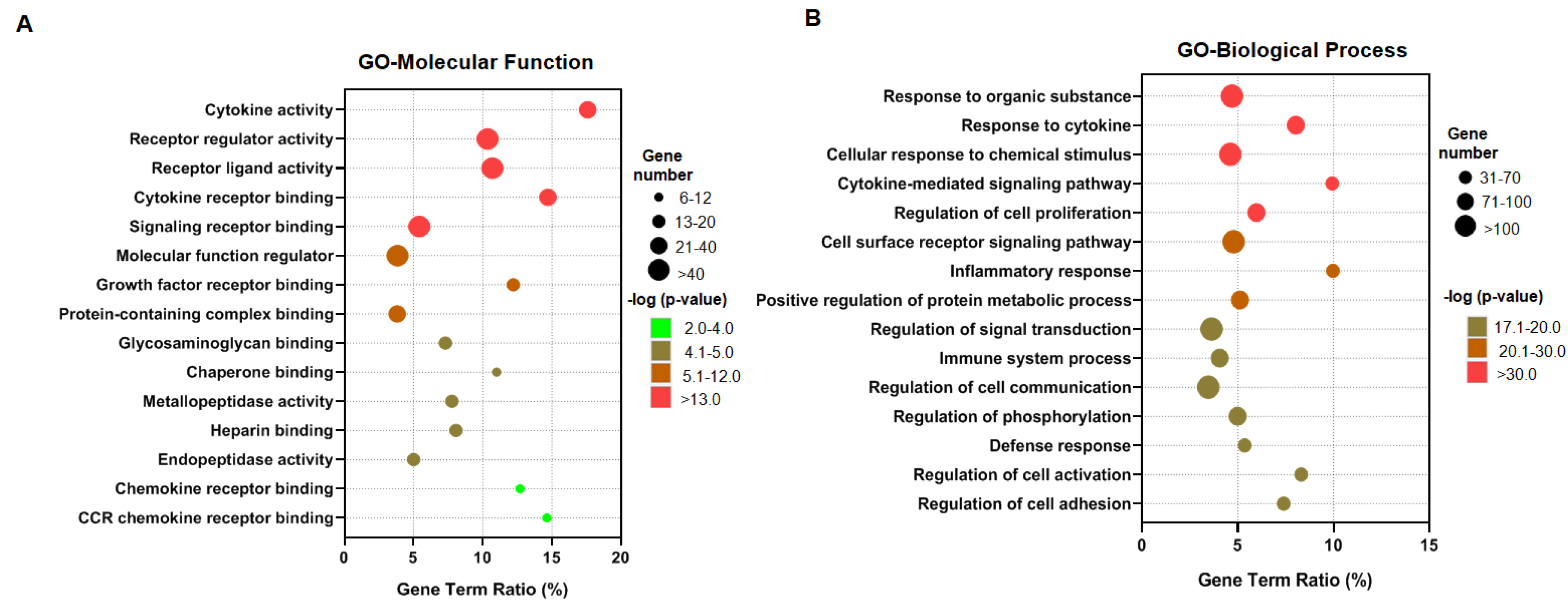
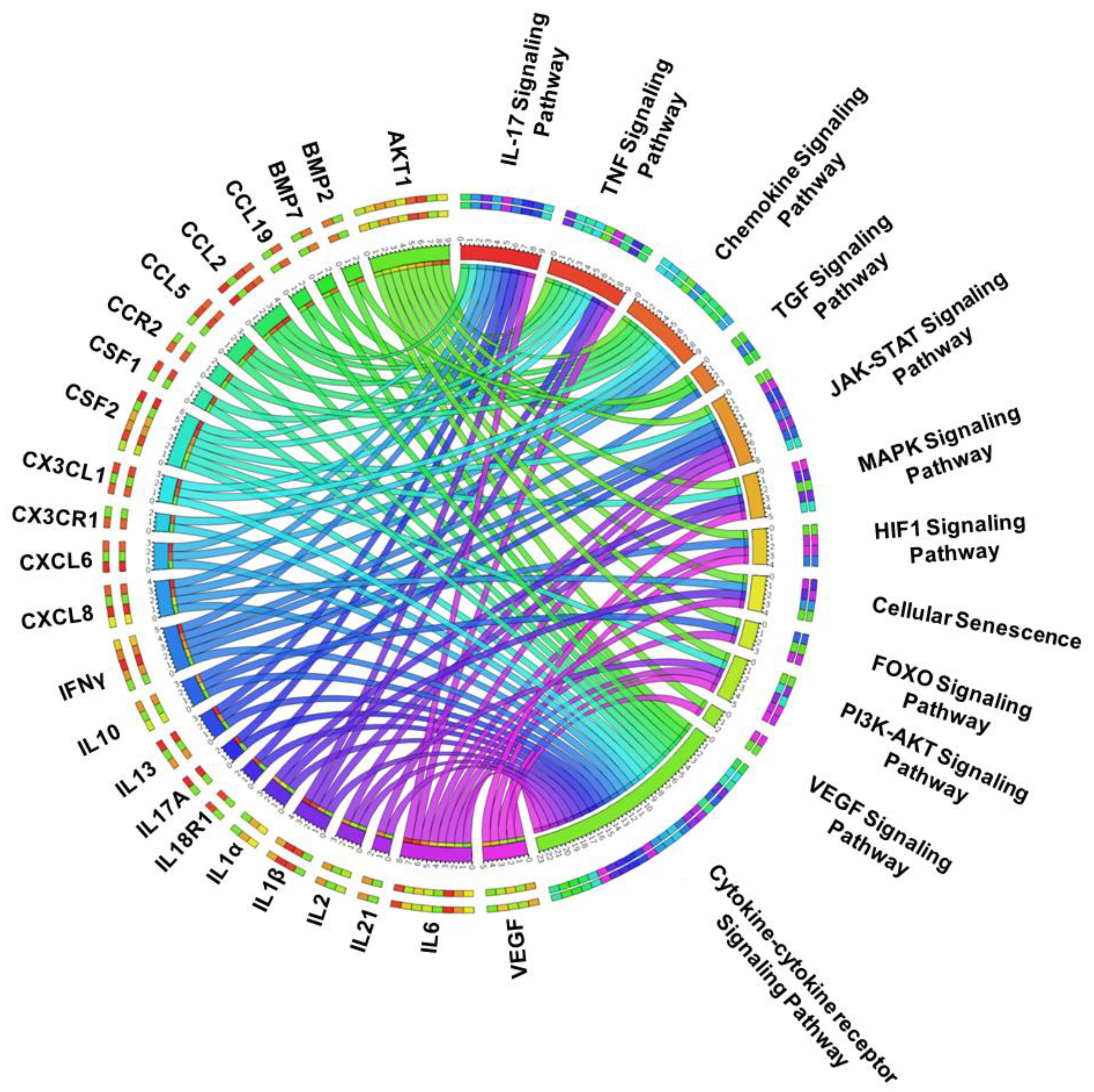
3.2. The Enrichment of Cytokine Receptor Signaling in IDD
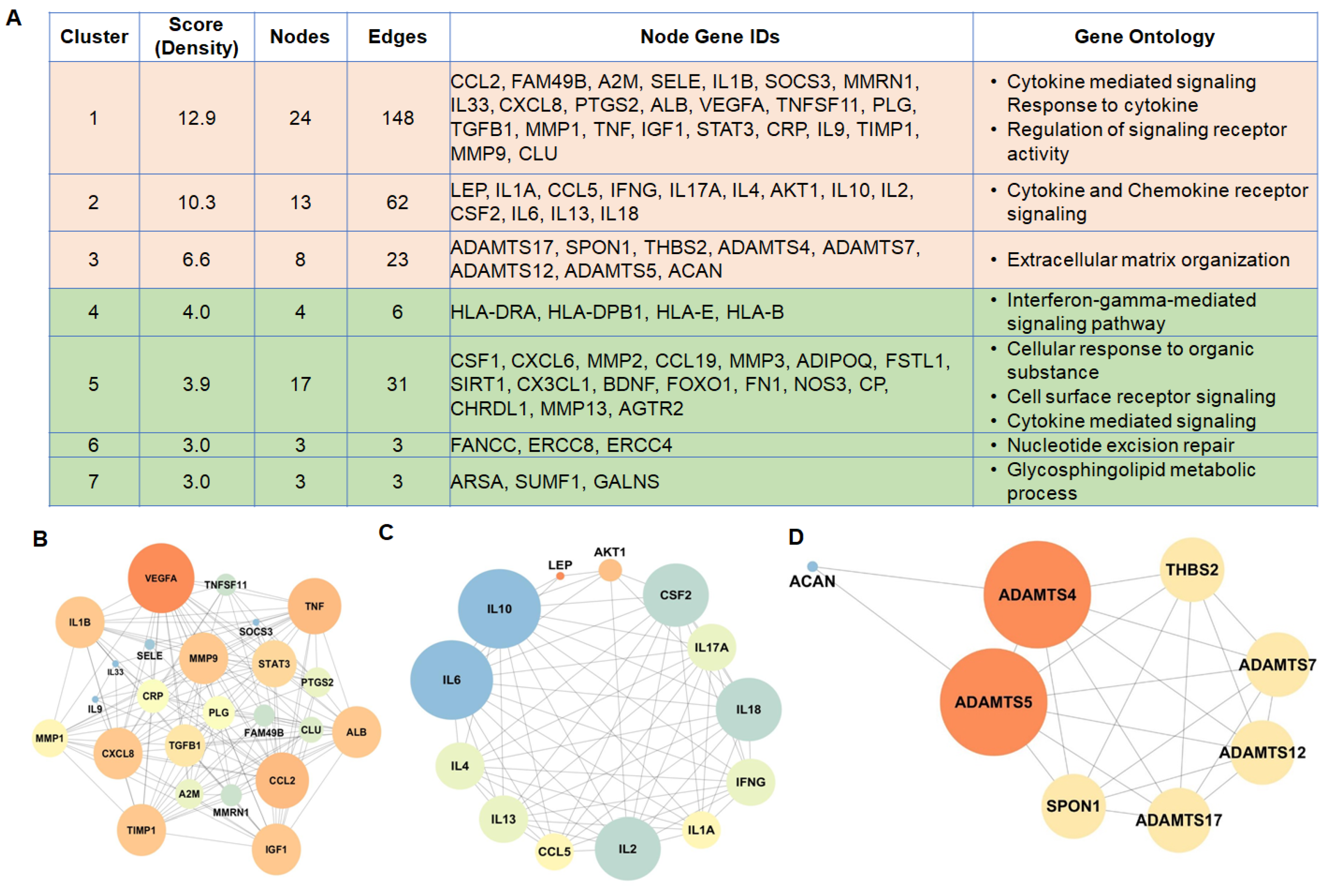
3.3. PPI Network Analysis Reveals an Abundance of Organized Networks of Cytokine and Chemokine Signaling in IDD
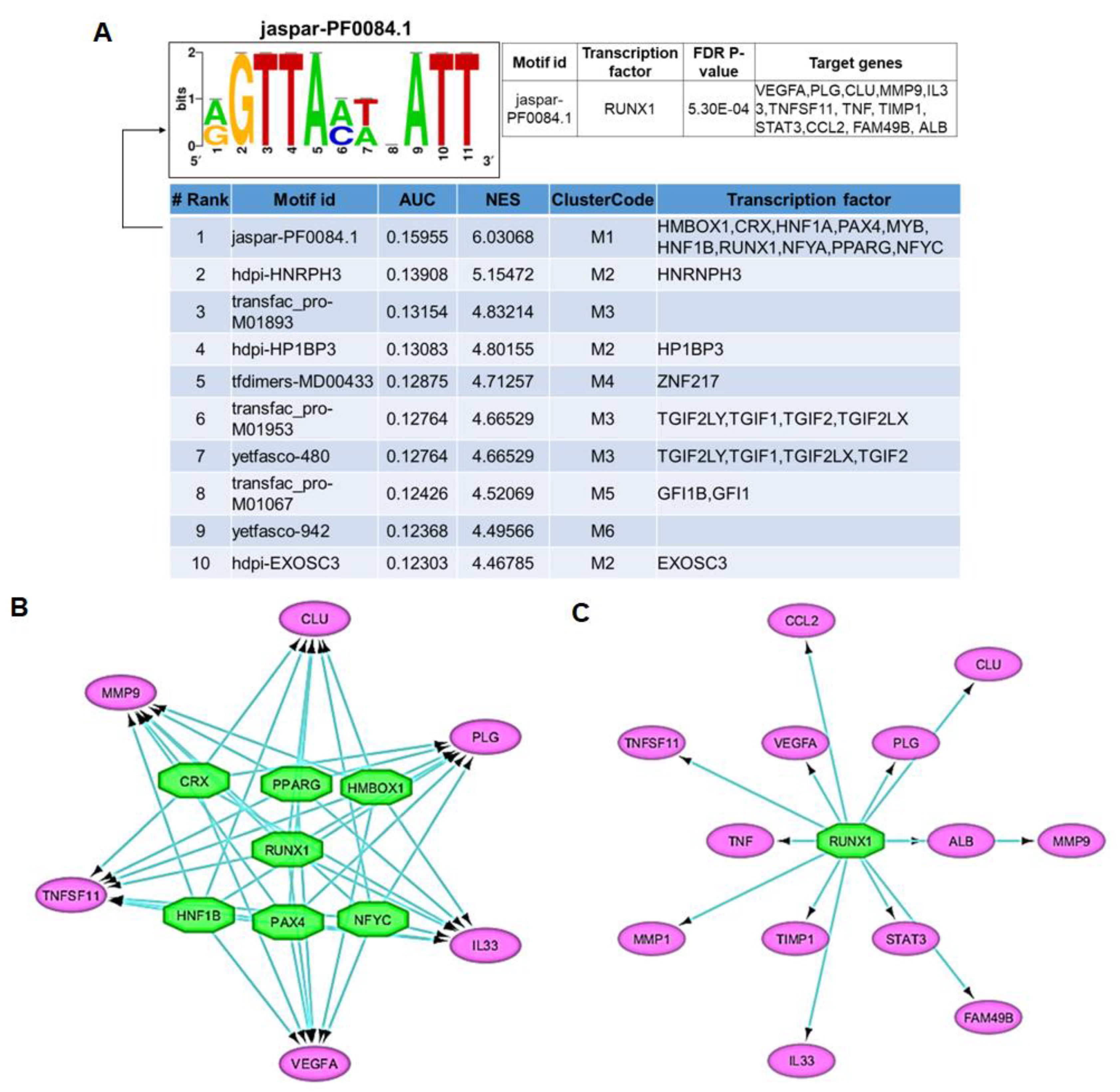
3.4. Transcription Factor–Target Gene Regulatory Network Analysis Predicts RUNX1 as a Major Driver of IDD
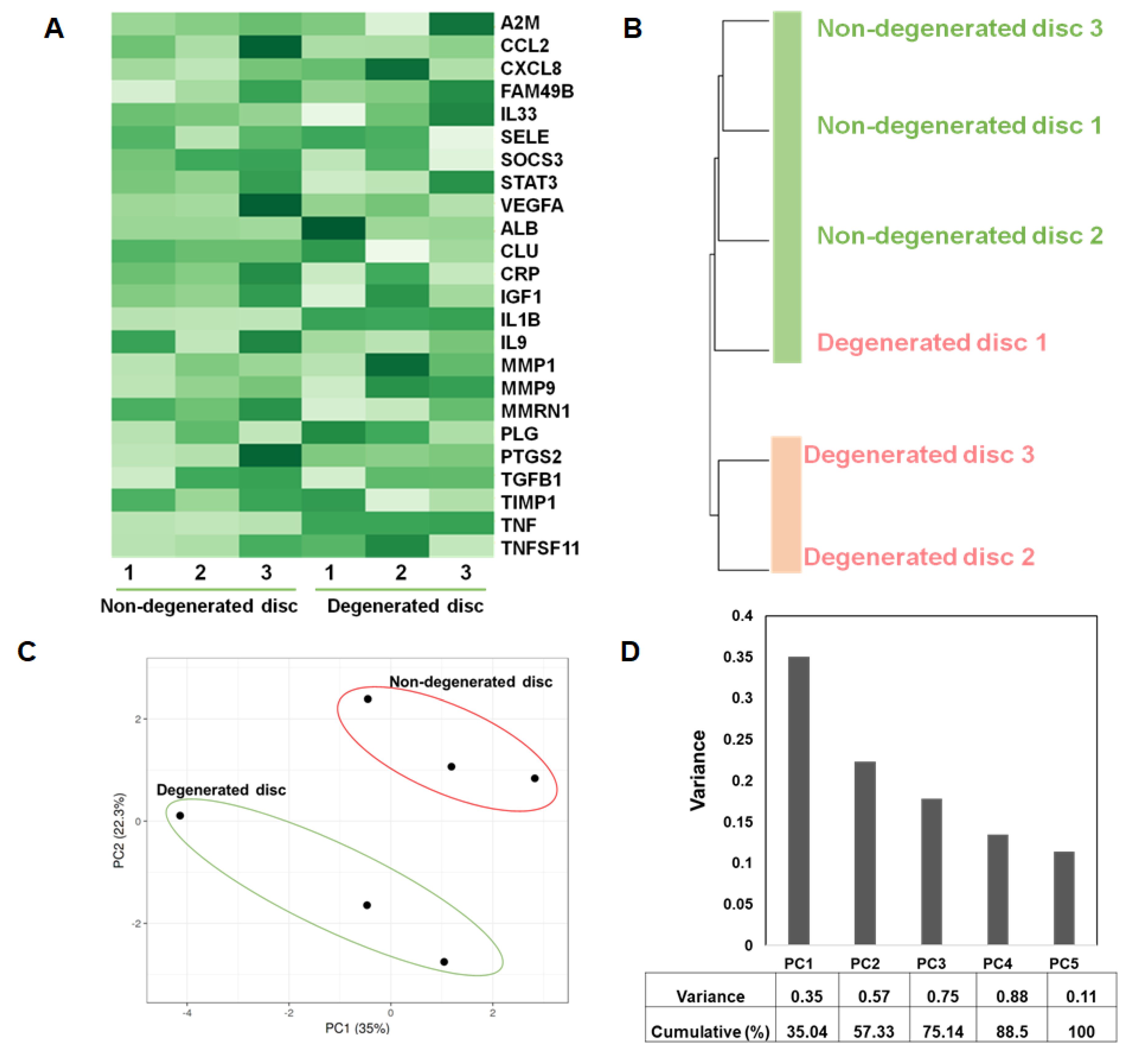
3.5. Microarray-Based Expression Profiling Validates the Genes Identified in Network Clusters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Clark, S.; Horton, R. Low back pain: A major global challenge. Lancet 2018, 391, 2302. [Google Scholar] [CrossRef]
- Buckwalter, J.A. Aging and degeneration of the human intervertebral disc. Spine 1995, 20, 1307–1314. [Google Scholar] [CrossRef] [PubMed]
- Hughes, S.P.; Freemont, A.J.; Hukins, D.W.; McGregor, A.H.; Roberts, S. The pathogenesis of degeneration of the intervertebral disc and emerging therapies in the management of back pain. J. Bone Joint Surg. Br. 2012, 94, 1298–1304. [Google Scholar] [CrossRef] [PubMed]
- Luoma, K.; Riihimaki, H.; Luukkonen, R.; Raininko, R.; Viikari-Juntura, E.; Lamminen, A. Low back pain in relation to lumbar disc degeneration. Spine 2000, 25, 487–492. [Google Scholar] [CrossRef]
