PPARG Pro12Ala Polymorphism with CKD in Asians: A Meta-Analysis Combined with a Case-Control Study—A Key for Reaching Null Association
Abstract
1. Introduction
2. Materials and Methods
2.1. Meta-Analysis
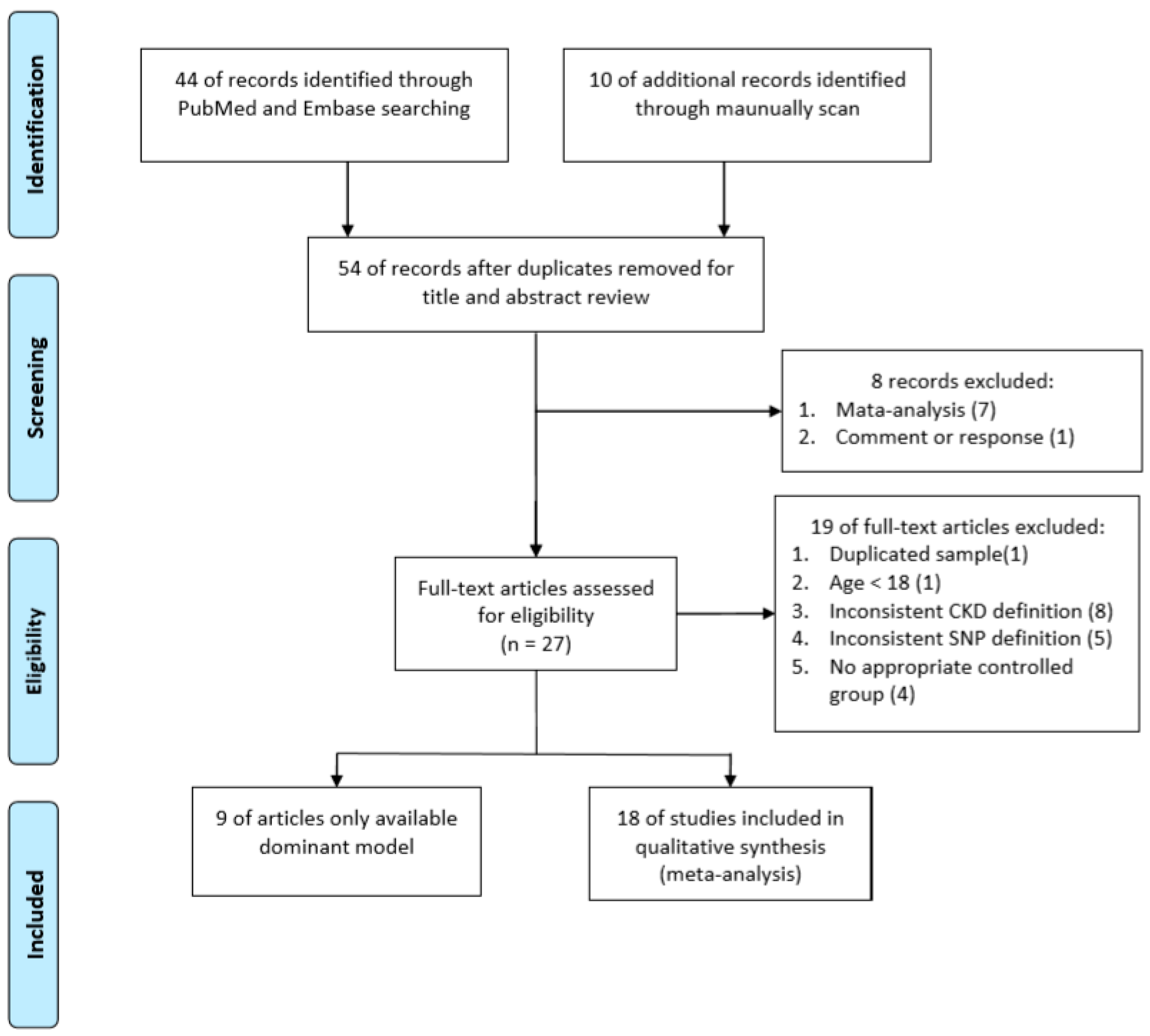
2.1.1. Search Methods and Criteria for Study Consideration
2.1.2. Data Extraction
2.1.3. Statistical Analysis
2.2. Case-Control Study
2.2.1. Ethical Issues
2.2.2. Subjects
2.2.3. Genomic DNA Extraction and Genotyping
2.2.4. Statistical Analysis
3. Results
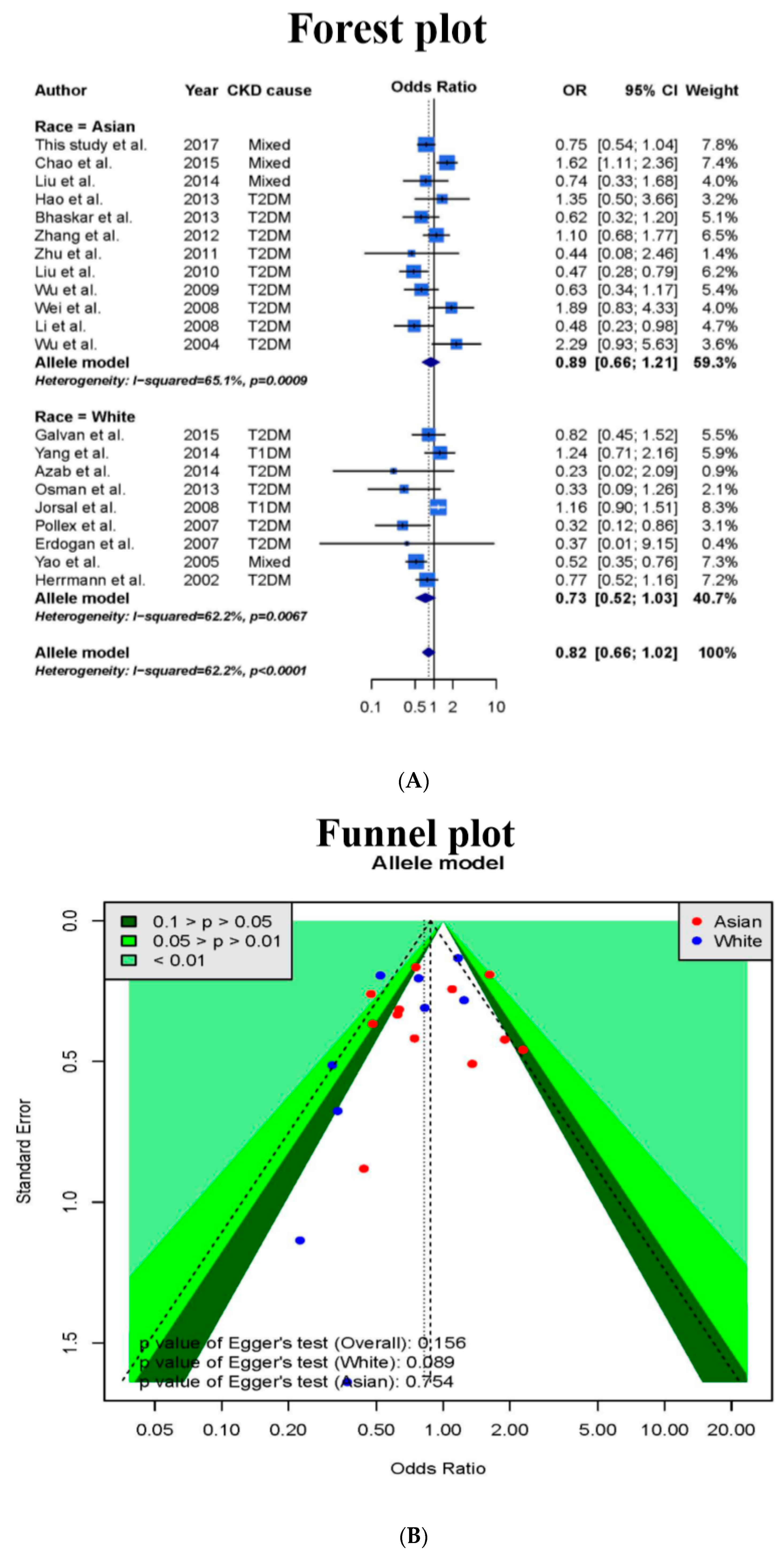
3.1. Meta-Analysis
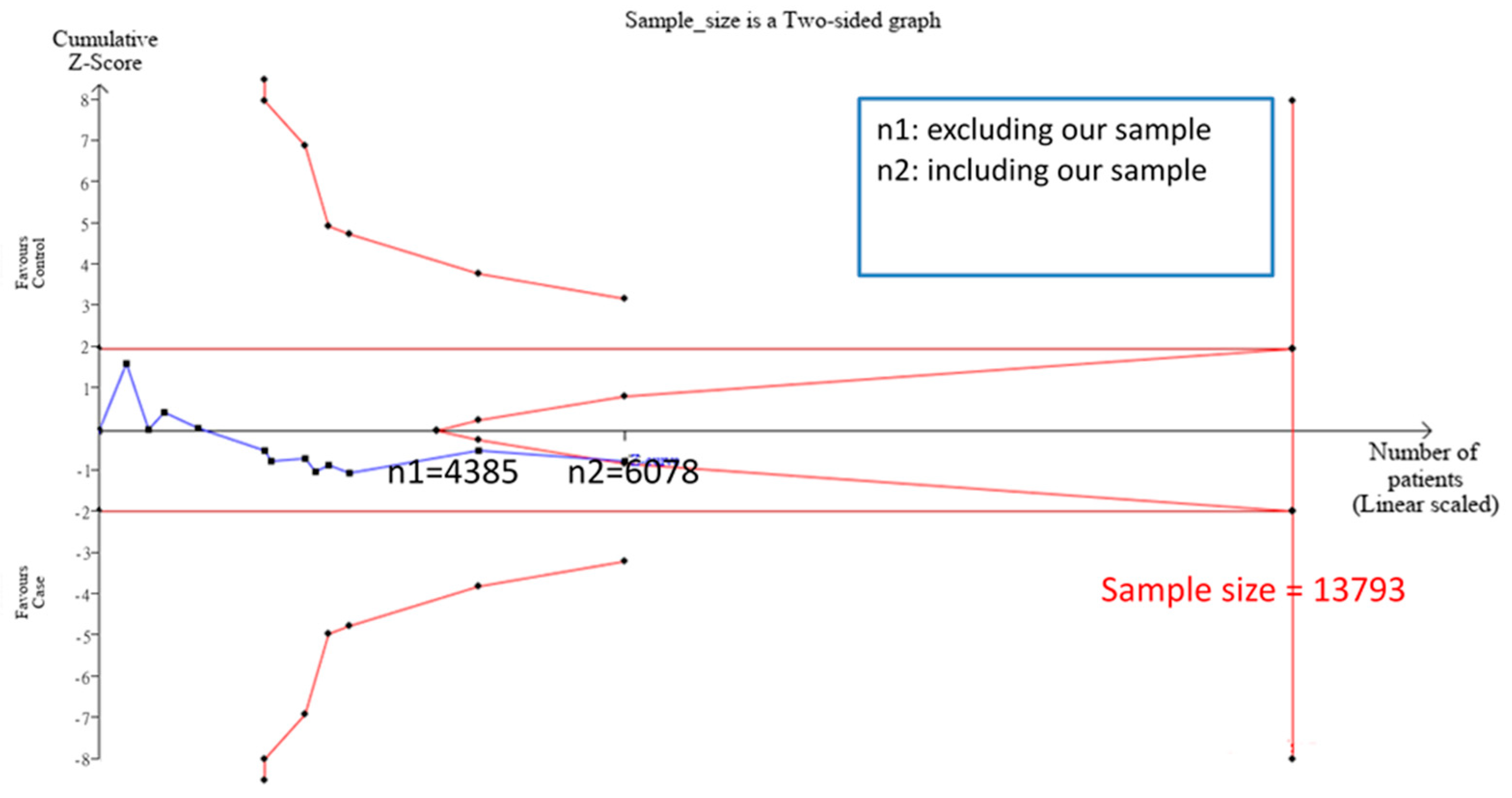
3.2. TSA Sample Estimation
3.3. Case-Control Study
3.4. Meta-Analysis Results after Addition of Case-Control Study and TSA Sample Estimation
3.5. Interactions between the Gene and Environmental Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Coresh, J.; Selvin, E.; Stevens, L.A.; Manzi, J.; Kusek, J.W.; Eggers, P.; Van Lente, F.; Levey, A.S. Prevalence of Chronic Kidney Disease in the United States. JAMA 2007, 298, 2038. [Google Scholar] [CrossRef] [PubMed]
- Imai, E.; Horio, M.; Watanabe, T.; Iseki, K.; Yamagata, K.; Hara, S.; Ura, N.; Kiyohara, Y.; Moriyama, T.; Ando, Y.; et al. Prevalence of chronic kidney disease in the Japanese general population. Clin. Exp. Nephrol. 2009, 13, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, F.; Wang, L.; Wang, W.; Liu, B.; Liu, J.; Chen, M.; He, Q.; Liao, Y.; Yu, X.; et al. Prevalence of chronic kidney disease in China: A cross-sectional survey. Lancet 2012, 379, 815–822. [Google Scholar] [CrossRef]