- Kalichman, L.; Hunter, D.J. The genetics of intervertebral disc degeneration. Familial predisposition and heritability estimation. Joint Bone Spine 2008, 75, 383–387. [Google Scholar] [CrossRef]
- Kepler, C.K.; Ponnappan, R.K.; Tannoury, C.A.; Risbud, M.V.; Anderson, D.G. The molecular basis of intervertebral disc degeneration. Spine J. 2013, 13, 318–330. [Google Scholar] [CrossRef]
- Gruber, H.E.; Hanley, E.N. Do we need biomarkers for disc degeneration? Biomark. Insights 2007, 1, 131–133. [Google Scholar] [CrossRef]
- Samartzis, D.; Karppinen, J.; Chan, D.; Luk, K.D.; Cheung, K.M. The association of lumbar intervertebral disc degeneration on magnetic resonance imaging with body mass index in overweight and obese adults: A population-based study. Arthritis Rheum. 2012, 64, 1488–1496. [Google Scholar] [CrossRef]
- Feng, Y.; Egan, B.; Wang, J. Genetic Factors in Intervertebral Disc Degeneration. Genes Dis 2016, 3, 178–185. [Google Scholar] [CrossRef]
- Gopal, D.; Ho, A.L.; Shah, A.; Chi, J.H. Molecular basis of intervertebral disc degeneration. Adv. Exp. Med. Biol. 2012, 760, 114–133. [Google Scholar]
- Cirincione, A.G.; Clark, K.L.; Kann, M.G. Pathway networks generated from human disease phenome. BMC Med. Genom. 2018, 11, 75. [Google Scholar] [CrossRef] [PubMed]
- Grimes, T.; Potter, S.S.; Datta, S. Integrating gene regulatory pathways into differential network analysis of gene expression data. Sci. Rep. 2019, 9, 5479. [Google Scholar] [CrossRef] [PubMed]
- Vidal, M.; Cusick, M.E.; Barabasi, A.L. Interactome networks and human disease. Cell 2011, 144, 986–998. [Google Scholar] [CrossRef]
- Barabasi, A.L.; Gulbahce, N.; Loscalzo, J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef]
- Chan, S.Y.; Loscalzo, J. The emerging paradigm of network medicine in the study of human disease. Circ. Res. 2012, 111, 359–374. [Google Scholar] [CrossRef]
- Zanzoni, A.; Soler-Lopez, M.; Aloy, P. A network medicine approach to human disease. FEBS Lett. 2009, 583, 1759–1765. [Google Scholar] [CrossRef] [PubMed]
- MacArthur, J.; Bowler, E.; Cerezo, M.; Gil, L.; Hall, P.; Hastings, E.; Junkins, H.; McMahon, A.; Milano, A.; Morales, J.; et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017, 45, D896–D901. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Haseeb, A.; Makki, M.S.; Khan, N.M.; Ahmad, I.; Haqqi, T.M. Deep sequencing and analyses of miRNAs, isomiRs and miRNA induced silencing. Sci. Rep. 2017, 7, 15178. [Google Scholar] [CrossRef]
- Martha, E.D.-H.; Nazir, M.K.; Camila, M.T.; Tim, Y.; Peter, M.; Steven, M.P.; Greg, G.; Hicham, D. Derivation of notochordal cells from human embryonic stem cells reveals unique. J. Cell Physiol. 2020, 235, 5241–5255. [Google Scholar]