- Go, A.S.; Chertow, G.M.; Fan, D.; McCulloch, C.E.; Hsu, C.Y. Chronic Kidney Disease and the Risks of Death, Cardiovascular Events, and Hospitalization. N. Engl. J. Med. 2004, 351, 1296–1305. [Google Scholar] [CrossRef]
- USRDS, Atlas ESRD. World Wide Web. 2012. Available online: http://www.usrds.org/atlas.aspx (accessed on 26 June 2017).
- Tsai, J.-C.; Chen, S.-C.; Hwang, S.-J.; Chang, J.-M.; Lin, M.-Y.; Chen, H.-C. Prevalence and Risk Factors for CKD in Spouses and Relatives of Hemodialysis Patients. Am. J. Kidney Dis. 2010, 55, 856–866. [Google Scholar] [CrossRef]
- Maccluer, J.W.; Scavini, M.; Shah, V.O.; Cole, S.A.; Laston, S.L.; Voruganti, V.S.; Paine, S.S.; Eaton, A.J.; Comuzzie, A.G.; Tentori, F.; et al. Heritability of Measures of Kidney Disease Among Zuni Indians: The Zuni Kidney Project. Am. J. Kidney Dis. 2010, 56, 289–302. [Google Scholar] [CrossRef]
- Parikh, N.I.; Hwang, S.-J.; Yang, Q.; Larson, M.G.; Guo, C.-Y.; Robins, S.J.; Sutherland, P.; Benjamin, E.J.; Levy, D.; Fox, C.S. Clinical Correlates and Heritability of Cystatin C (from the Framingham Offspring Study). Am. J. Cardiol. 2008, 102, 1194–1198. [Google Scholar] [CrossRef]
- Langefeld, C.D.; Beck, S.R.; Bowden, N.W.; Rich, S.S.; Wagenknecht, L.E.; Freedman, B.I. Heritability of GFR and albuminuria in Caucasians with type 2 diabetes mellitus. Am. J. Kidney Dis. 2004, 43, 796–800. [Google Scholar] [CrossRef]
- Imperatore, G.; Knowler, W.C.; Pettitt, D.J.; Kobes, S.; Bennett, P.H.; Hanson, R.L. Segregation analysis of diabetic nephropathy in Pima Indians. Diabetes 2000, 49, 1049–1056. [Google Scholar] [CrossRef]
- Wuttke, M.; Köttgen, A. Insights into kidney diseases from genome-wide association studies. Nat. Rev. Nephrol. 2016, 12, 549–562. [Google Scholar] [CrossRef]