- Otasek, D.; Morris, J.H.; Boucas, J.; Pico, A.R.; Demchak, B. Cytoscape Automation: Empowering workflow-based network analysis. Genome Biol. 2019, 20, 185. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Tang, Y.; Li, M.; Wang, J.; Pan, Y.; Wu, F.X. CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 2015, 127, 67–72. [Google Scholar] [CrossRef]
- Saito, R.; Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Lotia, S.; Pico, A.R.; Bader, G.D.; Ideker, T. A travel guide to Cytoscape plugins. Nat. Methods 2012, 9, 1069–1076. [Google Scholar] [CrossRef]
- Janky, R.; Verfaillie, A.; Imrichova, H.; Van de Sande, B.; Standaert, L.; Christiaens, V.; Hulselmans, G.; Herten, K.; Naval Sanchez, M.; Potier, D.; et al. iRegulon: From a gene list to a gene regulatory network using large motif and track collections. PLoS Comput. Biol. 2014, 10, e1003731. [Google Scholar] [CrossRef] [PubMed]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Qu, Y.; Liu, L.; Zhao, H.; Ma, H.; Si, M.; Cheng, L.; Nie, L. PPAR-gamma agonist pioglitazone protects against IL-17 induced intervertebral. Int. Immunopharmacol. 2019, 72, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Kepler, C.K.; Markova, D.Z.; Hilibrand, A.S.; Vaccaro, A.R.; Risbud, M.V.; Albert, T.J.; Anderson, D.G. Substance P stimulates production of inflammatory cytokines in human disc cells. Spine 2013, 38, E1291–E1299. [Google Scholar] [CrossRef]