- Kottgen, A.; Pattaro, C.; Böger, C.A.; Fuchsberger, C.; Olden, M.; Glazer, N.; Parsa, A.; Gao, X.; Yang, Q.; Smith, A.V.; et al. New loci associated with kidney function and chronic kidney disease. Nat. Genet. 2010, 42, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Kolehmainen, M.; Uusitupa, M.I.J.; Alhava, E.; Laakso, M.; Vidal, H. Effect of the Pro12Ala polymorphism in the peroxisome proliferator-activated receptor (PPAR) gamma2 gene on the expression of PPARgamma target genes in adipose tissue of massively obese subjects. J. Clin. Endocrinol. Metab. 2003, 88, 1717–1722. [Google Scholar] [CrossRef] [PubMed]
- Ahmadian, M.; Suh, J.M.; Hah, N.; Liddle, C.; Atkins, A.R.; Downes, M.; Evans, R.M. PPARgamma signaling and metabolism: The good, the bad and the future. Nat. Med. 2013, 19, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Cipolletta, D.; Feuerer, M.; Li, A.; Kamei, N.; Lee, J.; Shoelson, S.E.; Benoist, C.; Mathis, D. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature 2012, 486, 549–553. [Google Scholar] [CrossRef]
- Corzo, C.; Griffin, P.R. Targeting the Peroxisome Proliferator-Activated Receptor-γ to Counter the Inflammatory Milieu in Obesity. Diabetes Metab. J. 2013, 37, 395–403. [Google Scholar] [CrossRef]
- Yki-Jarvinen, H. Thiazolidinediones. N. Engl. J. Med. 2004, 351, 1106–1118. [Google Scholar] [CrossRef]
- Deeb, S.S.; Fajas, L.; Nemoto, M.; Pihlajamäki, J.; Mykkänen, L.; Kuusisto, J.; Laakso, M.; Fujimoto, W.; Auwerx, J. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat. Genet. 1998, 20, 284–287. [Google Scholar] [CrossRef]
- Vidal-Puig, A.; Jimenez-Linan, M.; Lowell, B.B.; Hamann, A.; Hu, E.; Spiegelman, B.; Flier, J.S.; Moller, D.E. Regulation of PPAR gamma gene expression by nutrition and obesity in rodents. J. Clin. Investig. 1996, 97, 2553–2561. [Google Scholar] [CrossRef]
- Jones, J.R.; Barrick, C.; Kim, K.A.; Lindner, J.; Blondeau, B.; Fujimoto, Y.; Shiota, M.; Kesterson, R.A.; Kahn, B.B.; Magnuson, M.A. Deletion of PPARgamma in adipose tissues of mice protects against high fat diet-induced obesity and insulin resistance. Proc. Natl. Acad Sci. USA 2005, 102, 6207–6212. [Google Scholar] [CrossRef]
- Parvanova, A.I.; Trevisan, R.; Iliev, I.P.; Dimitrov, B.D.; Vedovato, M.; Tiengo, A.; Remuzzi, G.; Ruggenenti, P. Insulin resistance and microalbuminuria: A cross-sectional, case-control study of 158 patients with type 2 diabetes and different degrees of urinary albumin excretion. Diabetes 2006, 55, 1456–1462. [Google Scholar] [CrossRef]
- De Cosmo, S.; Minenna, A.; Ludovico, O.; Mastroianno, S.; Di Giorgio, A.; Pirro, L.; Trischitta, V. Increased urinary albumin excretion, insulin resistance, and related cardiovascular risk factors in patients with type 2 diabetes: Evidence of a sex-specific association. Diabetes Care 2005, 28, 910–915. [Google Scholar] [CrossRef] [PubMed]
- Mykkänen, L.; Zaccaro, D.J.; Wagenknecht, L.E.; Robbins, D.C.; Gabriel, M.; Haffner, S.M. Microalbuminuria is associated with insulin resistance in nondiabetic subjects: The insulin resistance atherosclerosis study. Diabetes 1998, 47, 793–800. [Google Scholar] [CrossRef] [PubMed]
- GeneCards, PPARG gene. 2008. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PPARG (accessed on 26 June 2017).