- Purmessur, D.; Walter, B.A.; Roughley, P.J.; Laudier, D.M.; Hecht, A.C.; Iatridis, J. A role for TNFalpha in intervertebral disc degeneration: A non-recoverable catabolic shift. Biochem. Biophys. Res. Commun. 2013, 433, 151–156. [Google Scholar] [CrossRef]
- Rand, N.; Reichert, F.; Floman, Y.; Rotshenker, S. Murine nucleus pulposus-derived cells secrete interleukins-1-beta, -6, and -10 and granulocyte-macrophage colony-stimulating factor in cell culture. Spine 1997, 22, 2598–2602. [Google Scholar] [CrossRef]
- Yamamoto, J.; Maeno, K.; Takada, T.; Kakutani, K.; Yurube, T.; Zhang, Z.; Hirata, H.; Kurakawa, T.; Sakai, D.; Mochida, J.; et al. Fas ligand plays an important role for the production of pro-inflammatory cytokines in intervertebral disc nucleus pulposus cells. J. Orthop. Res. 2013, 31, 608–615. [Google Scholar] [CrossRef] [PubMed]
- Gruber, H.E.; Hoelscher, G.L.; Ingram, J.A.; Norton, H.J.; Hanley, E.N., Jr. Increased IL-17 expression in degenerated human discs and increased production in cultured annulus cells exposed to IL-1ss and TNF-alpha. Biotech Histochem. 2013, 88, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Shamji, M.F.; Setton, L.A.; Jarvis, W.; So, S.; Chen, J.; Jing, L.; Bullock, R.; Isaacs, R.E.; Brown, C.; Richardson, W.J. Proinflammatory cytokine expression profile in degenerated and herniated human intervertebral disc tissues. Arthritis Rheum. 2010, 62, 1974–1982. [Google Scholar] [PubMed]