- Algenabi, A.H.A.F. Study of PPARG2 Gene Polymorphism (Pro12Ala) in Iraqi Patients with Type 2 Diabetes Mellitus. J. Univ. Babylon 2016, 24, 1447–1454. [Google Scholar]
- De Cosmo, S.; Motterlini, N.; Prudente, S.; Pellegrini, F.; Trevisan, R.; Bossi, A.; Remuzzi, G.; Trischitta, V.; Ruggenenti, P. BENEDICT Study Group. Impact of the PPAR-gamma2 Pro12Ala polymorphism and ACE inhibitor therapy on new-onset microalbuminuria in type 2 diabetes: Evidence from BENEDICT. Diabets 2009, 58, 2920–2929. [Google Scholar] [CrossRef]
- Li, L.F.; Liu, L.M.; Zheng, T.S.; Wang, N.S.; Wang, F. Peroxisome proliferator activated receptor γ2 gene P12A polymorphism and type 2 diabetic nephropathy in Han population in Shanghai. J. Shanghai Jiaotong Univ. (Med. Sci.) 2008, 4, 008. [Google Scholar]
- Pollex, R.L.; Mamakeesick, M.; Zinman, B.; Harris, S.; Hegele, R.A.; Hanley, A.J. Peroxisome proliferator-activated receptor γ polymorphism Pro12Ala is associated with nephropathy in type 2 diabetes. J. Diabetes Its Complicat. 2007, 21, 166–171. [Google Scholar] [CrossRef]
- Caramori, M.L.; Canani, L.H.; Costa, L.A.; Gross, J.L. The human peroxisome proliferator-activated receptor gamma2 (PPARgamma2) Pro12Ala polymorphism is associated with decreased risk of diabetic nephropathy in patients with type 2 diabetes. Diabets 2003, 52, 3010–3013. [Google Scholar] [CrossRef]
- Liu, L.; Zheng, T.; Wang, F.; Wang, N.; Li, M. Pro12Ala Polymorphism in the PPARG Gene Contributes to the Development of Diabetic Nephropathy in Chinese Type 2 Diabetic Patients. Diabetes Care 2010, 33, 144–149. [Google Scholar] [CrossRef]
- Yao, Q.; Nordfors, L.; Axelsson, J.; Heimbürger, O.; Qureshi, A.R.; Bárány, P.; Lindholm, B.; Lonnqvist, F.; Schalling, M.; Stenvinkel, P. Peroxisome proliferator-activated receptor γ polymorphisms affect systemic inflammation and survival in end-stage renal disease patients starting renal replacement therapy. Atheroscler. 2005, 182, 105–111. [Google Scholar] [CrossRef]
- Cao, Y.; Zhao, H.; Yan, M.; Zhang, B.; Li, P. Meta-analysis of the Association of Prol2Ala Polymorphism of Peroxisome Proliferator Activated Receptor Gamma Gene with Diabetic Kidney Disease in Chinese Han Population. Hong Kong J. Nephrol. 2015, 17, S8. [Google Scholar] [CrossRef][Green Version]
- Li, T.; Shi, Y.; Yin, J.; Qin, Q.; Wei, S.; Nie, S.; Liu, L. The association between lipid metabolism gene polymorphisms and nephropathy in type 2 diabetes: A meta-analysis. Int. Urol. Nephrol. 2015, 47, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Zhu, C.; Mei, X.; Zhou, Y.; Feng, B.; Guo, Z. Peroxisome proliferator–activated receptor gamma Pro12Ala polymorphism decrease the risk of diabetic nephropathy in type 2 diabetes: A meta analysis. Int. J. Clin. Exp. Med. 2015, 8, 7655–7660. [Google Scholar] [PubMed]
- Zhou, T.B.; Guo, X.F.; Yin, S.S. Association of peroxisome proliferator–activated receptor gamma Pro12Ala gene polymorphism with type 2 diabetic nephropathy risk in Caucasian population. J. Recept. Signal Transduct. Res. 2014, 34, 180–184. [Google Scholar] [CrossRef]
- Liu, G.; Zhou, T.B.; Jiang, Z.; Zheng, D.; Yuan, F.; Li, Y.; Hu, H.; Chen, Z. Relationship between PPARgamma Pro12Ala gene polymorphism and type 2 diabetic nephropathy risk in Asian population: Results from a meta–analysis. J. Recept. Signal Transduct. Res. 2014, 34, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Teng, Z.; Cai, S.; Wang, D.; Zhao, X.; Yu, K. The association between the PPARγ2 Pro12Ala polymorphism and nephropathy susceptibility in type 2 diabetes: A meta-analysis based on 9,176 subjects. Diagn. Pathol. 2013, 8, 118. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, S.; Chen, J.; Tang, Y.; Hu, H.; Mohan, V.; Venkatesan, R.; Wang, J.; Chen, H. Peroxisome proliferator–activated receptor gamma polymorphism Pro12Ala Is associated with nephropathy in type 2 diabetes: Evidence from meta–analysis of 18 studies. Diabets Care 2012, 35, 1388–1393. [Google Scholar] [CrossRef]
- De Cosmo, S.; Prudente, S.; Lamacchia, O.; Lapice, E.; Morini, E.; Di Paola, R.; Copetti, M.; Ruggenenti, P.; Remuzzi, G.; Vaccaro, O.; et al. PPARgamma2 P12A polymorphism and albuminuria in patients with type 2 diabetes: A meta–analysis of case–control studies. Nephrol. Dial. Transplant. 2011, 26, 4011–4016. [Google Scholar] [CrossRef]
- Sterne, J.A.; Sutton, A.J.; Ioannidis, J.P.A.; Terrin, N.; Jones, D.R.; Lau, J.; Carpenter, J.R.; Rücker, G.; Harbord, R.; Schmid, C.H.; et al. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ 2011, 343, d4002. [Google Scholar] [CrossRef]
- Zambrano-Galván, G.; Reyes-Romero, M.; Lazalde, B.; Rodríguez-Morán, M.; Guerrero-Romero, F. Risk of microalbuminuria in relatives of subjects with diabetic nephropathy: A predictive model based on multivariable dimensionality reduction approach. Clin. Nephrol. 2015, 83, 86–92. [Google Scholar] [CrossRef]
- Chao, C.-T.; Chen, Y.-C.; Chiang, C.-K.; Huang, J.-W.; Hu, F.-C.; Fang, C.-C.; Chang, C.-C.; Yen, C.-J. Sequence Variants of Peroxisome Proliferator-Activated Receptor-Gamma Gene and the Clinical Courses of Patients with End-Stage Renal Disease. Dis. Markers 2015, 2015, 1–7. [Google Scholar] [CrossRef]
- Yang, B.; Zhao, H.; Millward, B.A.; Demaine, A.G. The Rate of Decline of Glomerular Filtration Rate May Not Be Associated with Polymorphism of the PPAR2 Gene in Patients with Type 1 Diabetes and Nephropathy. PPAR Res. 2014, 2014, 523584. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Mei, X.; Zhang, Y.; Qi, H.; Wang, J.; Wang, Y.; Jiang, W.; Zhang, X.; Yan, H.; Zhuang, S. Association of peroxisome proliferator-activated receptorγ gene Pro12Ala and C161T polymorphisms with cardiovascular risk factors in maintenance hemodialysis patients. Mol. Boil. Rep. 2014, 41, 7555–7565. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.X.; Su, X.L.; Bi, L.F. Association of p12a single nucleotide polymorphism in peroxisome proliferatorsactivated receptor—γ2 gene with type diabetic nephropathy. J. Inner. Mongolia. Med. Univ. 2013, 35, 290–294. [Google Scholar]
- Ahmed, A., N. Osman, M. NasrAllah and M. Kamal. The association between diabetic nephropathy and polymorphisms of PPAR PRO12ALA and CCR5 32 genes in type 2 diabetes. Nephrol. Dial. Transpl. 2013, 28, i382–i383. [Google Scholar]
- Moher, D., A. Liberati, J. Tetzlaff and D.G. Altman. Preferred reporting items for systematic reviews and meta–analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Thompson, S.G.; Deeks, J.; Altman, U.G. Measuring inconsistency in meta-analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef]
- Lin, C.; Chu, C.-M.; Lin, J.; Yang, H.-Y.; Su, S.-L. Gene-Gene and Gene-Environment Interactions in Meta-Analysis of Genetic Association Studies. PLoS ONE 2015, 10, e0124967. [Google Scholar] [CrossRef]
- Lin, C.; Yang, H.-Y.; Wu, C.-C.; Lee, H.-S.; Lin, Y.-F.; Lu, K.-C.; Chu, C.-M.; Lin, F.-H.; Kao, S.-Y.; Su, S.-L. Angiotensin-Converting Enzyme Insertion/Deletion Polymorphism Contributes High Risk for Chronic Kidney Disease in Asian Male with Hypertension–A Meta-Regression Analysis of 98 Observational Studies. PLoS ONE 2014, 9, e87604. [Google Scholar] [CrossRef]
- Viechtbauer, W. Conducting Meta-Analyses in R with the metafor Package. J. Stat. Softw. 2010, 36, 1–48. [Google Scholar] [CrossRef]
- Schwarzer, G. Meta: Meta–Analysis with R. 2012. Available online: https://cran.r-project.org/web/packages/meta/meta.pdf (accessed on 26 June 2017).
- Thorlund, K.; Engstrøm, J.; Wetterslev, J.; Brok, J.; Imberger, G.; Gluud, C. User Manual for Trial Sequential Analysis (TSA); Copenhagen Trial Unit, Centre for Clinical Intervention Research: Copenhagen, Denmark, 2011; pp. 1–15. [Google Scholar]
- Wacholder, S.; Chanock, S.; Garcia-Closas, M.; El Ghormli, L.; Rothman, N. Assessing the probability that a positive report is false: An approach for molecular epidemiology studies. J. Natl. Cancer Inst. 2004, 96, 434–442. [Google Scholar] [CrossRef]
- rs1801282. Available online: http://www.ensembl.org/Homo_sapiens/Variation/Explore?db=core;r=3:12351126–12352126;v=rs1801282;vdb=variation;vf=1245237 (accessed on 26 June 2017).
- Modification of Diet in Renal Disease (MDRD). 2004. Available online: https://repository.niddk.nih.gov/studies/mdrd/ (accessed on 26 June 2017).