- Le Maitre, C.L.; Hoyland, J.A.; Freemont, A.J. Catabolic cytokine expression in degenerate and herniated human intervertebral discs: IL-1beta and TNFalpha expression profile. Arthritis Res. Ther. 2007, 9, R77. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, S.; Yamashita, T.; Katahira, G.; Yokozawa, H.; Torigoe, T.; Sato, N. Chemokine profile of herniated intervertebral discs infiltrated with monocytes and macrophages. Spine 2002, 27, 1511–1516. [Google Scholar] [CrossRef] [PubMed]
- Risbud, M.V.; Shapiro, I.M. Role of cytokines in intervertebral disc degeneration: Pain and disc content. Nat. Rev. Rheumatol. 2014, 10, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Paglia, D.N.; Yang, X.; Kalinowski, J.; Jastrzebski, S.; Drissi, H.; Lorenzo, J. Runx1 Regulates Myeloid Precursor Differentiation Into Osteoclasts Without Affecting Differentiation Into Antigen Presenting or Phagocytic Cells in Both Males and Females. Endocrinology 2016, 157, 3058–3069. [Google Scholar] [CrossRef]
- Wang, Y.; Belflower, R.M.; Dong, Y.F.; Schwarz, E.M.; O’Keefe, R.J.; Drissi, H. Runx1/AML1/Cbfa2 mediates onset of mesenchymal cell differentiation toward chondrogenesis. J. Bone Miner. Res. 2005, 20, 1624–1636. [Google Scholar] [CrossRef]