- Tan, S.C.; Yiap, B.C. DNA, RNA, and Protein Extraction: The Past and The Present. J. Biomed. Biotechnol. 2009, 2009, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Perkel, J. SNP genotyping: Six technologies that keyed a revolution. Nat. Methods 2008, 5, 447–453. [Google Scholar] [CrossRef]
- Yang, H.-Y.; Lu, K.-C.; Fang, W.-H.; Lee, H.-S.; Wu, C.-C.; Huang, Y.-H.; Lin, C.-W.; Kao, S.; Lai, C.-H.; Chu, C.-M.; et al. Impact of interaction of cigarette smoking with angiotensin-converting enzyme polymorphisms on end-stage renal disease risk in a Han Chinese population. J. Renin-Angiotensin-Aldosterone Syst. 2015, 16, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Crow, J.F. Hardy, Weinberg and language impediments. Genetics 1999, 152, 821–825. [Google Scholar]
- Boitier, E.; Gautier, J.-C.; Roberts, R. Advances in understanding the regulation of apoptosis and mitosis by peroxisome-proliferator activated receptors in pre-clinical models: Relevance for human health and disease. Comp. Hepatol. 2003, 2, 3. [Google Scholar] [CrossRef]
- Pihlajamäki, J.; Schwab, U.; Kaminska, D.; Ågren, J.; Kuusisto, J.; Kolehmainen, M.; Paananen, J.; Laakso, M.; Uusitupa, M. Dietary polyunsaturated fatty acids and the Pro12Ala polymorphisms of PPARG regulate serum lipids through divergent pathways: A randomized crossover clinical trial. Genes Nutr. 2015, 10, 43. [Google Scholar] [CrossRef]
- Bhaskar, L.V.; Mahin, S.; Ginila, R.T.; Soundararajan, P. Role of the ACE ID and PPARG P12A Polymorphisms in Genetic Susceptibility of Diabetic Nephropathy in a South Indian Population. Nephro-Urology Mon. 2013, 5, 813–817. [Google Scholar] [CrossRef]
- Wu, L.S.H.; Hsieh, C.H.; Pei, D.; Hung, Y.J.; Kuo, S.W.; Lin, E. Association and interaction analyses of genetic variants in ADIPOQ, ENPP1, GHSR, PPARgamma and TCF7L2 genes for diabetic nephropathy in a Taiwanese population with type 2 diabetes. Nephrol. Dial. Transplant. 2009, 24, 3360–3366. [Google Scholar] [CrossRef][Green Version]
- Wei, F.; Wang, J.J.; Wang, C.L.; Huo, X.; Xue, Y.; Bai, L. The relationship between the pro12Ala polymorphism in PPAR–γ2 gene and diabetic nephropathy in Baotou. Chinese J. Diabets 2008, 16, 679–680. [Google Scholar]
- Wu, S. Study on the Relationship of Polymorphism of PPAR–g2 and Type 2 Diabetes and Diabetic Nephropathy; Tianjin Medical University: Tianjin, China, 2004. [Google Scholar]
- Wetterslev, J.; Thorlund, K.; Brok, J.; Gluud, C. Trial sequential analysis may establish when firm evidence is reached in cumulative meta-analysis. J. Clin. Epidemiol. 2008, 61, 64–75. [Google Scholar] [CrossRef]
- Whittemore, A.S.; Kolonel, L.N.; Wu, A.H.; John, E.M.; Gallagher, R.P.; Howe, G.R.; Burch, J.D.; Hankin, J.; Dreon, D.M.; West, D.W.; et al. Prostate Cancer in Relation to Diet, Physical Activity, and Body Size in Blacks, Whites, and Asians in the United States and Canada. J. Natl. Cancer Inst. 1995, 87, 652–661. [Google Scholar] [CrossRef] [PubMed]
- Morini, E.; Tassi, V.; Capponi, D.; Ludovico, O.; Dallapiccola, B.; Trischitta, V.; Prudente, S. Interaction between PPARgamma2 variants and gender on the modulation of body weight. Obesity 2008, 16, 1467–1470. [Google Scholar] [CrossRef]
- Tönjes, A.; Scholz, M.; Loeffler, M.; Stumvoll, M. Association of Pro12Ala polymorphism in peroxisome proliferator–activated receptor gamma with Pre–diabetic phenotypes: Meta–analysis of 57 studies on nondiabetic individuals. Diabets Care 2006, 29, 2489–2497. [Google Scholar] [CrossRef] [PubMed]
- Shende, V.R.; Singh, A.B.; Liu, J. A novel peroxisome proliferator response element modulates hepatic low–density lipoprotein receptor gene transcription in response to PPARdelta activation. Biochem. J. 2015, 472, 275–286. [Google Scholar] [CrossRef] [PubMed]
| Model | Race | n | Sample Size | Odds Ratio (95% CI) | I2 | Egger’s Test |
|---|---|---|---|---|---|---|
| All | All studies | 29 | 13,340 | 0.76 (0.63–0.92) | 61.6% | 0.065 |
| Asian | 12 | 6017 | 0.92 (0.67–1.26) | 60.8% | 0.354 | |
| Caucasian | 17 | 7323 | 0.67 (0.52–0.86) | 60.8% | 0.032 * | |
| Incomplete genotype | All studies | 9 | 6527 | 0.69 (0.50–0.95) | 63.9% | 0.262 |
| Asian | 1 | 1632 | 1.13 (0.69–1.84) | NA | NA | |
| Caucasian | 8 | 4895 | 0.64 (0.46–0.91) | 63.0% | 0.294 | |
| Complete genotype | All studies | 20 | 6813 | 0.80 (0.62–1.03) | 61.3% | 0.100 |
| Asian | 11 | 4385 | 0.90 (0.62–1.28) | 63.7% | 0.411 | |
| Caucasian | 9 | 2428 | 0.69 (0.48–1.01) | 61.3% | 0.087 |
| Dependent Variable Independent Variable | Control a (n = 846) | ESRD b (n = 847) | p-Value |
|---|---|---|---|
| Male, (%) | 377 (44.6%) | 430 (50.8%) | 0.011 * |
| Age, (mean ± SD, years) | 73.50 ± 7.21 | 71.84 ± 12.93 | 0.001 * |
| Diabetes, (%) | 105 (12.5%) | 372 (80.3%) | <0.001 * |
| Hypertension, (%) | 358 (42.7%) | 222 (81.3%) | <0.001 * |
| BMI, (mean ± SD), kg/m2 | 24.22 ± 3.37 | 24.66 ± 4.80 | 0.420 |
| Blood biochemistry values (mean ± SD) | |||
| Urea nitrogen, mg/dL | 15.90 ± 3.89 | 73.88 ± 24.72 | <0.001 * |
| Creatinine, mg/dL | 0.83 ± 0.72 | 9.42 ± 2.78 | <0.001 * |
| Preprandial blood glucose, mg/dL | 102.48 ± 25.20 | 148.90 ± 75.40 | <0.001 * |
| Triglycerides, mg/dL | 103.62 ± 40.82 | 157.02 ± 98.75 | <0.001 * |
| Cholesterol, mg/dL | 185.05 ± 33.35 | 162.42 ± 45.16 | <0.001 * |
| Glomerular filtration rate, mL/min/1.73 m2 | 93.81 ± 23.76 | 5.73 ± 2.45 | <0.001 * |
| Genotype | Control a (n = 846) | ESRD Patients (n = 847) | p-Value |
|---|---|---|---|
| Alleles | 0.083 | ||
| C Allele | 1604 (94.8%) | 1627 (96.0%) | |
| G Allele | 88 (5.2%) | 67 (4.0%) | |
| Co-dominant | 0.169 | ||
| CC | 762 (90.1%) | 781 (92.2%) | |
| CG | 80 (9.5%) | 65 (7.7%) | |
| GG | 4 (0.5%) | 1 (0.1%) | |
| Dominant Model | 0.122 | ||
| CC | 762 (90.1%) | 781 (92.2%) | |
| CG or GG | 84 (9.9%) | 66 (7.8%) | |
| Recessive Model | 0.218 | ||
| CC or CG | 842 (99.5%) | 846 (99.9%) | |
| GG | 4 (0.5%) | 1 (0.1%) |
| Genotype | ESRD/ Total | Crude-OR (95% CI) | p-Value | Adj-OR a (95% CI) | p-Value |
|---|---|---|---|---|---|
| Allele | |||||
| C Allele | 1627/3231 | 1.00 | 1.00 | ||
| G Allele | 67/155 | 0.75 (0.54–1.04) | 0.084 | 0.75 (0.54–1.05) | 0.092 |
| Co-dominant | 0.190 | 0.200 | |||
| CC | 781/1543 | 1.00 | 1.00 | ||
| CG | 65/145 | 0.79 (0.56–1.12) | 0.183 | 0.80 (0.57–1.13) | 0.201 |
| GG | 1/5 | 0.24 (0.03–2.18) | 0.207 | 0.24 (0.03–2.16) | 0.202 |
| Dominant Model | |||||
| CC | 781/1543 | 1.00 | 1.00 | ||
| CG or GG | 66/150 | 0.77 (0.55–1.07) | 0.123 | 0.77 (0.55–1.08) | 0.135 |
| Recessive Model | |||||
| CC or CG | 846/1688 | 1.00 | 1.00 | ||
| GG | 1/5 | 0.25 (0.03–2.23) | 0.214 | 0.24 (0.03–2.20) | 0.208 |
| Regulatory Factor | n | τ2 | Adjusted τ2 | OR | 95% CI | p-Value $ | Egger’s Test |
|---|---|---|---|---|---|---|---|
| Race | 20 | 0.134 | 0.149 | 0.91 | 0.58–1.44 | 0.695 | 0.212 |
| Gender | 15 | 0.155 | 0.103 | 0.04 | 0.00–0.67 | 0.025 | 0.793 |
| BMI | 11 | 0.131 | 0.162 | 1.40 | 0.38–5.08 | 0.610 | 0.655 |
| Diabetes | 19 | 0.104 | 0.113 | 0.65 | 0.19–2.28 | 0.502 | 0.280 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.-C.; Chen, W.-T.; Sung, T.-L.; Tsai, D.-J.; Lin, C.; Su, H.; Lin, Y.-F.; Chiu, H.-Y.; Su, S.-L. PPARG Pro12Ala Polymorphism with CKD in Asians: A Meta-Analysis Combined with a Case-Control Study—A Key for Reaching Null Association. Genes 2020, 11, 705. https://doi.org/10.3390/genes11060705
Chen H-C, Chen W-T, Sung T-L, Tsai D-J, Lin C, Su H, Lin Y-F, Chiu H-Y, Su S-L. PPARG Pro12Ala Polymorphism with CKD in Asians: A Meta-Analysis Combined with a Case-Control Study—A Key for Reaching Null Association. Genes. 2020; 11(6):705. https://doi.org/10.3390/genes11060705
Chicago/Turabian StyleChen, Hsiang-Cheng, Wei-Teing Chen, Tzu-Ling Sung, Dung-Jang Tsai, Chin Lin, Hao Su, Yuh-Feng Lin, Hung-Yi Chiu, and Sui-Lung Su. 2020. "PPARG Pro12Ala Polymorphism with CKD in Asians: A Meta-Analysis Combined with a Case-Control Study—A Key for Reaching Null Association" Genes 11, no. 6: 705. https://doi.org/10.3390/genes11060705
APA StyleChen, H.-C., Chen, W.-T., Sung, T.-L., Tsai, D.-J., Lin, C., Su, H., Lin, Y.-F., Chiu, H.-Y., & Su, S.-L. (2020). PPARG Pro12Ala Polymorphism with CKD in Asians: A Meta-Analysis Combined with a Case-Control Study—A Key for Reaching Null Association. Genes, 11(6), 705. https://doi.org/10.3390/genes11060705