- Sato, S.; Kimura, A.; Ozdemir, J.; Asou, Y.; Miyazaki, M.; Jinno, T.; Ae, K.; Liu, X.; Osaki, M.; Takeuchi, Y.; et al. The distinct role of the Runx proteins in chondrocyte differentiation and intervertebral disc degeneration: Findings in murine models and in human disease. Arthritis Rheum. 2008, 58, 2764–2775. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, N.M.; Diaz-Hernandez, M.E.; Presciutti, S.M.; Drissi, H. Network Analysis Identifies Gene Regulatory Network Indicating the Role of RUNX1 in Human Intervertebral Disc Degeneration. Genes 2020, 11, 771. https://doi.org/10.3390/genes11070771
Khan NM, Diaz-Hernandez ME, Presciutti SM, Drissi H. Network Analysis Identifies Gene Regulatory Network Indicating the Role of RUNX1 in Human Intervertebral Disc Degeneration. Genes. 2020; 11(7):771. https://doi.org/10.3390/genes11070771
Chicago/Turabian StyleKhan, Nazir M., Martha E Diaz-Hernandez, Steven M. Presciutti, and Hicham Drissi. 2020. "Network Analysis Identifies Gene Regulatory Network Indicating the Role of RUNX1 in Human Intervertebral Disc Degeneration" Genes 11, no. 7: 771. https://doi.org/10.3390/genes11070771
APA StyleKhan, N. M., Diaz-Hernandez, M. E., Presciutti, S. M., & Drissi, H. (2020). Network Analysis Identifies Gene Regulatory Network Indicating the Role of RUNX1 in Human Intervertebral Disc Degeneration. Genes, 11(7), 771. https://doi.org/10.3390/genes11070771




